

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146910.4 + phase: 0
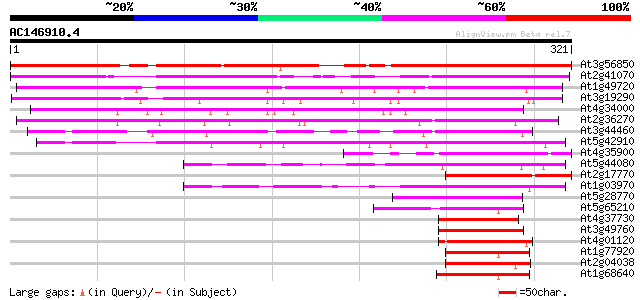
(321 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g56850 ABA-responsive element binding bZip transcription fact... 302 2e-82
At2g41070 bZIP transcription factor AtbZIP12 / DPBF4 241 5e-64
At1g49720 abscisic acid responsive elements-binding factor (ABRE... 164 5e-41
At3g19290 abscisic acid responsive elements-binding factor (ABRE... 159 2e-39
At4g34000 abscisic acid responsive elements-binding factor (ABRE... 146 1e-35
At2g36270 bZip transcription factor AtbZip39 139 2e-33
At3g44460 bZIP transcription factor DPBF2 / AtbZip67 134 7e-32
At5g42910 bzip protein AtbZip15 100 2e-21
At4g35900 bZIP transcription factor AtbZIP14 80 1e-15
At5g44080 bZIP protein AtbZIP13 79 3e-15
At2g17770 bZIP transcription factor atbzip27 75 6e-14
At1g03970 G-box binding bZip transcription factor GBF4 / AtbZip40 74 8e-14
At5g28770 bZip transcription factor BZO2H3 / AtbZip63 51 1e-06
At5g65210 bZip transcription factor TGA1 / AtbZip47 50 2e-06
At4g37730 bZip transcription factor AtbZip7 47 1e-05
At3g49760 bZIP transcription factor AtbZip5 47 1e-05
At4g01120 G-box binding bZip transcription factor GBF2 / AtbZip54 47 1e-05
At1g77920 bZip transcription factor AtbZip50 47 1e-05
At2g04038 bZip transcription factor AtbZip48 47 2e-05
At1g68640 bZip transcription factor PERIANTHIA (PAN) / AtbZip46 46 2e-05
>At3g56850 ABA-responsive element binding bZip transcription factor
AREB3 / AtbZip66
Length = 297
Score = 302 bits (773), Expect = 2e-82
Identities = 184/325 (56%), Positives = 214/325 (65%), Gaps = 32/325 (9%)
Query: 1 MGSQGGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGE 60
M SQ G V ++K+ L+R SLY+LTLDEVQNHLG+ GK LGSMNLDELLKSV SVEA +
Sbjct: 1 MDSQRGIVEQAKSQSLNRQSSLYSLTLDEVQNHLGSSGKALGSMNLDELLKSVCSVEANQ 60
Query: 61 VSDFGGSDVAATAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKKRGVDRDR 120
S +A G Q G + Q SLTL DLSKKTVDEVWKD+Q K G
Sbjct: 61 PSS-----MAVNGGAAAQE----GLSRQGSLTLPRDLSKKTVDEVWKDIQQNKNGGSAHE 111
Query: 121 KSREKQQTLGEMTLEDFLVKAGVVGESFHGKES----GLLRVDSNEDSRQKVSHGLHWMQ 176
+ R+KQ TLGEMTLED L+KAGVV E+ G G S Q ++ W+Q
Sbjct: 112 R-RDKQPTLGEMTLEDLLLKAGVVTETIPGSNHDGPVGGGSAGSGAGLGQNITQVGPWIQ 170
Query: 177 YPVHSVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAAISPSSLMGTLSDTQTLGR 236
Y H +P A P+ V+ QA+ +S SSLMG LSDTQT GR
Sbjct: 171 Y--------------HQLPSMPQPQAFM-PYPVSDMQAM---VSQSSLMGGLSDTQTPGR 212
Query: 237 KRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNE 296
KRVASG VVEKTVERRQKRMIKNRESAARSRAR+QAYT ELE+KVSRLEEENERLR+Q E
Sbjct: 213 KRVASGEVVEKTVERRQKRMIKNRESAARSRARKQAYTHELEIKVSRLEEENERLRKQKE 272
Query: 297 MEKEVPTAPPPEPKNQLRRTNSASF 321
+EK +P+ PPP+PK QLRRT+SA F
Sbjct: 273 VEKILPSVPPPDPKRQLRRTSSAPF 297
>At2g41070 bZIP transcription factor AtbZIP12 / DPBF4
Length = 262
Score = 241 bits (614), Expect = 5e-64
Identities = 161/321 (50%), Positives = 192/321 (59%), Gaps = 61/321 (19%)
Query: 1 MGSQGGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGE 60
MGS G + E + L+R SLY+L L EVQ HLG+ GKPLGSMNLDELLK+V A E
Sbjct: 1 MGSIRGNIEEPISQSLTRQNSLYSLKLHEVQTHLGSSGKPLGSMNLDELLKTVLP-PAEE 59
Query: 61 VSDFGGSDVAATAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKKRGVDRDR 120
G Q SLTL DLSKKTVDEVW+D+Q K G
Sbjct: 60 -----------------------GLVRQGSLTLPRDLSKKTVDEVWRDIQQDKNGNGTST 96
Query: 121 KSREKQQTLGEMTLEDFLVKAGVVGESFHGKESGLLRVDSNEDSRQKVSHGLHWMQYPVH 180
+ KQ TLGE+TLED L++AGVV E+ +E+ ++ + SN W++Y H
Sbjct: 97 TTTHKQPTLGEITLEDLLLRAGVVTETVVPQEN-VVNIASNGQ----------WVEYH-H 144
Query: 181 SVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAAISPSSLMGTLSDT-QTLGRKRV 239
QQQQ GF + V +MG LSDT Q GRKRV
Sbjct: 145 QPQQQQ---------GFMTYPVCEMQDMV--------------MMGGLSDTPQAPGRKRV 181
Query: 240 ASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEMEK 299
A G +VEKTVERRQKRMIKNRESAARSRAR+QAYT ELE+KVSRLEEENE+LRR E+EK
Sbjct: 182 A-GEIVEKTVERRQKRMIKNRESAARSRARKQAYTHELEIKVSRLEEENEKLRRLKEVEK 240
Query: 300 EVPTAPPPEPKNQLRRTNSAS 320
+P+ PPP+PK +LRRTNSAS
Sbjct: 241 ILPSEPPPDPKWKLRRTNSAS 261
>At1g49720 abscisic acid responsive elements-binding factor
(ABRE/ABF1)/ bZip transcription factor AtbZip35
Length = 392
Score = 164 bits (416), Expect = 5e-41
Identities = 143/385 (37%), Positives = 184/385 (47%), Gaps = 82/385 (21%)
Query: 5 GGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDF 64
G T +++ PL+R SLY+LT DE+Q+ LG GK GSMN+DELLK++W+ E +
Sbjct: 12 GDTSRGNESKPLARQSSLYSLTFDELQSTLGEPGKDFGSMNMDELLKNIWTAEDTQAFMT 71
Query: 65 GGSDVAA------TAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKK-RGVD 117
S VAA GGN G Q SLTL LS+KTVDEVWK + K+ +
Sbjct: 72 TTSSVAAPGPSGFVPGGN-------GLQRQGSLTLPRTLSQKTVDEVWKYLNSKEGSNGN 124
Query: 118 RDRKSREKQQTLGEMTLEDFLVKAGVVGE------------------------------- 146
+ E+QQTLGEMTLEDFL++AGVV E
Sbjct: 125 TGTDALERQQTLGEMTLEDFLLRAGVVKEDNTQQNENSSSGFYANNGAAGLEFGFGQPNQ 184
Query: 147 ---SFHGKESGLLRVDSNEDSRQKVSHGLHWMQYPVHSVQQQQHQ------YEKHTMPGF 197
SF+G S ++ ++ KV + Q P QQ HQ + K F
Sbjct: 185 NSISFNGNNSSMI-MNQAPGLGLKVGGTMQQQQQPHQQQLQQPHQRLPPTIFPKQANVTF 243
Query: 198 AA-VHAIQQPF-----------QVAGNQALDAAISP---SSLMGTLSD--TQTLGRKRVA 240
AA V+ + + + G A SP S+ T S GR R
Sbjct: 244 AAPVNMVNRGLFETSADGPANSNMGGAGGTVTATSPGTSSAENNTWSSPVPYVFGRGR-R 302
Query: 241 SGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQ------ 294
S +EK VERRQKRMIKNRESAARSRAR+QAYT ELE ++ L+ N+ L+++
Sbjct: 303 SNTGLEKVVERRQKRMIKNRESAARSRARKQAYTLELEAEIESLKLVNQDLQKKQAEIMK 362
Query: 295 --NEMEKEVPTAPPPEPKNQ-LRRT 316
N KE PP K Q LRRT
Sbjct: 363 THNSELKEFSKQPPLLAKRQCLRRT 387
>At3g19290 abscisic acid responsive elements-binding factor
(ABRE/ABF4) / bZip transcription factor AtbZip38
Length = 431
Score = 159 bits (402), Expect = 2e-39
Identities = 138/416 (33%), Positives = 189/416 (45%), Gaps = 110/416 (26%)
Query: 2 GSQGGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEV 61
GS +PL+R S+Y+LT DE+QN LG GK GSMN+DELLKS+W+ E +
Sbjct: 20 GSSNQMKPTGSVMPLARQSSVYSLTFDELQNTLGGPGKDFGSMNMDELLKSIWTAEEAQA 79
Query: 62 SDFGGSDVAATA-----------GGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWK--- 107
S AATA GGN+Q Q SLTL +S+KTVDEVWK
Sbjct: 80 MAM-TSAPAATAVAQPGAGIPPPGGNLQR--------QGSLTLPRTISQKTVDEVWKCLI 130
Query: 108 ----DMQGKKRGVDRDRKSREKQQTLGEMTLEDFLVKAGVVGE----------------S 147
+M+G G +QQTLGEMTLE+FL +AGVV E
Sbjct: 131 TKDGNMEGSSGGGGESNVPPGRQQTLGEMTLEEFLFRAGVVREDNCVQQMGQVNGNNNNG 190
Query: 148 FHGKESGL--------------LRVDSNEDSR---------QKVSHGLHWMQYPVHSVQQ 184
F+G + + + DS K+ + Q +QQ
Sbjct: 191 FYGNSTAAGGLGFGFGQPNQNSITFNGTNDSMILNQPPGLGLKMGGTMQQQQQQQQLLQQ 250
Query: 185 QQHQYEKHTMP-----------------GFAA-VHAIQQPFQVAGNQALD---------- 216
QQ Q ++ P F+A V+ + F A N +++
Sbjct: 251 QQQQMQQLNQPHPQQRLPQTIFPKQANVAFSAPVNITNKGFAGAANNSINNNNGLASYGG 310
Query: 217 -----AAISP---SSLMGTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRA 268
AA SP S+ +LS + + S +EK +ERRQ+RMIKNRESAARSRA
Sbjct: 311 TGVTVAATSPGTSSAENNSLSPVPYVLNRGRRSNTGLEKVIERRQRRMIKNRESAARSRA 370
Query: 269 RRQAYTQELELKVSRLEEENERLRRQN----EME----KEVPTAPPPEPKNQLRRT 316
R+QAYT ELE ++ +L++ N+ L+++ EM+ KE P + LRRT
Sbjct: 371 RKQAYTLELEAEIEKLKKTNQELQKKQAEMVEMQKNELKETSKRPWGSKRQCLRRT 426
>At4g34000 abscisic acid responsive elements-binding factor
(ABRE/ABF3) / bZip transcription factor AtbZip37
Length = 449
Score = 146 bits (369), Expect = 1e-35
Identities = 127/394 (32%), Positives = 181/394 (45%), Gaps = 112/394 (28%)
Query: 13 TLPLSRSGSLYNLTLDEVQNHLGN-LGKPLGSMNLDELLKSVWSVEAGE----------- 60
+LPL+R S+++LT DE QN G +GK GSMN+DELLK++W+ E
Sbjct: 24 SLPLTRQNSVFSLTFDEFQNSWGGGIGKDFGSMNMDELLKNIWTAEESHSMMGNNTSYTN 83
Query: 61 VSDFGGSDVAATAGGNM-------QHNQLGGF------NSQESLTLSGDLSKKTVDEVWK 107
+S+ + GGN + GGF Q SLTL +S+K VD+VWK
Sbjct: 84 ISNGNSGNTVINGGGNNIGGLAVGVGGESGGFFTGGSLQRQGSLTLPRTISQKRVDDVWK 143
Query: 108 DMQGKK---RGVDRDRKSR--EKQQTLGEMTLEDFLVKAGVVGE---------SFHG--- 150
++ + GV S ++QQTLGEMTLE+FLV+AGVV E +F+G
Sbjct: 144 ELMKEDDIGNGVVNGGTSGIPQRQQTLGEMTLEEFLVRAGVVREEPQPVESVTNFNGGFY 203
Query: 151 --KESGLLRVDSN-----------------------------EDSRQKVSHGLHWMQYPV 179
+G L SN + + ++ +Q P
Sbjct: 204 GFGSNGGLGTASNGFVANQPQDLSGNGVAVRQDLLTAQTQPLQMQQPQMVQQPQMVQQPQ 263
Query: 180 HSVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGN---------------------QALD-- 216
+Q Q+ + K T F+ + Q A QA+D
Sbjct: 264 QLIQTQERPFPKQTTIAFSNTVDVVNRSQPATQCQEVKPSILGIHNHPMNNNLLQAVDFK 323
Query: 217 -----AAISPSSLM-----------GTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNR 260
AA+SP S M +LS + + +G V+EK +ERRQKRMIKNR
Sbjct: 324 TGVTVAAVSPGSQMSPDLTPKSALDASLSPVPYMFGRVRKTGAVLEKVIERRQKRMIKNR 383
Query: 261 ESAARSRARRQAYTQELELKVSRLEEENERLRRQ 294
ESAARSRAR+QAYT ELE ++++L+E NE L+++
Sbjct: 384 ESAARSRARKQAYTMELEAEIAQLKELNEELQKK 417
>At2g36270 bZip transcription factor AtbZip39
Length = 442
Score = 139 bits (350), Expect = 2e-33
Identities = 134/406 (33%), Positives = 170/406 (41%), Gaps = 97/406 (23%)
Query: 5 GGTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGE---- 60
GG L R S+Y+LTLDE Q+ L GK GSMN+DE L S+W+ E
Sbjct: 26 GGGGENHPFTSLGRQSSIYSLTLDEFQHALCENGKNFGSMNMDEFLVSIWNAEENNNNQQ 85
Query: 61 --VSDFGGSDVAATAGGNMQHNQLGG----------------------------FNSQES 90
+ G V A G +N GG Q S
Sbjct: 86 QAAAAAGSHSVPANHNGFNNNNNNGGEGGVGVFSGGSRGNEDANNKRGIANESSLPRQGS 145
Query: 91 LTLSGDLSKKTVDEVWKDMQ---GKKRGVDRDRKSRE---------------KQQTLGEM 132
LTL L +KTVDEVW ++ G G D + +S +Q T GEM
Sbjct: 146 LTLPAPLCRKTVDEVWSEIHRGGGSGNGGDSNGRSSSSNGQNNAQNGGETAARQPTFGEM 205
Query: 133 TLEDFLVKAGVVGE-SFHGKESGLLRVDSNEDS------RQK---VSHGLHWMQYPVHSV 182
TLEDFLVKAGVV E + K + + N S +Q+ V G +P ++
Sbjct: 206 TLEDFLVKAGVVREHPTNPKPNPNPNQNQNPSSVIPAAAQQQLYGVFQGTGDPSFPGQAM 265
Query: 183 -QQQQHQYEKHT-MPGFAAVHAIQQPFQVAGNQALDAAISPSSLMGTLSDTQT------- 233
Y K T G+ +Q G A ++G LS +
Sbjct: 266 GVGDPSGYAKRTGGGGYQQAPPVQAGVCYGGGVGFGAGGQQMGMVGPLSPVSSDGLGHGQ 325
Query: 234 ---------------LGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELE 278
GRKRV G VEK VERRQ+RMIKNRESAARSRAR+QAYT ELE
Sbjct: 326 VDNIGGQYGVDMGGLRGRKRVVDG-PVEKVVERRQRRMIKNRESAARSRARKQAYTVELE 384
Query: 279 LKVSRLEEEN----------ERLRRQNEMEKEVPTAPPPEPKNQLR 314
++++L+EEN ER R+Q E A P PK+ R
Sbjct: 385 AELNQLKEENAQLKHALAELERKRKQQYFESLKSRAQPKLPKSNGR 430
>At3g44460 bZIP transcription factor DPBF2 / AtbZip67
Length = 331
Score = 134 bits (337), Expect = 7e-32
Identities = 113/311 (36%), Positives = 158/311 (50%), Gaps = 62/311 (19%)
Query: 11 SKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDFGGSDVA 70
S+ P+ R S+ +LTLDE+Q GK G+MN+DE L ++W+ ++ GG+
Sbjct: 30 SEEEPVGRQNSILSLTLDEIQM---KSGKSFGAMNMDEFLANLWTTVEENDNEGGGA--- 83
Query: 71 ATAGGNMQHN---QLGGFNSQESLTLSGDLSKKTVDEVWKDMQG-------KKRGVDRDR 120
HN + Q SL+L L KKTVDEVW ++Q
Sbjct: 84 --------HNDGEKPAVLPRQGSLSLPVPLCKKTVDEVWLEIQNGVQQHPPSSNSGQNSA 135
Query: 121 KSREKQQTLGEMTLEDFLVKAGVVGESFHGKESGLLRVDSNEDS-RQKVSHGLHWMQYPV 179
++ +QQTLGE+TLEDFLVKAGVV E +R+ S++ + GLH
Sbjct: 136 ENIRRQQTLGEITLEDFLVKAGVVQEPL----KTTMRMSSSDFGYNPEFGVGLHC----- 186
Query: 180 HSVQQQQHQYEKHTMPGFAAVHAIQQPF-QVAGNQALDAAISPSSLMGTLSDTQTLG--- 235
Q Q+ Y + +V++ +PF V G S S + G Q L
Sbjct: 187 ----QNQNNYGDNR-----SVYSENRPFYSVLGE-------SSSCMTGNGRSNQYLTGLD 230
Query: 236 ----RKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERL 291
+KR+ G E +ERRQ+RMIKNRESAARSRARRQAYT ELEL+++ L EEN +L
Sbjct: 231 AFRIKKRIIDG-PPEILMERRQRRMIKNRESAARSRARRQAYTVELELELNNLTEENTKL 289
Query: 292 R---RQNEMEK 299
+ +NE ++
Sbjct: 290 KEIVEENEKKR 300
>At5g42910 bzip protein AtbZip15
Length = 370
Score = 99.8 bits (247), Expect = 2e-21
Identities = 107/373 (28%), Positives = 160/373 (42%), Gaps = 95/373 (25%)
Query: 16 LSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDFGGSDVAATAGG 75
LSR GS+Y+ T+D+ Q +LG GSMN+DEL+K + S E +
Sbjct: 20 LSRQGSIYSWTVDQFQT---SLGLDCGSMNMDELVKHISSAEETQE-------------- 62
Query: 76 NMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKKR----GVDRDRKSREKQQTLGE 131
G Q S TL LSK+ V EVWK + +K G + + QQTLGE
Sbjct: 63 --------GSQRQGSTTLPPTLSKQNVGEVWKSITEEKHTNNNGGVTNITHLQGQQTLGE 114
Query: 132 MTLEDFLVKAG-----VVGESFHGKESGL---------------LRVDSNEDSRQKVSHG 171
+TLE+F ++AG G S H S + V ++ SH
Sbjct: 115 ITLEEFFIRAGARGGNTNGGSIHDSSSSISGNPHTSLGVQIQPKAMVSDFMNNMVPRSHD 174
Query: 172 LHWMQYPVHSVQQQQHQYEKHTMP-GFAAVHAIQ--QPFQVAGNQALD------------ 216
+ Q S+ Q Q +MP G++ I+ +GNQ+L
Sbjct: 175 SYLHQNVNGSMSTYQPQQSIMSMPNGYSYGKQIRFSNGSLGSGNQSLQDTKRSLVPSVAT 234
Query: 217 -----AAISPSSLMGTLSDTQTLGRK--------------------RVASGIVVEKT-VE 250
SP + TL+ Q + + ++ S I EK V+
Sbjct: 235 IPSEAITCSPVTPFPTLNGKQKINGESSLLSPSPYISNGSTSTRGGKINSEITAEKQFVD 294
Query: 251 RRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEMEKEVPTAP----- 305
++ +R IKNRESAARSRAR+QA T E+E+++ L+++ E L +Q+ ++ P
Sbjct: 295 KKLRRKIKNRESAARSRARKQAQTMEVEVELENLKKDYEELLKQHVELRKRQMEPGMISL 354
Query: 306 PPEPKNQLRRTNS 318
P+ +LRRT S
Sbjct: 355 HERPERKLRRTKS 367
>At4g35900 bZIP transcription factor AtbZIP14
Length = 285
Score = 80.5 bits (197), Expect = 1e-15
Identities = 55/130 (42%), Positives = 72/130 (55%), Gaps = 21/130 (16%)
Query: 192 HTMPGFAAVHAIQQPFQVAGNQALDAAISPSSLMGTLSDTQTLGRKRVASGIVVEKTVER 251
HT + HA F+ A+ PSS + G+KR E + R
Sbjct: 177 HTHHHLSNAHAFNTSFE---------ALVPSS---------SFGKKRGQDSN--EGSGNR 216
Query: 252 RQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEMEKEVPTAPPPEPKN 311
R KRMIKNRESAARSRAR+QAYT ELEL+V+ L+ EN RL+RQ + K P+ KN
Sbjct: 217 RHKRMIKNRESAARSRARKQAYTNELELEVAHLQAENARLKRQQDQLKMAAAIQQPK-KN 275
Query: 312 QLRRTNSASF 321
L+R+++A F
Sbjct: 276 TLQRSSTAPF 285
>At5g44080 bZIP protein AtbZIP13
Length = 315
Score = 79.3 bits (194), Expect = 3e-15
Identities = 81/236 (34%), Positives = 105/236 (44%), Gaps = 60/236 (25%)
Query: 100 KTVDEVWKDM-QGKKRGVDRDRKSREKQQTLGE-MTLEDFLVKAGVVGESFHGKESGLLR 157
K+VDE+W++M G+ +G+ K++T E MTLEDFL KA V E+
Sbjct: 120 KSVDEIWREMVSGEGKGM--------KEETSEEIMTLEDFLAKAAVEDET--------AV 163
Query: 158 VDSNEDSRQKVSHGLHWMQYPVHSVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDA 217
S ED K+ PV T GF PFQ+ +D
Sbjct: 164 TASAEDLDVKI---------PV-------------TNYGFDHSAPPHNPFQM-----IDK 196
Query: 218 AISPSSLMGTLSDTQTLGRKRVASGIVVE---KTVERRQKRMIKNRESAARSRARRQAYT 274
G D G + + ++VE K +RQ+RMIKNRESAARSR R+QAY
Sbjct: 197 VEGSIVAFGNGLDVYGGGARGKRARVMVEPLDKAAAQRQRRMIKNRESAARSRERKQAYQ 256
Query: 275 QELELKVSRLEEENERL----------RRQNEMEKEVPTA--PPPEPKNQLRRTNS 318
ELE ++LEEENE L R Q ME +P P +P LRR S
Sbjct: 257 VELEALAAKLEEENELLSKEIEDKRKERYQKLMEFVIPVVEKPKQQPPRFLRRIRS 312
>At2g17770 bZIP transcription factor atbzip27
Length = 234
Score = 74.7 bits (182), Expect = 6e-14
Identities = 40/72 (55%), Positives = 53/72 (73%), Gaps = 1/72 (1%)
Query: 250 ERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEMEKEVPTAPPPEP 309
+RR KRMIKNRESAARSRAR+QAYT ELEL+++ L+ EN RL+ Q E K + A +
Sbjct: 164 DRRYKRMIKNRESAARSRARKQAYTNELELEIAHLQTENARLKIQQEQLK-IAEATQNQV 222
Query: 310 KNQLRRTNSASF 321
K L+R+++A F
Sbjct: 223 KKTLQRSSTAPF 234
>At1g03970 G-box binding bZip transcription factor GBF4 / AtbZip40
Length = 270
Score = 74.3 bits (181), Expect = 8e-14
Identities = 67/231 (29%), Positives = 108/231 (46%), Gaps = 60/231 (25%)
Query: 100 KTVDEVWKDM-QGKKRGVDRDRKSREKQQTLGEMTLEDFLVKAGVVGESFHGKESGLLRV 158
K+VD+VWK++ G+++ + ++++ MTLEDFL KA + E +
Sbjct: 85 KSVDDVWKEIVSGEQKTI-----MMKEEEPEDIMTLEDFLAKAEM-------DEGASDEI 132
Query: 159 DSNEDSRQKVSHGLHWMQYPVHSVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAA 218
D + + + G + +P+ Q+H FQ+
Sbjct: 133 DVKIPTERLNNDGSYTFDFPM-----QRHS-----------------SFQM--------- 161
Query: 219 ISPSSLMGTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELE 278
+ G++ T G++ ++K +RQKRMIKNRESAARSR R+QAY ELE
Sbjct: 162 -----VEGSMGGGVTRGKRGRVMMEAMDKAAAQRQKRMIKNRESAARSRERKQAYQVELE 216
Query: 279 LKVSRLEEENERLRRQNE----------MEKEVPTAPPPEPKNQ-LRRTNS 318
++LEEENE+L ++ E ME +P P P ++ L R++S
Sbjct: 217 TLAAKLEEENEQLLKEIEESTKERYKKLMEVLIPVDEKPRPPSRPLSRSHS 267
>At5g28770 bZip transcription factor BZO2H3 / AtbZip63
Length = 307
Score = 50.8 bits (120), Expect = 1e-06
Identities = 30/75 (40%), Positives = 43/75 (57%), Gaps = 1/75 (1%)
Query: 220 SPSSLMGTLSDTQTLGRKRVASGIV-VEKTVERRQKRMIKNRESAARSRARRQAYTQELE 278
+P S ++ G + A G + T +R KRM+ NRESA RSR R+QA+ ELE
Sbjct: 114 TPMMSSAITSGSELSGDEEEADGETNMNPTNVKRVKRMLSNRESARRSRRRKQAHLSELE 173
Query: 279 LKVSRLEEENERLRR 293
+VS+L EN +L +
Sbjct: 174 TQVSQLRVENSKLMK 188
>At5g65210 bZip transcription factor TGA1 / AtbZip47
Length = 368
Score = 49.7 bits (117), Expect = 2e-06
Identities = 30/89 (33%), Positives = 51/89 (56%), Gaps = 8/89 (8%)
Query: 209 VAGNQALDAAISPSSLMGTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRA 268
+ NQ LD +S + GT ++ S + ++ Q+R+ +NRE+A +SR
Sbjct: 47 IPNNQKLDNNVSEDTSHGTAGTPHMFDQEASTS-----RHPDKIQRRLAQNREAARKSRL 101
Query: 269 RRQAYTQELE---LKVSRLEEENERLRRQ 294
R++AY Q+LE LK+ +LE+E +R R+Q
Sbjct: 102 RKKAYVQQLETSRLKLIQLEQELDRARQQ 130
>At4g37730 bZip transcription factor AtbZip7
Length = 305
Score = 47.4 bits (111), Expect = 1e-05
Identities = 24/46 (52%), Positives = 32/46 (69%)
Query: 246 EKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERL 291
E T ER++KRM NRESA RSR R+Q++ L +V+RL+ EN L
Sbjct: 192 EMTDERKRKRMESNRESAKRSRMRKQSHIDNLREQVNRLDLENREL 237
>At3g49760 bZIP transcription factor AtbZip5
Length = 156
Score = 47.4 bits (111), Expect = 1e-05
Identities = 19/49 (38%), Positives = 37/49 (74%)
Query: 246 EKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQ 294
+ T ER++KR + NRESA RSR ++Q + +E+ +++++L+ +N+ L+ Q
Sbjct: 67 DNTDERKKKRKLSNRESAKRSREKKQKHLEEMSIQLNQLKIQNQELKNQ 115
>At4g01120 G-box binding bZip transcription factor GBF2 / AtbZip54
Length = 360
Score = 47.0 bits (110), Expect = 1e-05
Identities = 30/59 (50%), Positives = 37/59 (61%), Gaps = 6/59 (10%)
Query: 246 EKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQ-----NEMEK 299
EK V +R+KR NRESA RSR R+QA T++L +KV L EN LR + NE EK
Sbjct: 247 EKEV-KREKRKQSNRESARRSRLRKQAETEQLSVKVDALVAENMSLRSKLGQLNNESEK 304
>At1g77920 bZip transcription factor AtbZip50
Length = 368
Score = 47.0 bits (110), Expect = 1e-05
Identities = 21/51 (41%), Positives = 40/51 (78%), Gaps = 3/51 (5%)
Query: 250 ERRQKRMIKNRESAARSRARRQAYTQELE---LKVSRLEEENERLRRQNEM 297
++ ++R+ +NRE+A +SR R++AY Q+LE LK+S+LE+E E++++Q +
Sbjct: 92 DKMKRRLAQNREAARKSRLRKKAYVQQLEESRLKLSQLEQELEKVKQQGHL 142
>At2g04038 bZip transcription factor AtbZip48
Length = 166
Score = 46.6 bits (109), Expect = 2e-05
Identities = 25/53 (47%), Positives = 35/53 (65%), Gaps = 4/53 (7%)
Query: 250 ERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEEN----ERLRRQNEME 298
ER+Q+RM+ NRESA RSR R+Q + EL +V RL EN ++L R +E +
Sbjct: 73 ERKQRRMLSNRESARRSRMRKQRHLDELWSQVIRLRNENNCLIDKLNRVSETQ 125
>At1g68640 bZip transcription factor PERIANTHIA (PAN) / AtbZip46
Length = 452
Score = 46.2 bits (108), Expect = 2e-05
Identities = 22/56 (39%), Positives = 41/56 (72%), Gaps = 3/56 (5%)
Query: 245 VEKTVERRQKRMIKNRESAARSRARRQAYTQELE---LKVSRLEEENERLRRQNEM 297
V+ + +R +R+ +NRE+A +SR R++AY Q+LE +++++LEEE +R R+Q +
Sbjct: 160 VKSSDQRTLRRLAQNREAARKSRLRKKAYVQQLENSRIRLAQLEEELKRARQQGSL 215
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.127 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,034,160
Number of Sequences: 26719
Number of extensions: 298461
Number of successful extensions: 1612
Number of sequences better than 10.0: 137
Number of HSP's better than 10.0 without gapping: 88
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 1396
Number of HSP's gapped (non-prelim): 222
length of query: 321
length of database: 11,318,596
effective HSP length: 99
effective length of query: 222
effective length of database: 8,673,415
effective search space: 1925498130
effective search space used: 1925498130
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146910.4


