

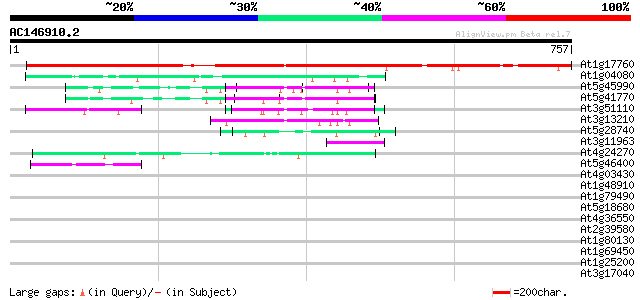
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146910.2 + phase: 1 /pseudo/partial
(757 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g17760 unknown protein 935 0.0
At1g04080 unknown protein 77 4e-14
At5g45990 CRN (crooked neck) protein 69 7e-12
At5g41770 cell cycle control crn (crooked neck) protein-like 65 1e-10
At3g51110 crooked neck-like protein 65 1e-10
At3g13210 crn-like protein 50 6e-06
At5g28740 putative protein 49 1e-05
At3g11963 pre-rRNA processing protein like 47 5e-05
At4g24270 unknown protein 46 9e-05
At5g46400 putative protein 43 6e-04
At4g03430 putative pre-mRNA splicing factor 34 0.27
At1g48910 hypothetical protein 34 0.35
At1g79490 unknown protein 33 0.45
At5g18680 tub family-like protein 33 0.77
At4g36550 putative protein 32 1.3
At2g39580 unknown protein 32 1.7
At1g80130 unknown protein 31 2.2
At1g69450 hypothetical protein 31 2.2
At1g25200 31 2.9
At3g17040 unknown protein 30 3.8
>At1g17760 unknown protein
Length = 734
Score = 935 bits (2416), Expect = 0.0
Identities = 489/765 (63%), Positives = 577/765 (74%), Gaps = 83/765 (10%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
IA+ATPIYEQLL LYPT+A+FWKQYVEA MAVNNDDA KQIFSRCLL CLQVPLW+CYIR
Sbjct: 22 IAQATPIYEQLLSLYPTSARFWKQYVEAQMAVNNDDATKQIFSRCLLTCLQVPLWQCYIR 81
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
FIRKV DKKGAEGQEET KAFEFML+Y+G+DIASGP+W EYIAFLKSLPA + E+ HR
Sbjct: 82 FIRKVYDKKGAEGQEETTKAFEFMLNYIGTDIASGPIWTEYIAFLKSLPALNLNEDLHRK 141
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T +RKVY RAI+TPTHH+EQLWKDY++FE++V+++LAKGL++EYQPK+NSARAVYRERKK
Sbjct: 142 TALRKVYHRAILTPTHHVEQLWKDYENFENTVNRQLAKGLVNEYQPKFNSARAVYRERKK 201
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
+ +EIDWNMLAVPPTG+ K K +LS F N
Sbjct: 202 YIEEIDWNMLAVPPTGTSKEETQWVAWKKFLS---------FEKGNP------------- 239
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSIDAA 322
+RID ASS KR+I+ YEQCLM LYHYPDVWYDYA WH K+GS DAA
Sbjct: 240 --------------QRIDTASSTKRIIYAYEQCLMCLYHYPDVWYDYAEWHVKSGSTDAA 285
Query: 323 IKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRFL 382
IKVFQR+LKA+PDSEML+YA+AE+EESRGAIQ+AKK+YEN+LG S N+ LAHIQ++RFL
Sbjct: 286 IKVFQRALKAIPDSEMLKYAFAEMEESRGAIQSAKKLYENILGASTNS--LAHIQYLRFL 343
Query: 383 RRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPV 442
RR EGVE ARKYFLDARKSPSCTYHVY+A+A++AFC+DK+PK+AHN+FE GLK +M EPV
Sbjct: 344 RRAEGVEAARKYFLDARKSPSCTYHVYIAFATMAFCIDKEPKVAHNIFEEGLKLYMSEPV 403
Query: 443 YILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQR 502
YIL+YADFL RLNDD+NIRALFERALS+LP+EDS EVWKRF++FEQTYGDLAS+LKVEQR
Sbjct: 404 YILKYADFLTRLNDDRNIRALFERALSTLPVEDSAEVWKRFIQFEQTYGDLASILKVEQR 463
Query: 503 RKEAF---GEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQEWLVKNTKKVEKSI 559
KEA GEE ++ ESSLQDVVSRYS+MDLWPC+SNDLD+L+RQE LVKN K
Sbjct: 464 MKEALSGKGEEGSSPPESSLQDVVSRYSYMDLWPCTSNDLDHLARQELLVKNLNKKA--- 520
Query: 560 MLNGTTFIDKGPVASIS-TTSSKVVYPDTSKMLIYDP--------KHNPGTGAA------ 604
G T + P A S +SSKVVYPDTS+M++ DP NP +A
Sbjct: 521 ---GKTNLPHVPAAIGSVASSSKVVYPDTSQMVVQDPTKKSEFASSANPVAASASNTFPS 577
Query: 605 ---------GTNAFDEILKATPPALVAFLANLPSVDGPTPNVDIVLSICLQSDLPTGQSV 655
+ FDEI K TPPALVAFLANLP VDGPTPNVD+VLSICLQSD PTGQ+V
Sbjct: 578 TVTATATHGSASTFDEIPKTTPPALVAFLANLPIVDGPTPNVDVVLSICLQSDFPTGQTV 637
Query: 656 KVGIPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQ 715
K ++ G P+ ++ SG ++ G+S P ++ KRK D QEEDDT +VQ
Sbjct: 638 KQSFAAK---GNPPSQNDPSGPTR------GVSQRLPRDRRATKRKDSDRQEEDDTATVQ 688
Query: 716 SQPLPQDAFRIRQFQKARAGSTSQ---TGSVSYGSALSGDLSGST 757
SQPLP D FR+RQ +KAR +TS TGS SYGSA SG+LSGST
Sbjct: 689 SQPLPTDVFRLRQMRKARGIATSSQTPTGSTSYGSAFSGELSGST 733
>At1g04080 unknown protein
Length = 768
Score = 77.0 bits (188), Expect = 4e-14
Identities = 105/536 (19%), Positives = 203/536 (37%), Gaps = 86/536 (16%)
Query: 22 SIAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCL-QVPLWRCY 80
+IA+ +Y+ L +P +WK++ + V D + +++ R +L V +W Y
Sbjct: 117 NIAKIRKVYDAFLAEFPLCYGYWKKFADHEARVGAMDKVVEVYERAVLGVTYSVDIWLHY 176
Query: 81 IRFIRKVNDKKGAEGQEET-KKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEET 139
F +N G ET ++ FE L YVG+D S P+W +YI + + Q++
Sbjct: 177 CTFA--IN----TYGDPETIRRLFERALVYVGTDFLSSPLWDKYIEY------EYMQQDW 224
Query: 140 HRMTVVRKVYQRAIITPTHHIEQLWKDYDSF-------------ESSVSQKLAKGLISEY 186
R+ + +Y R + P ++++ + + ES+ + G SE
Sbjct: 225 SRVAL---IYTRILENPIQNLDRYFSSFKELAETRPLSELRSAEESAAAAVAVAGDASES 281
Query: 187 QPKYNSARA-VYRERKKFFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFW 245
+ +A R + E + + T + K++ + + F +K+
Sbjct: 282 AASESGEKADEGRSQVDGSTEQSPKLESASSTEPEELKKYVGIREAMYIKSKEFESKIIG 341
Query: 246 NDN-------HFPPKSLCLFLLTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMY 298
+ H P ++ L N +D +E F +V+ YE+C++
Sbjct: 342 YEMAIRRPYFHVRPLNVA-ELENWHNYLDFIERDGDF----------NKVVKLYERCVVT 390
Query: 299 LYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAY-AELEESRGAIQAAK 357
+YP+ W Y T +GS D A R+ + + + + A L+E G I A+
Sbjct: 391 CANYPEYWIRYVTNMEASGSADLAENALARATQVFVKKQPEIHLFAARLKEQNGDIAGAR 450
Query: 358 KIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYH------VYVA 411
Y+ + + A I+ R ++ A + H +Y
Sbjct: 451 AAYQLVHSEISPGLLEAVIKHANMEYRLGNLDDAFSLYEQVIAVEKGKEHSTILPLLYAQ 510
Query: 412 YASVAFCLDKDPKMAHNVFEAGLKHF---------------MHEPVYILEYADFLIRL-- 454
Y+ ++ + +D + A + L H + P ++Y + L+
Sbjct: 511 YSRFSYLVSRDAEKARRIIVEALDHVQPSKPLMEALIHFEAIQPPPREIDYLEPLVEKVI 570
Query: 455 ---NDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAF 507
D QNI + ER SL +++F +GD+ S+ K E + + F
Sbjct: 571 KPDADAQNIASSTEREELSL----------IYIEFLGIFGDVKSIKKAEDQHVKLF 616
>At5g45990 CRN (crooked neck) protein
Length = 673
Score = 69.3 bits (168), Expect = 7e-12
Identities = 53/206 (25%), Positives = 96/206 (45%), Gaps = 10/206 (4%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRG 351
+E+ L Y +W YA + K ++ A V+ RS+ LP + L Y +EE G
Sbjct: 101 WERALEGEYRNHTLWVKYAEFEMKNKFVNNARNVWDRSVTLLPRVDQLWEKYIYMEEKLG 160
Query: 352 AIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHV--Y 409
+ A++I+E + S + A + FI+F R +E AR + + C V +
Sbjct: 161 NVTGARQIFERWMNWSPDQK--AWLCFIKFELRYNEIERARSIY---ERFVLCHPKVSAF 215
Query: 410 VAYASVAFCLDKDPKMAHNVFEAGLKHFMHE---PVYILEYADFLIRLNDDQNIRALFER 466
+ YA K+A V+E + ++ + + +A+F R + + R +++
Sbjct: 216 IRYAKFEMKRGGQVKLAREVYERAVDKLANDEEAEILFVSFAEFEERCKEVERARFIYKF 275
Query: 467 ALSSLPLEDSVEVWKRFVKFEQTYGD 492
AL + + E++K+FV FE+ YGD
Sbjct: 276 ALDHIRKGRAEELYKKFVAFEKQYGD 301
Score = 51.2 bits (121), Expect = 2e-06
Identities = 77/341 (22%), Positives = 132/341 (38%), Gaps = 75/341 (21%)
Query: 76 LWRCYIRFIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPV----WMEYIAFLKSLP 131
L++ ++ F ++ DK+G E KK FE+ +++ P+ W +Y+
Sbjct: 288 LYKKFVAFEKQYGDKEGIEDAIVGKKRFEYE-----DEVSKNPLNYDSWFDYVRL----- 337
Query: 132 AAHPQEETHRMTVVRKVYQRAIIT-PTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPK- 189
+E +R++Y+RAI P ++ W+ Y + + L E + K
Sbjct: 338 ----EESVGNKDRIREIYERAIANVPPAQEKRFWQRY------IYLWINYALYEEIETKD 387
Query: 190 YNSARAVYRERKKFFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNH 249
R VYRE K +P T KF F K WL + +L
Sbjct: 388 VERTRDVYRECLKL----------IPHT------KFSF-AKIWLLAAEYEIRQLN----- 425
Query: 250 FPPKSLCLFLLTGR-----NAVDVMEETSIF*KRIDIAS---SNKRVIFTYEQCLMYLYH 301
LTG NA+ + IF K I++ + R YE+ L +
Sbjct: 426 ----------LTGARQILGNAIGKAPKVKIFKKYIEMELKLVNIDRCRKLYERFLEWSPE 475
Query: 302 YPDVWYDYATWHAKAGSIDAAIKVFQRSLK--ALPDSEMLRYAYAELEESRGAIQAAKKI 359
W +YA + + A +F+ ++ AL E+L Y + E S G + + +
Sbjct: 476 NCYAWRNYAEFEISLAETERARAIFELAISQPALDMPELLWKTYIDFEISEGEFEKTRAL 535
Query: 360 YENLLGDSENATALAHIQFIRF-----LRRTEGVEPARKYF 395
YE LL +++ I F +F + +G++ AR F
Sbjct: 536 YERLLDRTKHCK--VWISFAKFEASASEHKEDGIKSARVIF 574
Score = 45.1 bits (105), Expect = 2e-04
Identities = 49/196 (25%), Positives = 87/196 (44%), Gaps = 23/196 (11%)
Query: 306 WYDYATWHAKAGSIDAAIKVFQRSLKALPD-SEMLRYAYAELEESRGAIQAAKKIYENL- 363
W + + + I+ A +++R + P S +RYA E++ G ++ A+++YE
Sbjct: 182 WLCFIKFELRYNEIERARSIYERFVLCHPKVSAFIRYAKFEMKRG-GQVKLAREVYERAV 240
Query: 364 --LGDSENATALAHIQFIRFLRRTEGVEPAR---KYFLDARKSPSCTYHVYVAYASVAFC 418
L + E A L + F F R + VE AR K+ LD + +Y + VAF
Sbjct: 241 DKLANDEEAEIL-FVSFAEFEERCKEVERARFIYKFALDHIRKGRAE-ELYKKF--VAFE 296
Query: 419 LDKDPKMAHNVFEAGLKHFMHE------PVYILEYADFLIRLNDD----QNIRALFERAL 468
K G K F +E P+ + D+ +RL + IR ++ERA+
Sbjct: 297 KQYGDKEGIEDAIVGKKRFEYEDEVSKNPLNYDSWFDY-VRLEESVGNKDRIREIYERAI 355
Query: 469 SSLPLEDSVEVWKRFV 484
+++P W+R++
Sbjct: 356 ANVPPAQEKRFWQRYI 371
Score = 42.4 bits (98), Expect = 0.001
Identities = 45/217 (20%), Positives = 91/217 (41%), Gaps = 25/217 (11%)
Query: 304 DVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRY------------AYAELEESRG 351
D W+DY G+ D ++++R++ +P ++ R+ Y E+E
Sbjct: 329 DSWFDYVRLEESVGNKDRIREIYERAIANVPPAQEKRFWQRYIYLWINYALYEEIETKD- 387
Query: 352 AIQAAKKIYEN---LLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDA-RKSPSCT-Y 406
++ + +Y L+ ++ + A + + R + AR+ +A K+P +
Sbjct: 388 -VERTRDVYRECLKLIPHTKFSFAKIWLLAAEYEIRQLNLTGARQILGNAIGKAPKVKIF 446
Query: 407 HVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILE-YADFLIRLNDDQNIRALFE 465
Y+ +D+ K+ E + E Y YA+F I L + + RA+FE
Sbjct: 447 KKYIEMELKLVNIDRCRKLYERFLE-----WSPENCYAWRNYAEFEISLAETERARAIFE 501
Query: 466 RALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQR 502
A+S L+ +WK ++ FE + G+ + +R
Sbjct: 502 LAISQPALDMPELLWKTYIDFEISEGEFEKTRALYER 538
Score = 37.0 bits (84), Expect = 0.041
Identities = 38/200 (19%), Positives = 77/200 (38%), Gaps = 33/200 (16%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
+ A ++++ + L P + W++Y+ + N +QIF R + W C+I+
Sbjct: 128 VNNARNVWDRSVTLLPRVDQLWEKYIYMEEKLGNVTGARQIFERWMNWSPDQKAWLCFIK 187
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
F + N+ + A E F+L + P +I + K + ++
Sbjct: 188 FELRYNEIERARSIYE-----RFVLCH--------PKVSAFIRYAK-----FEMKRGGQV 229
Query: 143 TVVRKVYQRAI--ITPTHHIEQLWKDYDSFESSVSQ-------------KLAKGLISEYQ 187
+ R+VY+RA+ + E L+ + FE + + KG E
Sbjct: 230 KLAREVYERAVDKLANDEEAEILFVSFAEFEERCKEVERARFIYKFALDHIRKGRAEELY 289
Query: 188 PKYNSARAVYRERKKFFDEI 207
K+ + Y +++ D I
Sbjct: 290 KKFVAFEKQYGDKEGIEDAI 309
Score = 34.7 bits (78), Expect = 0.20
Identities = 41/193 (21%), Positives = 78/193 (40%), Gaps = 13/193 (6%)
Query: 302 YPDVWYDYATWHA-KAGSIDAAIKVFQRSLKALPDSEM----LRYAYAELEESRGAIQAA 356
Y +W +YA + + ++ V++ LK +P ++ + AE E + + A
Sbjct: 370 YIYLWINYALYEEIETKDVERTRDVYRECLKLIPHTKFSFAKIWLLAAEYEIRQLNLTGA 429
Query: 357 KKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVA 416
++I N +G + ++I + ++ RK + + + + YA
Sbjct: 430 RQILGNAIGKAPKVKIFK--KYIEMELKLVNIDRCRKLYERFLEWSPENCYAWRNYAEFE 487
Query: 417 FCLDKDPKMAHNVFEAGLKHFMHEPVYIL--EYADFLIRLNDDQNIRALFERALSSLPLE 474
L + + A +FE + + +L Y DF I + + RAL+ER L
Sbjct: 488 ISL-AETERARAIFELAISQPALDMPELLWKTYIDFEISEGEFEKTRALYERLLDR---T 543
Query: 475 DSVEVWKRFVKFE 487
+VW F KFE
Sbjct: 544 KHCKVWISFAKFE 556
Score = 32.7 bits (73), Expect = 0.77
Identities = 30/136 (22%), Positives = 59/136 (43%), Gaps = 19/136 (13%)
Query: 21 LSIAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLN-CLQVP--LW 77
++I +YE+ L+ P W+ Y E +++ + + IF + L +P LW
Sbjct: 457 VNIDRCRKLYERFLEWSPENCYAWRNYAEFEISLAETERARAIFELAISQPALDMPELLW 516
Query: 78 RCYIRFIRKVNDKKGAEGQ-EETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQ 136
+ YI F + +EG+ E+T+ +E +L VW+ + F A+ +
Sbjct: 517 KTYIDF-------EISEGEFEKTRALYERLLDRT----KHCKVWISFAKF----EASASE 561
Query: 137 EETHRMTVVRKVYQRA 152
+ + R ++ RA
Sbjct: 562 HKEDGIKSARVIFDRA 577
Score = 29.6 bits (65), Expect = 6.5
Identities = 16/55 (29%), Positives = 24/55 (43%), Gaps = 3/55 (5%)
Query: 30 YEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLN---CLQVPLWRCYI 81
YE + P W YV +V N D I++I+ R + N + W+ YI
Sbjct: 317 YEDEVSKNPLNYDSWFDYVRLEESVGNKDRIREIYERAIANVPPAQEKRFWQRYI 371
>At5g41770 cell cycle control crn (crooked neck) protein-like
Length = 705
Score = 65.1 bits (157), Expect = 1e-10
Identities = 50/206 (24%), Positives = 96/206 (46%), Gaps = 11/206 (5%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRG 351
+E+ + Y +W YA + K +++A V+ R++ LP + L Y Y +EE G
Sbjct: 115 WERAIEGDYRNHTLWLKYAEFEMKNKFVNSARNVWDRAVTLLPRVDQLWYKYIHMEEILG 174
Query: 352 AIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHV--Y 409
I A++I+E + S + + FI+F R +E AR + + C V Y
Sbjct: 175 NIAGARQIFERWMDWSPDQQ--GWLSFIKFELRYNEIERARTIY---ERFVLCHPKVSAY 229
Query: 410 VAYASVAFCLDKDPKMAHNVFEAGLKHFMHE---PVYILEYADFLIRLNDDQNIRALFER 466
+ YA + +V+E + + + + +A+F R + + R +++
Sbjct: 230 IRYAKFEM-KGGEVARCRSVYERATEKLADDEEAEILFVAFAEFEERCKEVERARFIYKF 288
Query: 467 ALSSLPLEDSVEVWKRFVKFEQTYGD 492
AL +P + +++++FV FE+ YGD
Sbjct: 289 ALDHIPKGRAEDLYRKFVAFEKQYGD 314
Score = 47.8 bits (112), Expect = 2e-05
Identities = 70/307 (22%), Positives = 113/307 (36%), Gaps = 72/307 (23%)
Query: 76 LWRCYIRFIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHP 135
L+R ++ F ++ DK+G E K+ F++ V ++ W +Y+
Sbjct: 301 LYRKFVAFEKQYGDKEGIEDAIVGKRRFQYE-DEVRKSPSNYDSWFDYVRL--------- 350
Query: 136 QEETHRMTVVRKVYQRAIITPTHHIEQ--------LWKDYDSFESSVSQKLAKGLISEYQ 187
+E +R++Y+RAI E+ LW +Y FE ++ + +
Sbjct: 351 EESVGNKDRIREIYERAIANVPPAEEKRYWQRYIYLWINYALFEEIETEDIER------- 403
Query: 188 PKYNSARAVYRERKKFFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWND 247
R VYRE K SKF F K WL + +L
Sbjct: 404 -----TRDVYRECLKLIPH----------------SKFSF-AKIWLLAAQFEIRQLN--- 438
Query: 248 NHFPPKSLCLFLLTGR-----NAVDVMEETSIF*KRIDIA---SSNKRVIFTYEQCLMYL 299
LTG NA+ + IF K I+I + R YE+ L +
Sbjct: 439 ------------LTGARQILGNAIGKAPKDKIFKKYIEIELQLGNMDRCRKLYERYLEWS 486
Query: 300 YHYPDVWYDYATWHAKAGSIDAAIKVFQRSLK--ALPDSEMLRYAYAELEESRGAIQAAK 357
W YA + A +F+ ++ AL E+L AY + E S G ++ +
Sbjct: 487 PENCYAWSKYAELERSLVETERARAIFELAISQPALDMPELLWKAYIDFEISEGELERTR 546
Query: 358 KIYENLL 364
+YE LL
Sbjct: 547 ALYERLL 553
Score = 44.3 bits (103), Expect = 3e-04
Identities = 47/204 (23%), Positives = 88/204 (43%), Gaps = 17/204 (8%)
Query: 304 DVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRY---------AYAELEESRGA-I 353
D W+DY G+ D ++++R++ +P +E RY YA EE I
Sbjct: 342 DSWFDYVRLEESVGNKDRIREIYERAIANVPPAEEKRYWQRYIYLWINYALFEEIETEDI 401
Query: 354 QAAKKIYEN---LLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDA-RKSPSCTYHVY 409
+ + +Y L+ S+ + A + +F R + AR+ +A K+P ++
Sbjct: 402 ERTRDVYRECLKLIPHSKFSFAKIWLLAAQFEIRQLNLTGARQILGNAIGKAPKDK--IF 459
Query: 410 VAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS 469
Y + L + ++E L+ +YA+ L + + RA+FE A+S
Sbjct: 460 KKYIEIELQLGNMDR-CRKLYERYLEWSPENCYAWSKYAELERSLVETERARAIFELAIS 518
Query: 470 SLPLEDSVEVWKRFVKFEQTYGDL 493
L+ +WK ++ FE + G+L
Sbjct: 519 QPALDMPELLWKAYIDFEISEGEL 542
Score = 40.4 bits (93), Expect = 0.004
Identities = 28/111 (25%), Positives = 55/111 (49%), Gaps = 3/111 (2%)
Query: 392 RKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFL 451
RK F D + V+V YA KD A +V+E ++ L+YA+F
Sbjct: 78 RKEFEDQIRRARWNIQVWVKYAQWEES-QKDYARARSVWERAIEGDYRNHTLWLKYAEFE 136
Query: 452 IRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQR 502
++ + R +++RA++ LP D ++W +++ E+ G++A ++ +R
Sbjct: 137 MKNKFVNSARNVWDRAVTLLPRVD--QLWYKYIHMEEILGNIAGARQIFER 185
Score = 36.2 bits (82), Expect = 0.070
Identities = 44/203 (21%), Positives = 83/203 (40%), Gaps = 13/203 (6%)
Query: 298 YLYHYPDVWYDYATWHA-KAGSIDAAIKVFQRSLKALPDSEM----LRYAYAELEESRGA 352
Y Y +W +YA + + I+ V++ LK +P S+ + A+ E +
Sbjct: 379 YWQRYIYLWINYALFEEIETEDIERTRDVYRECLKLIPHSKFSFAKIWLLAAQFEIRQLN 438
Query: 353 IQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAY 412
+ A++I N +G + ++I + ++ RK + + + + Y
Sbjct: 439 LTGARQILGNAIGKAPKDKIFK--KYIEIELQLGNMDRCRKLYERYLEWSPENCYAWSKY 496
Query: 413 ASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYIL--EYADFLIRLNDDQNIRALFERALSS 470
A + L + + A +FE + + +L Y DF I + + RAL+ER L
Sbjct: 497 AELERSL-VETERARAIFELAISQPALDMPELLWKAYIDFEISEGELERTRALYERLLDR 555
Query: 471 LPLEDSVEVWKRFVKFEQTYGDL 493
+VW F KFE + +L
Sbjct: 556 ---TKHYKVWVSFAKFEASAAEL 575
Score = 30.8 bits (68), Expect = 2.9
Identities = 16/55 (29%), Positives = 26/55 (47%)
Query: 30 YEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIRFI 84
YE ++ P+ W YV +V N D I++I+ R + N R + R+I
Sbjct: 330 YEDEVRKSPSNYDSWFDYVRLEESVGNKDRIREIYERAIANVPPAEEKRYWQRYI 384
Score = 30.4 bits (67), Expect = 3.8
Identities = 29/114 (25%), Positives = 50/114 (43%), Gaps = 17/114 (14%)
Query: 29 IYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLN-CLQVP--LWRCYIRFIR 85
+YE+ L+ P W +Y E ++ + + IF + L +P LW+ YI F
Sbjct: 478 LYERYLEWSPENCYAWSKYAELERSLVETERARAIFELAISQPALDMPELLWKAYIDF-- 535
Query: 86 KVNDKKGAEGQ-EETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEE 138
+ +EG+ E T+ +E +L VW+ + F S AA +E+
Sbjct: 536 -----EISEGELERTRALYERLLDRT----KHYKVWVSFAKFEAS--AAELEED 578
>At3g51110 crooked neck-like protein
Length = 599
Score = 65.1 bits (157), Expect = 1e-10
Identities = 55/201 (27%), Positives = 96/201 (47%), Gaps = 16/201 (7%)
Query: 300 YHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKI 359
Y +W YA + + S++ A V+ R++K LP + Y Y +EE G I A+KI
Sbjct: 97 YRNHTLWLKYAEFEMRNKSVNHARNVWDRAVKILPRVDQFWYKYIHMEEILGNIDGARKI 156
Query: 360 YENLLGDSENATALAHIQFIRFLRRTEGVEPAR----KYFLDARKSPSCTYHVYVAYASV 415
+E + S + A + FI+F R +E +R ++ L K+ S ++ YA
Sbjct: 157 FERWMDWSPDQQ--AWLCFIKFELRYNEIERSRSIYERFVLCHPKASS-----FIRYAKF 209
Query: 416 AFCLDKDPKMAHNVFEAG---LKHFMHEPVYI-LEYADFLIRLNDDQNIRALFERALSSL 471
+ +A V+E LK E I + +A+F + + R L++ AL +
Sbjct: 210 EM-KNSQVSLARIVYERAIEMLKDVEEEAEMIFVAFAEFEELCKEVERARFLYKYALDHI 268
Query: 472 PLEDSVEVWKRFVKFEQTYGD 492
P + +++K+FV FE+ YG+
Sbjct: 269 PKGRAEDLYKKFVAFEKQYGN 289
Score = 50.8 bits (120), Expect = 3e-06
Identities = 59/266 (22%), Positives = 100/266 (37%), Gaps = 58/266 (21%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEM--------LRY-- 341
+++ + L WY Y G+ID A K+F+R + PD + LRY
Sbjct: 123 WDRAVKILPRVDQFWYKYIHMEEILGNIDGARKIFERWMDWSPDQQAWLCFIKFELRYNE 182
Query: 342 ----------------------AYAELEESRGAIQAAKKIYE---NLLGDSENATALAHI 376
YA+ E + A+ +YE +L D E + +
Sbjct: 183 IERSRSIYERFVLCHPKASSFIRYAKFEMKNSQVSLARIVYERAIEMLKDVEEEAEMIFV 242
Query: 377 QFIRFLRRTEGVEPAR---KYFLD--ARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFE 431
F F + VE AR KY LD + Y +VA+ +K + +
Sbjct: 243 AFAEFEELCKEVERARFLYKYALDHIPKGRAEDLYKKFVAF-------EKQYGNKEGIDD 295
Query: 432 A--GLKHFMHE------PVYILEYADFLI---RLNDDQNIRALFERALSSLPLEDSVEVW 480
A G + +E P+ + D++ L D IR ++ERA++++PL + W
Sbjct: 296 AIVGRRKLQYEGEVRKNPLNYDSWFDYISLEETLGDKDRIREVYERAIANVPLAEEKRYW 355
Query: 481 KRFVKFEQTYGDLASMLKVEQRRKEA 506
+R++ Y +L + R A
Sbjct: 356 QRYIYLWIDYALFEEILAEDVERTRA 381
Score = 43.1 bits (100), Expect = 6e-04
Identities = 37/177 (20%), Positives = 73/177 (40%), Gaps = 22/177 (12%)
Query: 22 SIAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYI 81
S+ A ++++ +++ P +FW +Y+ + N D ++IF R + W C+I
Sbjct: 115 SVNHARNVWDRAVKILPRVDQFWYKYIHMEEILGNIDGARKIFERWMDWSPDQQAWLCFI 174
Query: 82 RFIRKVNDKKGAEGQEET-----KKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQ 136
+F + N+ + + E KA F + Y ++ + V + I + +++
Sbjct: 175 KFELRYNEIERSRSIYERFVLCHPKASSF-IRYAKFEMKNSQVSLARIVYERAIEMLKDV 233
Query: 137 EETHRMTVV---------------RKVYQRAI-ITPTHHIEQLWKDYDSFESSVSQK 177
EE M V R +Y+ A+ P E L+K + +FE K
Sbjct: 234 EEEAEMIFVAFAEFEELCKEVERARFLYKYALDHIPKGRAEDLYKKFVAFEKQYGNK 290
Score = 37.7 bits (86), Expect = 0.024
Identities = 44/191 (23%), Positives = 73/191 (38%), Gaps = 28/191 (14%)
Query: 304 DVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENL 363
D W+DY + G D +V++R++ +P +E RY + + ++E +
Sbjct: 317 DSWFDYISLEETLGDKDRIREVYERAIANVPLAEEKRYWQRYIY-----LWIDYALFEEI 371
Query: 364 LGDSENATALAHIQFI---RFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLD 420
L + T + R L G P K F Y+ +D
Sbjct: 372 LAEDVERTRAVQLNLSGARRILGNAIGKAPKHKIFKK-----------YIEIELHLGNID 420
Query: 421 KDPKMAHNVFEAGLKHFMHEPVYI-LEYADFLIRLNDDQNIRALFERALSSLPLEDSV-- 477
+ K+ E + E Y ++A+F L + + RA+FE A+S L D
Sbjct: 421 RCRKLYARYLE-----WSPESCYAWTKFAEFERSLAETERARAIFELAISQPRLLDRTKH 475
Query: 478 -EVWKRFVKFE 487
+VW F KFE
Sbjct: 476 YKVWLSFAKFE 486
Score = 36.6 bits (83), Expect = 0.054
Identities = 38/182 (20%), Positives = 74/182 (39%), Gaps = 5/182 (2%)
Query: 324 KVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRFLR 383
K F+ ++ + + YA+ EES+ A+ ++E L D +++ F
Sbjct: 52 KEFEDQIRGAKTNSQVWVRYADWEESQKDHDRARSVWERALEDESYRNHTLWLKYAEFEM 111
Query: 384 RTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVY 443
R + V AR + A K + Y + L + A +FE + + +
Sbjct: 112 RNKSVNHARNVWDRAVKILPRVDQFWYKYIHMEEILG-NIDGARKIFERWMDWSPDQQAW 170
Query: 444 ILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRR 503
+ + F +R N+ + R+++ER + P S + R+ KFE ++ V +R
Sbjct: 171 LC-FIKFELRYNEIERSRSIYERFVLCHPKASS---FIRYAKFEMKNSQVSLARIVYERA 226
Query: 504 KE 505
E
Sbjct: 227 IE 228
>At3g13210 crn-like protein
Length = 657
Score = 49.7 bits (117), Expect = 6e-06
Identities = 59/268 (22%), Positives = 110/268 (41%), Gaps = 46/268 (17%)
Query: 272 TSIF*KRIDIASSNKRVIFT---YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQR 328
T ++ K D NK V +++ + L +WY + K G+I A ++ +R
Sbjct: 91 TQVWVKYADFEMKNKSVNEARNVWDRAVSLLPRVDQLWYKFIHMEEKLGNIAGARQILER 150
Query: 329 SLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGV 388
+ PD + + + E I+ A+ IYE + + A+I++ +F + V
Sbjct: 151 WIHCSPDQQAW-LCFIKFELKYNEIECARSIYERFVLCHPKVS--AYIRYAKFEMKHGQV 207
Query: 389 EPARKYFLDARKSPSCTYHVYVAYASVA-------FCLDKDPK-MAHNVFE--------- 431
E A K F A+K + + + + A F LD+ PK A N++
Sbjct: 208 ELAMKVFERAKKELADDEEAEILFVAFAEFEEQYKFALDQIPKGRAENLYSKFVAFEKQN 267
Query: 432 ----------AGLKHFMHE------PVYILEYADFLIRLND----DQNIRALFERALSSL 471
G + +E P+ + DF +RL + IR ++ERA++++
Sbjct: 268 GDKEGIEDAIIGKRRCQYEDEVRKNPLNYDSWFDF-VRLEETVGNKDRIREIYERAVANV 326
Query: 472 PLEDSVE--VWKRFVKFEQTYGDLASML 497
P ++ E W+R++ Y A M+
Sbjct: 327 PPPEAQEKRYWQRYIYLWINYAFFAEMV 354
Score = 40.4 bits (93), Expect = 0.004
Identities = 80/412 (19%), Positives = 146/412 (35%), Gaps = 76/412 (18%)
Query: 22 SIAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYI 81
S+ EA ++++ + L P + W +++ + N +QI R + W C+I
Sbjct: 106 SVNEARNVWDRAVSLLPRVDQLWYKFIHMEEKLGNIAGARQILERWIHCSPDQQAWLCFI 165
Query: 82 RFIRKVNDKKGAEGQEE----TKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQE 137
+F K N+ + A E + Y ++ G V + F ++ E
Sbjct: 166 KFELKYNEIECARSIYERFVLCHPKVSAYIRYAKFEMKHGQVELAMKVFERAKKELADDE 225
Query: 138 ETHRMTVV----RKVYQRAI-ITPTHHIEQLWKDYDSFESSVSQK--LAKGLISEYQPKY 190
E + V + Y+ A+ P E L+ + +FE K + +I + + +Y
Sbjct: 226 EAEILFVAFAEFEEQYKFALDQIPKGRAENLYSKFVAFEKQNGDKEGIEDAIIGKRRCQY 285
Query: 191 -NSARAVYRERKKFFD---------------EIDWNMLA-VPPTGSHKASKFLFLCKYWL 233
+ R +FD EI +A VPP + + +YW
Sbjct: 286 EDEVRKNPLNYDSWFDFVRLEETVGNKDRIREIYERAVANVPPPEAQEK-------RYWQ 338
Query: 234 SLLSVFVNKLFWN-------DNHFPPKSLCLFL------------------------LTG 262
+ +++N F+ ++ CL L LTG
Sbjct: 339 RYIYLWINYAFFAEMVTEDVESTRDVYRACLKLIPHSKFSFAKIWLLAAQHEIRQLNLTG 398
Query: 263 R-----NAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPD---VWYDYATWHA 314
NA+ + IF K I+I + + + YL P W YA +
Sbjct: 399 ARQILGNAIGKAPKDKIFKKYIEIELQLRNIDRCRKLYERYLEWSPGNCYAWRKYAEFEM 458
Query: 315 KAGSIDAAIKVFQRSLK--ALPDSEMLRYAYAELEESRGAIQAAKKIYENLL 364
+ +F+ ++ AL E+L Y + E S G ++ + +YE LL
Sbjct: 459 SLAETERTRAIFELAISQPALDMPELLWKTYIDFEISEGELERTRALYERLL 510
Score = 40.0 bits (92), Expect = 0.005
Identities = 47/210 (22%), Positives = 87/210 (41%), Gaps = 27/210 (12%)
Query: 304 DVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEML------RYAYA------------- 344
D W+D+ G+ D ++++R++ +P E RY Y
Sbjct: 297 DSWFDFVRLEETVGNKDRIREIYERAVANVPPPEAQEKRYWQRYIYLWINYAFFAEMVTE 356
Query: 345 ELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDA-RKSPS 403
++E +R +A K+ + LA IR L T AR+ +A K+P
Sbjct: 357 DVESTRDVYRACLKLIPHSKFSFAKIWLLAAQHEIRQLNLTG----ARQILGNAIGKAPK 412
Query: 404 CTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRAL 463
++ Y + L ++ ++E L+ +YA+F + L + + RA+
Sbjct: 413 DK--IFKKYIEIELQL-RNIDRCRKLYERYLEWSPGNCYAWRKYAEFEMSLAETERTRAI 469
Query: 464 FERALSSLPLEDSVEVWKRFVKFEQTYGDL 493
FE A+S L+ +WK ++ FE + G+L
Sbjct: 470 FELAISQPALDMPELLWKTYIDFEISEGEL 499
Score = 38.5 bits (88), Expect = 0.014
Identities = 32/130 (24%), Positives = 59/130 (44%), Gaps = 16/130 (12%)
Query: 22 SIAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLN-CLQVP--LWR 78
+I +YE+ L+ P W++Y E M++ + + IF + L +P LW+
Sbjct: 428 NIDRCRKLYERYLEWSPGNCYAWRKYAEFEMSLAETERTRAIFELAISQPALDMPELLWK 487
Query: 79 CYIRFIRKVNDKKGAEGQ-EETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQE 137
YI F + +EG+ E T+ +E +L VW+++ F ++ A H ++
Sbjct: 488 TYIDF-------EISEGELERTRALYERLLDRT----KHCKVWVDFAKF-EASAAEHKED 535
Query: 138 ETHRMTVVRK 147
E + RK
Sbjct: 536 EEEEDAIERK 545
Score = 36.2 bits (82), Expect = 0.070
Identities = 17/58 (29%), Positives = 35/58 (60%), Gaps = 2/58 (3%)
Query: 445 LEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQR 502
++YADF ++ R +++RA+S LP D ++W +F+ E+ G++A ++ +R
Sbjct: 95 VKYADFEMKNKSVNEARNVWDRAVSLLPRVD--QLWYKFIHMEEKLGNIAGARQILER 150
Score = 32.7 bits (73), Expect = 0.77
Identities = 17/67 (25%), Positives = 35/67 (51%), Gaps = 3/67 (4%)
Query: 447 YADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEA 506
+ F ++ N+ + R+++ER + P V + R+ KFE +G + +KV +R K+
Sbjct: 164 FIKFELKYNEIECARSIYERFVLCHP---KVSAYIRYAKFEMKHGQVELAMKVFERAKKE 220
Query: 507 FGEEATA 513
++ A
Sbjct: 221 LADDEEA 227
Score = 32.3 bits (72), Expect = 1.0
Identities = 41/197 (20%), Positives = 78/197 (38%), Gaps = 13/197 (6%)
Query: 298 YLYHYPDVWYDYATW-HAKAGSIDAAIKVFQRSLKALPDSEM----LRYAYAELEESRGA 352
Y Y +W +YA + +++ V++ LK +P S+ + A+ E +
Sbjct: 336 YWQRYIYLWINYAFFAEMVTEDVESTRDVYRACLKLIPHSKFSFAKIWLLAAQHEIRQLN 395
Query: 353 IQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAY 412
+ A++I N +G + ++I + ++ RK + + + + Y
Sbjct: 396 LTGARQILGNAIGKAPKDKIFK--KYIEIELQLRNIDRCRKLYERYLEWSPGNCYAWRKY 453
Query: 413 ASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYIL--EYADFLIRLNDDQNIRALFERALSS 470
A L + + +FE + + +L Y DF I + + RAL+ER L
Sbjct: 454 AEFEMSL-AETERTRAIFELAISQPALDMPELLWKTYIDFEISEGELERTRALYERLLDR 512
Query: 471 LPLEDSVEVWKRFVKFE 487
+VW F KFE
Sbjct: 513 ---TKHCKVWVDFAKFE 526
>At5g28740 putative protein
Length = 917
Score = 48.9 bits (115), Expect = 1e-05
Identities = 45/228 (19%), Positives = 92/228 (39%), Gaps = 39/228 (17%)
Query: 301 HYPDVWYDYATWHAKA---GSIDAAIKVFQRSLKALPDSEM--LRYAYAELEESRGAIQA 355
H D+W Y T K ++ A ++F+ ++ P + L YA+LEE G +
Sbjct: 583 HVKDIWVTYLTKFVKRYGKTKLERARELFEHAVSMAPSDAVRTLYLQYAKLEEDYGLAKR 642
Query: 356 AKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASV 415
A K+YE T+ V +K + Y +Y++ A+
Sbjct: 643 AMKVYEEA---------------------TKKVPEGQKLEM---------YEIYISRAAE 672
Query: 416 AFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPLED 475
F + + ++ E+GL H + +++A+ L + RAL++ +
Sbjct: 673 IFGVPRTREIYEQAIESGLPH-KDVKIMCIKFAELERSLGEIDRARALYKYSSQFADPRS 731
Query: 476 SVEVWKRFVKFEQTYGD---LASMLKVEQRRKEAFGEEATAASESSLQ 520
E W ++ +FE +G+ ML++++ ++ + E+ +Q
Sbjct: 732 DPEFWNKWHEFEVQHGNEDTYREMLRIKRSVSASYSQTHFILPENMMQ 779
Score = 44.7 bits (104), Expect = 2e-04
Identities = 55/243 (22%), Positives = 94/243 (38%), Gaps = 65/243 (26%)
Query: 285 NKRVIFTYEQCLMY--LYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYA 342
N RVIF + Y + H VW ++A + + A+++ +R+ A+P E+ R
Sbjct: 441 NTRVIFDKAVQVNYKTVDHLASVWCEWAEMELRHKNFKGALELMRRAT-AVPTVEVRRRV 499
Query: 343 ---------------------YAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRF 381
Y +LEES G +++ + +YE +L D AT + +
Sbjct: 500 AADGNEPVQMKLHRSLRLWSFYVDLEESLGTLESTRAVYEKIL-DLRIATPQIIMNYAFL 558
Query: 382 LRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMH-- 439
L KYF DA K V+E G+K F +
Sbjct: 559 LEEN-------KYFEDAFK----------------------------VYERGVKIFKYPH 583
Query: 440 -EPVYILEYADFLIRLNDD--QNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASM 496
+ +++ F+ R + R LFE A+S P + ++ ++ K E+ YG
Sbjct: 584 VKDIWVTYLTKFVKRYGKTKLERARELFEHAVSMAPSDAVRTLYLQYAKLEEDYGLAKRA 643
Query: 497 LKV 499
+KV
Sbjct: 644 MKV 646
Score = 32.3 bits (72), Expect = 1.0
Identities = 38/170 (22%), Positives = 72/170 (42%), Gaps = 11/170 (6%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGS-IDAAIKVFQRSLKALPDSEMLRYAY-AELEES 349
YE+ L+ +W+ Y AKA S +++R+LKALP S L YAY E +
Sbjct: 16 YEEELLRNQFSLKLWWRYLI--AKAESPFKKRFIIYERALKALPGSYKLWYAYLRERLDI 73
Query: 350 RGAIQAAKKIYENLLGDSENATALAH------IQFIRFLRRTEGVEPARKYFLDARKSPS 403
+ Y++L E H + +++ L + + R+ F A +
Sbjct: 74 VRNLPVTHPQYDSLNNTFERGLVTMHKMPRIWVMYLQTLTVQQLITRTRRTFDRALCALP 133
Query: 404 CTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIR 453
T H + + F + ++ ++ M++P +I E+ +FL++
Sbjct: 134 VTQHDRIWEPYLVF-VSQNGIPIETSLRVYRRYLMYDPSHIEEFIEFLVK 182
Score = 32.0 bits (71), Expect = 1.3
Identities = 63/302 (20%), Positives = 120/302 (38%), Gaps = 44/302 (14%)
Query: 291 TYEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRY--AYAELEE 348
T+E+ L+ ++ P +W Y I + F R+L ALP ++ R Y
Sbjct: 90 TFERGLVTMHKMPRIWVMYLQTLTVQQLITRTRRTFDRALCALPVTQHDRIWEPYLVFVS 149
Query: 349 SRG-AIQAAKKIYENLLGDSENATALAHI-QFIRFLRRTEGVEPAR----------KYFL 396
G I+ + ++Y L +HI +FI FL ++E + + K++
Sbjct: 150 QNGIPIETSLRVYRRYLMYDP-----SHIEEFIEFLVKSERWQESAERLASVLNDDKFYS 204
Query: 397 DARKSPSCTY----HVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYI-LEYADFL 451
K+ + + V +A+V L+ D + G++ F E + AD+
Sbjct: 205 IKGKTKHKLWLELCELLVHHANVISGLNVDA-----IIRGGIRKFTDEVGMLWTSLADYY 259
Query: 452 IRLNDDQNIRALFERALSS-LPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFGEE 510
IR N + R ++E + + + D ++ + +FE++ +A +++ E E
Sbjct: 260 IRKNLLEKARDIYEEGMMKVVTVRDFSVIFDVYSRFEES--TVAKKMEMMSSSDEEDENE 317
Query: 511 ATAASESSLQDV----------VSRYSFMDLWPCSSNDLD-NLSRQEWLVKNTKKVEKSI 559
E +DV + R W ND+D L+R E L+ + S+
Sbjct: 318 ENGV-EDDEEDVRLNFNLSVKELQRKILNGFWLNDDNDVDLRLARLEELMNRRPALANSV 376
Query: 560 ML 561
+L
Sbjct: 377 LL 378
>At3g11963 pre-rRNA processing protein like
Length = 1126
Score = 46.6 bits (109), Expect = 5e-05
Identities = 29/79 (36%), Positives = 47/79 (58%), Gaps = 1/79 (1%)
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS-SLPLEDSVEVWKRFVKF 486
++FE L+ + Y D IRL +D IR+LFERA+S SLP + ++K+F+++
Sbjct: 1041 SLFEGVLREYPKRTDLWSVYLDQEIRLGEDDVIRSLFERAISLSLPPKKMKFLFKKFLEY 1100
Query: 487 EQTYGDLASMLKVEQRRKE 505
E++ GD + V+QR E
Sbjct: 1101 EKSVGDEERVEYVKQRAME 1119
Score = 37.4 bits (85), Expect = 0.031
Identities = 19/87 (21%), Positives = 40/87 (45%), Gaps = 2/87 (2%)
Query: 29 IYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIRFIRKVN 88
++E +L+ YP W Y++ + + DD I+ +F R + L +P + F + +
Sbjct: 1042 LFEGVLREYPKRTDLWSVYLDQEIRLGEDDVIRSLFERAI--SLSLPPKKMKFLFKKFLE 1099
Query: 89 DKKGAEGQEETKKAFEFMLSYVGSDIA 115
+K +E + + + Y S +A
Sbjct: 1100 YEKSVGDEERVEYVKQRAMEYANSTLA 1126
Score = 32.7 bits (73), Expect = 0.77
Identities = 54/252 (21%), Positives = 96/252 (37%), Gaps = 26/252 (10%)
Query: 110 VGSDIASGPVWMEYIAFLKSLPAAHPQEETHRMTVVRKVYQRAIITPTHHIEQ----LWK 165
V S S VW++Y+AF+ SL R + +RA+ T E+ +W
Sbjct: 870 VRSSPNSSFVWIKYMAFMLSLADIEK---------ARSIAERALRTINIREEEEKLNIWV 920
Query: 166 DYDSFESSVSQKLAKGL--ISEYQPKYNSARAVYRERKKFFDEIDWNMLAVPPTGSHKAS 223
Y + E+ + + + E +Y + VY ++ + LA
Sbjct: 921 AYFNLENEHGNPPEESVKKVFERARQYCDPKKVYLALLGVYERTEQYKLA-DKLLDEMIK 979
Query: 224 KFLFLCKYWLSLLSVFVNKLFWNDNHFPPK-SLCLFLLTGRNAVDVMEETSIF*KRIDIA 282
KF CK WL + + L N+ + L L + + +T+I + +A
Sbjct: 980 KFKQSCKIWLRKIQ---SSLKQNEEAIQSVVNRALLCLPRHKHIKFISQTAILEFKCGVA 1036
Query: 283 SSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLK-ALPDSEM--L 339
+ + +E L D+W Y + G D +F+R++ +LP +M L
Sbjct: 1037 DRGRSL---FEGVLREYPKRTDLWSVYLDQEIRLGEDDVIRSLFERAISLSLPPKKMKFL 1093
Query: 340 RYAYAELEESRG 351
+ E E+S G
Sbjct: 1094 FKKFLEYEKSVG 1105
>At4g24270 unknown protein
Length = 816
Score = 45.8 bits (107), Expect = 9e-05
Identities = 94/503 (18%), Positives = 195/503 (38%), Gaps = 92/503 (18%)
Query: 31 EQLLQLYPTAAKFWKQYV--EAHMAVN-NDDAIKQIFSRCLLNCLQVPLWRCYIRFIRKV 87
E + ++P + W ++ EA +A + N I ++ R L + V LW Y+ F+ +
Sbjct: 90 EAMSAIFPLSPSLWLEWARDEASLAASENVPEIVMLYERGLSDYQSVSLWCDYLSFMLEF 149
Query: 88 NDK-KG--AEGQEETKKAFEFMLSYVGSDIASG-PVWMEYIAF----LKSLPAAHPQEET 139
+ +G +EG + + FE + G + G +W Y F L ++ A +E
Sbjct: 150 DPSVRGYPSEGISKMRSLFERAIPAAGFHVTEGNRIWEGYREFEQGVLATIDEADIEERN 209
Query: 140 HRMTVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRE 199
++ +R ++ R + P ++ Y ++E + Q + + S+ K + AV +
Sbjct: 210 KQIQRIRSIFHRHLSVPLENLSSTLIAYKTWE--LEQGIDLDIGSDDLSKVSHQVAVANK 267
Query: 200 R-KKFFDE---IDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSL 255
+ ++ + E ++ N+ + + K +F+ K+
Sbjct: 268 KAQQMYSERAHLEENISKQDLSDTEKFQEFMNYIKF------------------------ 303
Query: 256 CLFLLTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAK 315
E+TS RV YE+ + D+W DY + K
Sbjct: 304 --------------EKTS---------GDPTRVQAIYERAVAEYPVSSDLWIDYTVYLDK 340
Query: 316 AGSIDAAI-KVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALA 374
+ AI + R+ ++ P + L Y L RG+ + K+IY+ + S T +
Sbjct: 341 TLKVGKAITHAYSRATRSCPWTGDLWARYL-LALERGS-ASEKEIYD-VFEKSLQCTFSS 397
Query: 375 HIQFI-RFLRRTEGV----------------------EPARKYFLDARKSPSCTYHVYVA 411
+++ +L R +G+ + A Y ++ H++
Sbjct: 398 FEEYLDLYLTRVDGLRRRMLSTRMLEALDYSLIRETFQQASDYLTPHMQNTDSLLHLHTY 457
Query: 412 YASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS-S 470
+A++ + KD A V+++ LK Y D + L + R+++ R +
Sbjct: 458 WANLELNIGKDLAGARGVWDSFLKKSGGMLAAWHAYIDMEVHLGHIKEARSIYRRCYTRK 517
Query: 471 LPLEDSVEVWKRFVKFEQTYGDL 493
S ++ K +++FE+ +GDL
Sbjct: 518 FDGTGSEDICKGWLRFEREHGDL 540
Score = 35.0 bits (79), Expect = 0.16
Identities = 32/149 (21%), Positives = 68/149 (45%), Gaps = 22/149 (14%)
Query: 374 AHIQFIRFLRRTEGVEPARKYFLDARKSPSCTY----HVYVAYASVAFCLDKDPKMAHNV 429
A++Q+I+ LR+T +E R+ AR++ S + +++ +A L + V
Sbjct: 68 AYVQYIKLLRKTANLEKLRQ----AREAMSAIFPLSPSLWLEWARDEASLAASENVPEIV 123
Query: 430 --FEAGLKHFMHEPVYILEYADFLIRLNDD---------QNIRALFERAL--SSLPLEDS 476
+E GL + ++ +Y F++ + +R+LFERA+ + + +
Sbjct: 124 MLYERGLSDYQSVSLW-CDYLSFMLEFDPSVRGYPSEGISKMRSLFERAIPAAGFHVTEG 182
Query: 477 VEVWKRFVKFEQTYGDLASMLKVEQRRKE 505
+W+ + +FEQ +E+R K+
Sbjct: 183 NRIWEGYREFEQGVLATIDEADIEERNKQ 211
>At5g46400 putative protein
Length = 1022
Score = 43.1 bits (100), Expect = 6e-04
Identities = 29/151 (19%), Positives = 64/151 (42%), Gaps = 15/151 (9%)
Query: 29 IYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCL-QVPLWRCYIRFIRKV 87
+Y+ L +P +W++Y + + + ++F R + V +W Y F
Sbjct: 70 VYDAFLLEFPLCHGYWRKYAYHKIKLCTLEDAVEVFERAVQAATYSVAVWLDYCAFA--- 126
Query: 88 NDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRMTVVRK 147
E + + FE LS++G D + +W +YI +L + + +
Sbjct: 127 --VAAYEDPHDVSRLFERGLSFIGKDYSCCTLWDKYIEYLLG---------QQQWSSLAN 175
Query: 148 VYQRAIITPTHHIEQLWKDYDSFESSVSQKL 178
VY R + P+ ++ +K++ +S+ +K+
Sbjct: 176 VYLRTLKYPSKKLDLYYKNFRKIAASLKEKI 206
Score = 34.3 bits (77), Expect = 0.27
Identities = 49/245 (20%), Positives = 93/245 (37%), Gaps = 18/245 (7%)
Query: 292 YEQCLMYLYHYPDVWYDYATW-HAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESR 350
YE+CL+ +Y + W+ Y + +K G A + + S + + ++ A +E
Sbjct: 321 YERCLIPCANYTEFWFRYVDFVESKGGRELANFALARASQTFVKSASVIHLFNARFKEHV 380
Query: 351 GAIQAA----KKIYENL-LGDSENATALAHIQ--FIRFLRRTEGVEPARKYFLDARKSPS 403
G AA + E L G EN T A+++ F A L +++
Sbjct: 381 GDASAASVALSRCGEELGFGFVENVTKKANMEKRLGNFEAAVTTYREALNKTLIGKENLE 440
Query: 404 CTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRAL 463
T +YV ++ + + + A + G ++ H + + E L+ + + L
Sbjct: 441 TTARLYVQFSRLKYVITNSADDAAQILLEGNENVPHCKLLLEELMRLLMMHGGSRQVDLL 500
Query: 464 ---FERALS-------SLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFGEEATA 513
++ LS L ED E+ +++F G + + K R + F A A
Sbjct: 501 DPIIDKELSHQADSSDGLSAEDKEEISNLYMEFIDLSGTIHDVRKALGRHIKLFPHSARA 560
Query: 514 ASESS 518
S
Sbjct: 561 KLRGS 565
>At4g03430 putative pre-mRNA splicing factor
Length = 1029
Score = 34.3 bits (77), Expect = 0.27
Identities = 37/172 (21%), Positives = 65/172 (37%), Gaps = 5/172 (2%)
Query: 316 AGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALAH 375
AG + AA + Q + A+P+SE + A +LE + A+ + T
Sbjct: 706 AGDVPAARAILQEAYAAIPNSEEIWLAAFKLEFENKEPERARMLLAK--ARERGGTERVW 763
Query: 376 IQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLK 435
++ R VE R+ + K + +++ + K + A ++ GLK
Sbjct: 764 MKSAIVERELGNVEEERRLLNEGLKQFPTFFKLWLMLGQLEERF-KHLEQARKAYDTGLK 822
Query: 436 HFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFE 487
H H L AD ++N RA+ A P E+W ++ E
Sbjct: 823 HCPHCIPLWLSLADLEEKVNGLNKARAILTTARKKNP--GGAELWLAAIRAE 872
>At1g48910 hypothetical protein
Length = 383
Score = 33.9 bits (76), Expect = 0.35
Identities = 22/69 (31%), Positives = 34/69 (48%), Gaps = 5/69 (7%)
Query: 536 SNDLDNLSRQEWLVKNTKK--VEKSIMLNGTTFIDKGPVASIST---TSSKVVYPDTSKM 590
S DL N ++ T + V K ++ G T + PVA + T T +K++Y D SK
Sbjct: 186 SFDLCNFGANTTILIRTPRHVVTKEVIHLGMTLLKYAPVAMVDTLVTTMAKILYGDLSKY 245
Query: 591 LIYDPKHNP 599
++ PK P
Sbjct: 246 GLFRPKQGP 254
>At1g79490 unknown protein
Length = 836
Score = 33.5 bits (75), Expect = 0.45
Identities = 29/112 (25%), Positives = 47/112 (41%), Gaps = 6/112 (5%)
Query: 313 HAKAGSIDAAIKVFQRSLKA--LPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENA 370
HAK+G ++ A+ VF+ KA LP E+ G + +A KIY ++
Sbjct: 428 HAKSGKLEVAMTVFKDMEKAGFLPTPSTYS-CLLEMHAGSGQVDSAMKIYNSMTNAGLRP 486
Query: 371 TALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKD 422
++I + L V+ A K L+ + + Y V V + V KD
Sbjct: 487 GLSSYISLLTLLANKRLVDVAGKILLEMK---AMGYSVDVCASDVLMIYIKD 535
Score = 29.3 bits (64), Expect = 8.5
Identities = 20/89 (22%), Positives = 41/89 (45%), Gaps = 1/89 (1%)
Query: 314 AKAGSIDAAIKVFQR-SLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATA 372
AK+G +DAA K+FQ+ + L S + + + G + + K+Y + G +A
Sbjct: 324 AKSGRLDAAFKLFQQMKERKLRPSFSVFSSLVDSMGKAGRLDTSMKVYMEMQGFGHRPSA 383
Query: 373 LAHIQFIRFLRRTEGVEPARKYFLDARKS 401
+ I + ++ A + + + +KS
Sbjct: 384 TMFVSLIDSYAKAGKLDTALRLWDEMKKS 412
>At5g18680 tub family-like protein
Length = 389
Score = 32.7 bits (73), Expect = 0.77
Identities = 28/114 (24%), Positives = 47/114 (40%), Gaps = 16/114 (14%)
Query: 588 SKMLIYDPKHNPGTGAAGTNAFDEILKATPPALVAFLANLPSVDGPTPNVDIVLSI---- 643
+K ++D P TGAA ++ PA V+ A +P P ++ L++
Sbjct: 182 TKFTVFDGNLLPSTGAAKLRKS----RSYNPAKVS--AKVPLGSYPVAHITYELNVLGSR 235
Query: 644 ------CLQSDLPTGQSVKVGIPSQLPAGPAPATSELSGSSKSHPVQSGLSHMQ 691
CL +PT G+ S+ P T S+S P++S SH++
Sbjct: 236 GPRKMQCLMDTIPTSTMEPQGVASEPSEFPLLGTRSTLSRSQSKPLRSSSSHLK 289
>At4g36550 putative protein
Length = 680
Score = 32.0 bits (71), Expect = 1.3
Identities = 25/83 (30%), Positives = 44/83 (52%), Gaps = 2/83 (2%)
Query: 611 EILKATPPALVAFLA-NLPSVDGPTPNVDIVLSICLQSDLPTGQSVKV-GIPSQLPAGPA 668
EI+K L+AFL+ N +++ V + S+ L+S++ +++ + + S P GP+
Sbjct: 400 EIIKGGLDLLLAFLSGNRRAIESLEEEVFKMFSVFLESEVVAEEALNILEVLSNHPHGPS 459
Query: 669 PATSELSGSSKSHPVQSGLSHMQ 691
TS S SS V+S H+Q
Sbjct: 460 KITSSGSLSSLLKIVESQAEHLQ 482
>At2g39580 unknown protein
Length = 1567
Score = 31.6 bits (70), Expect = 1.7
Identities = 18/60 (30%), Positives = 29/60 (48%), Gaps = 5/60 (8%)
Query: 47 YVEAHMAVNNDDAIKQIFSRCLLNCLQV-----PLWRCYIRFIRKVNDKKGAEGQEETKK 101
+VEA V++ + K R L L V LWRCY + + +++G E +E +K
Sbjct: 1502 WVEAGEIVSDINGFKTRAERFLKKALSVYPMSVKLWRCYWSLCKSIEERRGIEIEEAARK 1561
>At1g80130 unknown protein
Length = 305
Score = 31.2 bits (69), Expect = 2.2
Identities = 27/116 (23%), Positives = 51/116 (43%), Gaps = 1/116 (0%)
Query: 351 GAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEG-VEPARKYFLDARKSPSCTYHVY 409
G + A Y + DS +L + +FL+ +G ++ A +Y A + +V
Sbjct: 159 GRSEDATDTYYREMIDSNPGNSLLTGNYAKFLKEVKGDMKKAEEYCERAILGNTNDGNVL 218
Query: 410 VAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFE 465
YA + +D + AH+ ++ +K + YA FL +++D+ AL E
Sbjct: 219 SLYADLILHNHQDRQRAHSYYKQAVKMSPEDCYVQASYARFLWDVDEDEEDEALGE 274
>At1g69450 hypothetical protein
Length = 646
Score = 31.2 bits (69), Expect = 2.2
Identities = 22/76 (28%), Positives = 36/76 (46%), Gaps = 2/76 (2%)
Query: 612 ILKATPPALVAFLANLPSVDGPTPNVDIVLSICLQSDLPTGQSVKVGIPSQLPAGPAPAT 671
IL P LV L NLP+++ P + ++LS+ + S + TG + + + L P T
Sbjct: 297 ILFLVPVVLVQGLTNLPALEFMFPFLSLILSMKVVSQIITGYLPSLILQTSLKV--VPPT 354
Query: 672 SELSGSSKSHPVQSGL 687
E S + H S +
Sbjct: 355 MEFLSSIQGHICHSDI 370
>At1g25200
Length = 248
Score = 30.8 bits (68), Expect = 2.9
Identities = 41/177 (23%), Positives = 71/177 (39%), Gaps = 23/177 (12%)
Query: 573 ASISTTSSKVVYPDTSKMLIYDPKHNPGTGAAGTNAF-----DEILKATPPALVAFLANL 627
AS S ++S +P + + PK+ + +++ D L + + L ++
Sbjct: 79 ASRSVSNSSSAHPSQAARQLTRPKYPASRSVSSSSSAHPSQADRQLTRLKQLVSSPLRSI 138
Query: 628 PSVDGPTP------NVDIVLSICLQSDLPTGQSVKVGIPSQLPAGPAPATSELSGSSKSH 681
P GP+ ++ ++S L+S P G S QL PA+ +S SS +H
Sbjct: 139 PPA-GPSQAARQLTRLNQLVSSPLRSIPPAGPSQTA---RQLTRPKYPASRFVSSSSSAH 194
Query: 682 PVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQPLPQDAFRIRQFQKARAGSTS 738
P Q+ +Q + KQL S + S + RI+Q + STS
Sbjct: 195 PSQAA--------RQLTRLKQLVSSPVSSSSSAHPYQAARQLTRIKQLVSSPVQSTS 243
>At3g17040 unknown protein
Length = 652
Score = 30.4 bits (67), Expect = 3.8
Identities = 15/45 (33%), Positives = 28/45 (61%)
Query: 315 KAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKI 359
+AG++ AA ++F+ SL S + +A+LEE +G + A++I
Sbjct: 490 RAGNLSAARRLFRSSLNINSQSYVTWMTWAQLEEDQGDTERAEEI 534
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,612,144
Number of Sequences: 26719
Number of extensions: 704333
Number of successful extensions: 2181
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 2044
Number of HSP's gapped (non-prelim): 113
length of query: 757
length of database: 11,318,596
effective HSP length: 107
effective length of query: 650
effective length of database: 8,459,663
effective search space: 5498780950
effective search space used: 5498780950
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146910.2


