

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146866.3 + phase: 0
(482 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
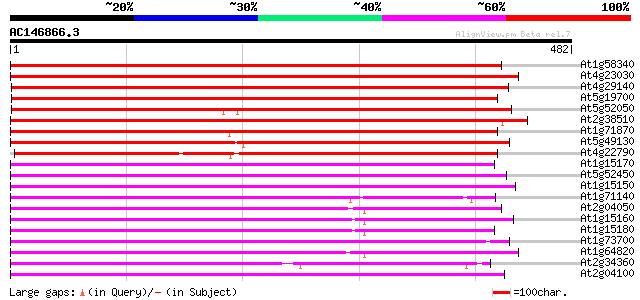
Score E
Sequences producing significant alignments: (bits) Value
At1g58340 putative protein 677 0.0
At4g23030 unknown protein 570 e-163
At4g29140 unknown protein 514 e-146
At5g19700 putative protein 504 e-143
At5g52050 integral membrane protein-like 482 e-136
At2g38510 hypothetical protein 453 e-128
At1g71870 unknown protein 440 e-124
At5g49130 unknown protein 426 e-119
At4g22790 predicted protein 307 7e-84
At1g15170 unknown protein 279 3e-75
At5g52450 unknown protein 276 2e-74
At1g15150 unknown protein (At1g15150) 275 5e-74
At1g71140 hypothetical protein 272 3e-73
At2g04050 hypothetical protein 270 1e-72
At1g15160 hypothetical protein 270 1e-72
At1g15180 unknown protein 267 8e-72
At1g73700 putative integral membrane protein 266 2e-71
At1g64820 hypothetical protein 259 3e-69
At2g34360 hypothetical protein 258 5e-69
At2g04100 unknown protein 256 3e-68
>At1g58340 putative protein
Length = 532
Score = 677 bits (1746), Expect = 0.0
Identities = 328/422 (77%), Positives = 376/422 (88%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
MISM+FLGYLGE+ELAGGSLSIGFANITGYSVISGL+MGMEPICGQAYGAKQ K+LGLTL
Sbjct: 81 MISMLFLGYLGELELAGGSLSIGFANITGYSVISGLSMGMEPICGQAYGAKQMKLLGLTL 140
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
QRTVLLLLS S+PISF W+NM+RILL+ GQD EISS+AQ F+LF +PDLFLLS+LHPLRI
Sbjct: 141 QRTVLLLLSCSVPISFSWLNMRRILLWCGQDEEISSVAQQFLLFAIPDLFLLSLLHPLRI 200
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
YLRTQ ITLP+TY +AVSVLLH+PLN+LLVV +MG+AGV+IAMVLTNLNLV+LLSSF+Y
Sbjct: 201 YLRTQNITLPVTYSTAVSVLLHVPLNYLLVVKLEMGVAGVAIAMVLTNLNLVVLLSSFVY 260
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIAS 240
F+SV+ +W+ ++D +KGWS+LLSLAIPTCVSVCLEWWWYEFMI++CGLL NP+AT+AS
Sbjct: 261 FTSVHSDTWVPITIDSLKGWSALLSLAIPTCVSVCLEWWWYEFMIILCGLLANPRATVAS 320
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFT 300
MGILIQTT+LVYVFPSSLSLGVSTRI NELGA RP KAR+SMI+SLF A+ LGL AM+F
Sbjct: 321 MGILIQTTALVYVFPSSLSLGVSTRISNELGAKRPAKARVSMIISLFCAIALGLMAMVFA 380
Query: 301 TLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLG 360
L+R+ WG+ FT D EIL+LTSI LPIVGLCELGNCPQTTGCGVLRG ARPT+GANINLG
Sbjct: 381 VLVRHHWGRLFTTDAEILQLTSIALPIVGLCELGNCPQTTGCGVLRGCARPTLGANINLG 440
Query: 361 SFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTK 420
SFY VGMPVAI GFV K GFPGLW GLLAAQ +CA LML L RTDW +Q +RA+ELT
Sbjct: 441 SFYFVGMPVAILFGFVFKQGFPGLWFGLLAAQATCASLMLCALLRTDWKVQAERAEELTS 500
Query: 421 SS 422
+
Sbjct: 501 QT 502
>At4g23030 unknown protein
Length = 502
Score = 570 bits (1469), Expect = e-163
Identities = 274/438 (62%), Positives = 350/438 (79%), Gaps = 1/438 (0%)
Query: 1 MISMIFLGYLGEME-LAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLT 59
MISM+FLG L ++ L+GGSL++GFANITGYS++SGL++GMEPIC QA+GAK++K+LGL
Sbjct: 55 MISMLFLGRLNDLSALSGGSLALGFANITGYSLLSGLSIGMEPICVQAFGAKRFKLLGLA 114
Query: 60 LQRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLR 119
LQRT LLLL S+PIS +W+N+K+ILLF GQD EIS+ A+ FILF +PDL L S LHP+R
Sbjct: 115 LQRTTLLLLLCSLPISILWLNIKKILLFFGQDEEISNQAEIFILFSLPDLILQSFLHPIR 174
Query: 120 IYLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFL 179
IYLR+Q ITLPLTY + +VLLHIP+N+LLV +G+ GV++ + TN+NL+ L ++
Sbjct: 175 IYLRSQSITLPLTYSAFFAVLLHIPINYLLVSSLGLGLKGVALGAIWTNVNLLGFLIIYI 234
Query: 180 YFSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIA 239
FS VY+K+W S+DC KGW SL+ LAIP+CVSVCLEWWWYE MI++CGLL+NP+AT+A
Sbjct: 235 VFSGVYQKTWGGFSMDCFKGWRSLMKLAIPSCVSVCLEWWWYEIMILLCGLLLNPQATVA 294
Query: 240 SMGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLF 299
SMGILIQTT+L+Y+FPSSLS+ VSTR+GNELGAN+P KARI+ L L++ LGL AM F
Sbjct: 295 SMGILIQTTALIYIFPSSLSISVSTRVGNELGANQPDKARIAARTGLSLSLGLGLLAMFF 354
Query: 300 TTLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINL 359
++RN W + FT++ EI++LTS+VLPI+GLCELGNCPQTT CGVLRGSARP +GANINL
Sbjct: 355 ALMVRNCWARLFTDEEEIVKLTSMVLPIIGLCELGNCPQTTLCGVLRGSARPKLGANINL 414
Query: 360 GSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELT 419
FY VGMPVA++L F + F GLW+GL AAQGSC + MLVVL RTDW ++V RAKEL
Sbjct: 415 CCFYFVGMPVAVWLSFFSGFDFKGLWLGLFAAQGSCLISMLVVLARTDWEVEVHRAKELM 474
Query: 420 KSSTTSDDVDAKLPIFME 437
S D+ D P ++
Sbjct: 475 TRSCDGDEDDGNTPFLLD 492
>At4g29140 unknown protein
Length = 532
Score = 514 bits (1323), Expect = e-146
Identities = 236/427 (55%), Positives = 327/427 (76%)
Query: 2 ISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTLQ 61
+SM FLG LG++ELA GSL+I FANITGYSV+SGLA+GMEP+C QA+GA ++K+L LTL
Sbjct: 80 VSMFFLGQLGDLELAAGSLAIAFANITGYSVLSGLALGMEPLCSQAFGAHRFKLLSLTLH 139
Query: 62 RTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRIY 121
RTV+ LL +PIS +W N+ +I ++ QD +I+ +AQ++++F +PDL ++LHP+RIY
Sbjct: 140 RTVVFLLVCCVPISVLWFNVGKISVYLHQDPDIAKLAQTYLIFSLPDLLTNTLLHPIRIY 199
Query: 122 LRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYF 181
LR QGI P+T S + H+P N LV + ++G+ GV++A +TN+ +V L +++
Sbjct: 200 LRAQGIIHPVTLASLSGAVFHLPANLFLVSYLRLGLTGVAVASSITNIFVVAFLVCYVWA 259
Query: 182 SSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIASM 241
S ++ +W P+ DC +GW+ LL LA P+CVSVCLEWWWYE MI++CGLLVNP++T+A+M
Sbjct: 260 SGLHAPTWTDPTRDCFRGWAPLLRLAGPSCVSVCLEWWWYEIMIVLCGLLVNPRSTVAAM 319
Query: 242 GILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTT 301
G+LIQTTS +YVFPSSLS VSTR+GNELGANRP+ A+++ V++ A V G+ A F
Sbjct: 320 GVLIQTTSFLYVFPSSLSFAVSTRVGNELGANRPKTAKLTATVAIVFAAVTGIIAAAFAY 379
Query: 302 LMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGS 361
+RN WG+ FT D+EIL+LT+ LPI+GLCE+GNCPQT GCGV+RG+ARP+ AN+NLG+
Sbjct: 380 SVRNAWGRIFTGDKEILQLTAAALPILGLCEIGNCPQTVGCGVVRGTARPSTAANVNLGA 439
Query: 362 FYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTKS 421
FYLVGMPVA+ LGF A +GF GLW+GLLAAQ SCA LM+ V+ TDW + ++A+ LT +
Sbjct: 440 FYLVGMPVAVGLGFWAGIGFNGLWVGLLAAQISCAGLMMYVVGTTDWESEAKKAQTLTCA 499
Query: 422 STTSDDV 428
T +D+
Sbjct: 500 ETVENDI 506
>At5g19700 putative protein
Length = 508
Score = 504 bits (1299), Expect = e-143
Identities = 240/418 (57%), Positives = 319/418 (75%)
Query: 2 ISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTLQ 61
ISM+FLG++GE+ELAGGSL+I FANITGYSV++GLA+GM+P+C QA+GA + K+L LTLQ
Sbjct: 60 ISMLFLGHIGELELAGGSLAIAFANITGYSVLAGLALGMDPLCSQAFGAGRPKLLSLTLQ 119
Query: 62 RTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRIY 121
RTVL LL++S+ I +W+N+ +I+++ QD ISS+AQ++IL +PDL S LHPLRIY
Sbjct: 120 RTVLFLLTSSVVIVALWLNLGKIMIYLHQDPSISSLAQTYILCSIPDLLTNSFLHPLRIY 179
Query: 122 LRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYF 181
LR QGIT PLT + + HIP+NF LV + G GVS+A +NL +VI L + ++
Sbjct: 180 LRAQGITSPLTLATLAGTIFHIPMNFFLVSYLGWGFMGVSMAAAASNLLVVIFLVAHVWI 239
Query: 182 SSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIASM 241
+ +++ +W PS +C K W +++LAIP+C+ VCLEWWWYE M ++CGLL++P +ASM
Sbjct: 240 AGLHQPTWTRPSSECFKDWGPVVTLAIPSCIGVCLEWWWYEIMTVLCGLLIDPSTPVASM 299
Query: 242 GILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTT 301
GILIQTTSL+Y+FPSSL L VSTR+GNELG+NRP KAR+S IV++ A V+GL A F
Sbjct: 300 GILIQTTSLLYIFPSSLGLAVSTRVGNELGSNRPNKARLSAIVAVSFAGVMGLTASAFAW 359
Query: 302 LMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGS 361
+ + WG FTND I++LT+ LPI+GLCELGNCPQT GCGV+RG+ARP++ ANINLG+
Sbjct: 360 GVSDVWGWIFTNDVAIIKLTAAALPILGLCELGNCPQTVGCGVVRGTARPSMAANINLGA 419
Query: 362 FYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELT 419
FYLVG PVA+ L F A GF GLW+GLLAAQ CA +ML V+ TDW + RA++LT
Sbjct: 420 FYLVGTPVAVGLTFWAAYGFCGLWVGLLAAQICCAAMMLYVVATTDWEKEAIRARKLT 477
>At5g52050 integral membrane protein-like
Length = 505
Score = 482 bits (1240), Expect = e-136
Identities = 238/437 (54%), Positives = 321/437 (72%), Gaps = 7/437 (1%)
Query: 2 ISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTLQ 61
+S+ FLG LG+ LAGGSL+ FANITGYS+ SGL MG+E IC QA+GA+++ + +++
Sbjct: 59 VSLSFLGGLGDATLAGGSLAAAFANITGYSLFSGLTMGVESICSQAFGARRYNYVCASVK 118
Query: 62 RTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRIY 121
R ++LLL TS+P++ +W+NM++ILL QD +++S A F+L+ VPDL S LHPLR+Y
Sbjct: 119 RGIILLLVTSLPVTLLWMNMEKILLILKQDKKLASEAHIFLLYSVPDLVAQSFLHPLRVY 178
Query: 122 LRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYF 181
LRTQ TLPL+ C+ ++ LH+P+ F LV + +GI G++++ V++N NLV L ++ F
Sbjct: 179 LRTQSKTLPLSICTVIASFLHLPITFFLVSYLGLGIKGIALSGVVSNFNLVAFLFLYICF 238
Query: 182 S----SVYKKSWISPSL--DCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPK 235
SV + I+ D ++ W LL LAIP+C+SVCLEWW YE MI++CG L++PK
Sbjct: 239 FEDKLSVNEDEKITEETCEDSVREWKKLLCLAIPSCISVCLEWWCYEIMILLCGFLLDPK 298
Query: 236 ATIASMGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLG 295
A++ASMGILIQ TSLVY+FP SLSLGVSTR+GNELG+N+P++AR + IV L L++ LG
Sbjct: 299 ASVASMGILIQITSLVYIFPHSLSLGVSTRVGNELGSNQPKRARRAAIVGLGLSIALGFT 358
Query: 296 AMLFTTLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGA 355
A FT +RN W FFT+D+EI++LT++ LPIVGLCELGNCPQTTGCGVLRGSARP IGA
Sbjct: 359 AFAFTVSVRNTWAMFFTDDKEIMKLTAMALPIVGLCELGNCPQTTGCGVLRGSARPKIGA 418
Query: 356 NINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRA 415
NIN +FY VG+PV L F GF GLW+G+LAAQ +C + M+ CRTDW L+ +RA
Sbjct: 419 NINGVAFYAVGIPVGAVLAFWFGFGFKGLWLGMLAAQITCVIGMMAATCRTDWELEAERA 478
Query: 416 KELTKS-STTSDDVDAK 431
K LT + S D DAK
Sbjct: 479 KVLTTAVDCGSSDDDAK 495
>At2g38510 hypothetical protein
Length = 486
Score = 453 bits (1166), Expect = e-128
Identities = 219/449 (48%), Positives = 321/449 (70%), Gaps = 4/449 (0%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+ISM FL +LG++ELAGG+L++GF NITG SV+ GL++GM+PICGQA+GAK+W +L T
Sbjct: 29 IISMWFLSHLGKVELAGGALAMGFGNITGVSVLKGLSVGMDPICGQAFGAKRWTVLSHTF 88
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
Q+ LL+ S+PI+ W+N++ I L GQD +I+ +A++++LF VP+L ++LHPLR
Sbjct: 89 QKMFCLLIVVSVPIAVTWLNIEPIFLRLGQDPDITKVAKTYMLFFVPELLAQAMLHPLRT 148
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
+LRTQG+T PLT + VS+LLH N++ VV ++G+ GV+IAM +N+ + L +
Sbjct: 149 FLRTQGLTSPLTISAIVSILLHPLFNYVFVVRMRLGVKGVAIAMAFNTMNIDVGLLVYTC 208
Query: 181 FSSVYKKSWISPSLDCI-KGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIA 239
FS K W +L + +GW LLSLA P+ +SVCLE+WWYE M+ +CGLL NPKA++A
Sbjct: 209 FSDSLIKPWEGLALRSLFRGWWPLLSLAAPSAISVCLEYWWYEIMLFLCGLLGNPKASVA 268
Query: 240 SMGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLF 299
+MGILIQTT ++YV P ++S ++TR+G+ LG +P +A+ + ++ L LA+ GL A +F
Sbjct: 269 AMGILIQTTGILYVVPFAISSAIATRVGHALGGGQPTRAQCTTVIGLILAVAYGLAAAVF 328
Query: 300 TTLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINL 359
T +R+ WGK FT++ EIL L S LPI+GLCE+GN PQT CGVL G+ARP GA +NL
Sbjct: 329 VTALRSVWGKMFTDEPEILGLISAALPILGLCEIGNSPQTAACGVLTGTARPKDGARVNL 388
Query: 360 GSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELT 419
+FY+VG+PVA+ F K+GF GLW GLL+AQ +C ++ML L RTDW+ QV+RA+ELT
Sbjct: 389 CAFYIVGLPVAVTTTFGFKVGFRGLWFGLLSAQMTCLVMMLYTLIRTDWSHQVKRAEELT 448
Query: 420 KSS---TTSDDVDAKLPIFMEGNVNKNNV 445
++ + S+D + + +V+ N++
Sbjct: 449 SAAADKSHSEDETVHAEVQDDDDVSSNDL 477
>At1g71870 unknown protein
Length = 510
Score = 440 bits (1131), Expect = e-124
Identities = 212/437 (48%), Positives = 303/437 (68%), Gaps = 18/437 (4%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
++S++FLG LG +ELAGG+LSIGF NITGYSV+ GLA G+EP+C QAYG+K W +L L+L
Sbjct: 47 VVSVLFLGRLGSLELAGGALSIGFTNITGYSVMVGLASGLEPVCSQAYGSKNWDLLTLSL 106
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
R V++LL S+PIS +WIN+ I+LF GQ+ EI++ A + L+ +PDL ++L PLR+
Sbjct: 107 HRMVVILLMASLPISLLWINLGPIMLFMGQNPEITATAAEYCLYALPDLLTNTLLQPLRV 166
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
YLR+Q +T P+ +C+ +V H+PLN+ LV+ G+ GV+IA V+TNL +V+LL +++
Sbjct: 167 YLRSQRVTKPMMWCTLAAVAFHVPLNYWLVMVKHWGVPGVAIASVVTNLIMVVLLVGYVW 226
Query: 181 FSSVYKK-----------------SWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEF 223
S + +K + S ++ + G L+ +A+P+C+ +CLEWWWYE
Sbjct: 227 VSGMLQKRVSGDGDGGSTTMVAVVAQSSSVMELVGGLGPLMRVAVPSCLGICLEWWWYEI 286
Query: 224 MIMMCGLLVNPKATIASMGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMI 283
+I+M G L NPK +A+ GILIQTTSL+Y P +L+ VS R+GNELGA RP KAR++
Sbjct: 287 VIVMGGYLENPKLAVAATGILIQTTSLMYTVPMALAGCVSARVGNELGAGRPYKARLAAN 346
Query: 284 VSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCG 343
V+L A V+G + +T +++ +W FT + L + V+PIVGLCELGNCPQTTGCG
Sbjct: 347 VALACAFVVGALNVAWTVILKERWAGLFTGYEPLKVLVASVMPIVGLCELGNCPQTTGCG 406
Query: 344 VLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAM-LMLVV 402
+LRG+ RP +GA++NLGSFY VG PVA+ L F K+GF GLW GLL+AQ +C + ++ V
Sbjct: 407 ILRGTGRPAVGAHVNLGSFYFVGTPVAVGLAFWLKIGFSGLWFGLLSAQAACVVSILYAV 466
Query: 403 LCRTDWNLQVQRAKELT 419
L RTDW + +A LT
Sbjct: 467 LARTDWEGEAVKAMRLT 483
>At5g49130 unknown protein
Length = 502
Score = 426 bits (1096), Expect = e-119
Identities = 211/444 (47%), Positives = 295/444 (65%), Gaps = 16/444 (3%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
M S++ +G LG +ELAGG+L+IGF NITGYSV+SGLA GMEP+CGQA G+K + LTL
Sbjct: 49 MTSVVCMGRLGSLELAGGALAIGFTNITGYSVLSGLATGMEPLCGQAIGSKNPSLASLTL 108
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
+RT+ LLL S+PIS +W+N+ ++L Q +I+ +A + F +PDL S LHPLRI
Sbjct: 109 KRTIFLLLLASLPISLLWLNLAPLMLMLRQQHDITRVASLYCSFSLPDLLANSFLHPLRI 168
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
YLR +G T PL +C+ VSVLLH+P+ + +G+ GV+++ LTN + LL ++Y
Sbjct: 169 YLRCKGTTWPLMWCTLVSVLLHLPITAFFTFYISLGVPGVAVSSFLTNFISLSLLLCYIY 228
Query: 181 FSSVYKKSWISPSLDCIKG---------------WSSLLSLAIPTCVSVCLEWWWYEFMI 225
+ S SL C+ WS+L+ A+P+C++VCLEWWWYEFM
Sbjct: 229 LENNNNDKTTSKSL-CLDTPLMLYGSRDSGENDVWSTLVKFAVPSCIAVCLEWWWYEFMT 287
Query: 226 MMCGLLVNPKATIASMGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVS 285
++ G L PK +A+ I+IQTTSL+Y P++LS VSTR+ NELGA RP+KA+ + V+
Sbjct: 288 VLAGYLPEPKVALAAAAIVIQTTSLMYTIPTALSAAVSTRVSNELGAGRPEKAKTAATVA 347
Query: 286 LFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVL 345
+ A+ + + ++ TT+ R WGK FT D+ +LELT+ V+P++G CEL NCPQT CG+L
Sbjct: 348 VGAAVAVSVFGLVGTTVGREAWGKVFTADKVVLELTAAVIPVIGACELANCPQTISCGIL 407
Query: 346 RGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCR 405
RGSARP IGA IN +FY+VG PVA+ L FV LGF GL GLL AQ +CA+ +L V+
Sbjct: 408 RGSARPGIGAKINFYAFYVVGAPVAVVLAFVWGLGFMGLCYGLLGAQLACAISILTVVYN 467
Query: 406 TDWNLQVQRAKELTKSSTTSDDVD 429
TDWN + +A +L + S +VD
Sbjct: 468 TDWNKESLKAHDLVGKNVISPNVD 491
>At4g22790 predicted protein
Length = 491
Score = 307 bits (787), Expect = 7e-84
Identities = 164/422 (38%), Positives = 258/422 (60%), Gaps = 12/422 (2%)
Query: 5 IFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTLQRTV 64
+FLG GE+ LAGGSL FAN+TG+SV+ G++ MEPICGQA+GAK +K+L TL V
Sbjct: 57 VFLGRQGELNLAGGSLGFSFANVTGFSVLYGISAAMEPICGQAFGAKNFKLLHKTLFMAV 116
Query: 65 LLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRIYLRT 124
LLLL S+PISF+W+N+ +IL GQ +IS +A+ ++L+L+P+L +LS L PL+ YL +
Sbjct: 117 LLLLLISVPISFLWLNVHKILTGFGQREDISFIAKKYLLYLLPELPILSFLCPLKAYLSS 176
Query: 125 QGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYFSSV 184
QG+TLP+ + +A + LHIP+N +V+ GI GV++A+ +T+ +VILL+ ++
Sbjct: 177 QGVTLPIMFTTAAATSLHIPIN--IVLSKARGIEGVAMAVWITDFIVVILLTGYVIVVER 234
Query: 185 YKKS------WISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATI 238
K++ W++ S + W +L+ L+ P C++VCLEWW YE ++++ G L NP +
Sbjct: 235 MKENKWKQGGWLNQS---AQDWLTLIKLSGPCCLTVCLEWWCYEILVLLTGRLPNPVQAV 291
Query: 239 ASMGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAML 298
+ + I+ L+Y SL V+TR+ NELGAN P+ A + +L + ++ G L
Sbjct: 292 SILIIVFNFDYLLYAVMLSLGTCVATRVSNELGANNPKGAYRAAYTTLIVGIISGCIGAL 351
Query: 299 FTTLMRNQWGKFFT-NDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANI 357
R WG +T +D+ IL ++ I+ + E+ N P ++RG+A+P++G
Sbjct: 352 VMIAFRGFWGSLYTHHDQLILNGVKKMMLIMAVIEVVNFPLMVCGEIVRGTAKPSLGMYA 411
Query: 358 NLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKE 417
NL FYL+ +P+ L F AK G G IGL C ++L+ + R DW + +A+
Sbjct: 412 NLSGFYLLALPLGATLAFKAKQGLQGFLIGLFVGISLCLSILLIFIARIDWEKEAGKAQI 471
Query: 418 LT 419
LT
Sbjct: 472 LT 473
>At1g15170 unknown protein
Length = 481
Score = 279 bits (713), Expect = 3e-75
Identities = 146/416 (35%), Positives = 237/416 (56%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
++SM+ +G+LG + LA SL+ F N+TG+S I GL+ ++ + GQAYGAK ++ LG+
Sbjct: 57 IVSMMMVGHLGNLSLASASLASSFCNVTGFSFIIGLSCALDTLSGQAYGAKLYRKLGVQT 116
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
+ L +P+S IW NM+++LL GQD I+ A + +L+P LF ++L PL
Sbjct: 117 YTAMFCLALVCLPLSLIWFNMEKLLLILGQDPSIAHEAGKYATWLIPGLFAYAVLQPLTR 176
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
Y + Q + PL S V +H+PL + LV + +G G ++A+ L+N I L SF+Y
Sbjct: 177 YFQNQSLITPLLITSYVVFCIHVPLCWFLVYNSGLGNLGGALAISLSNWLYAIFLGSFMY 236
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIAS 240
+SS ++ S++ G A+P+ +CLEWW YE +I++ GLL NP+ +
Sbjct: 237 YSSACSETRAPLSMEIFDGIGEFFKYALPSAAMICLEWWSYELIILLSGLLPNPQLETSV 296
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFT 300
+ + +QT S +Y P +++ STRI NELGA + A I + ++ LA++ L +
Sbjct: 297 LSVCLQTISTMYSIPLAIAAAASTRISNELGAGNSRAAHIVVYAAMSLAVIDALIVSMSL 356
Query: 301 TLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLG 360
+ RN +G F++D+E ++ + + P+V + + + Q G+ RG IGA INLG
Sbjct: 357 LIGRNLFGHIFSSDKETIDYVAKMAPLVSISLMLDALQGVLSGIARGCGWQHIGAYINLG 416
Query: 361 SFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAK 416
+FYL G+P+A L F L GLWIG+ A +L+ +V T+W Q +A+
Sbjct: 417 AFYLWGIPIAASLAFWIHLKGVGLWIGIQAGAVLQTLLLALVTGCTNWESQADKAR 472
>At5g52450 unknown protein
Length = 486
Score = 276 bits (706), Expect = 2e-74
Identities = 142/427 (33%), Positives = 251/427 (58%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+IS++F+G+LG + L+ S++ FA++TG+S + G A ++ +CGQAYGAK++ +LG+ +
Sbjct: 49 VISVMFVGHLGSLPLSAASIATSFASVTGFSFLMGTASALDTLCGQAYGAKKYGMLGIQM 108
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
QR + +L SIP+S IW N + +L+F GQ+ I+++A S+ F++P +F +L
Sbjct: 109 QRAMFVLTLASIPLSIIWANTEHLLVFFGQNKSIATLAGSYAKFMIPSIFAYGLLQCFNR 168
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
+L+ Q P+ +CS V+ LH+ L ++LV +G G ++A ++ V+LL ++
Sbjct: 169 FLQAQNNVFPVVFCSGVTTSLHVLLCWVLVFKSGLGFQGAALANSISYWLNVVLLFCYVK 228
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIAS 240
FS +W S + ++ L LA+P+ + VCLE W +E ++++ GLL NP +
Sbjct: 229 FSPSCSLTWTGFSKEALRDILPFLRLAVPSALMVCLEMWSFELLVLLSGLLPNPVLETSV 288
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFT 300
+ I + T+ +++ P LS STRI NELGA P+ A++++ V + +A+ +
Sbjct: 289 LSICLNTSGTMWMIPFGLSGAASTRISNELGAGNPKVAKLAVRVVICIAVAESIVIGSVL 348
Query: 301 TLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLG 360
L+RN WG ++++ E++ + ++PI+ L + Q GV RG IGA INLG
Sbjct: 349 ILIRNIWGLAYSSELEVVSYVASMMPILALGNFLDSLQCVLSGVARGCGWQKIGAIINLG 408
Query: 361 SFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTK 420
S+YLVG+P + L F +G GLW+G++ A + +V T+W+ + ++A +
Sbjct: 409 SYYLVGVPSGLLLAFHFHVGGRGLWLGIICALVVQVFGLGLVTIFTNWDEEAKKATNRIE 468
Query: 421 SSTTSDD 427
SS++ D
Sbjct: 469 SSSSVKD 475
>At1g15150 unknown protein (At1g15150)
Length = 487
Score = 275 bits (702), Expect = 5e-74
Identities = 146/434 (33%), Positives = 245/434 (55%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+ISM+ +G+LG + LA S ++ F N+TG+S I GL+ ++ + GQAYGAK ++ LG+
Sbjct: 54 IISMVMVGHLGRLSLASASFAVSFCNVTGFSFIIGLSCALDTLSGQAYGAKLYRKLGVQA 113
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
+ L +P+S +W NM ++++ GQD I+ A + +L+P LF ++L PL
Sbjct: 114 YTAMFCLTLVCLPLSLLWFNMGKLIVILGQDPAIAHEAGRYAAWLIPGLFAYAVLQPLIR 173
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
Y + Q + PL S+V +H+PL +LLV +G G ++A+ L+ I L SF+Y
Sbjct: 174 YFKNQSLITPLLVTSSVVFCIHVPLCWLLVYKSGLGHIGGALALSLSYWLYAIFLGSFMY 233
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIAS 240
+SS ++ +++ +G + A+P+ +CLEWW YE +I++ GLL NP+ +
Sbjct: 234 YSSACSETRAPLTMEIFEGVREFIKYALPSAAMLCLEWWSYELIILLSGLLPNPQLETSV 293
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFT 300
+ I +T S+ Y P +++ STRI NELGA + A I + ++ LA++ L +
Sbjct: 294 LSICFETLSITYSIPLAIAAAASTRISNELGAGNSRAAHIVVYAAMSLAVMDALMVSMSL 353
Query: 301 TLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLG 360
R+ +G F++D++ +E + + P+V + + + Q GV G IGA IN G
Sbjct: 354 LAGRHVFGHVFSSDKKTIEYVAKMAPLVSISIILDSLQGVLSGVASGCGWQHIGAYINFG 413
Query: 361 SFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTK 420
+FYL G+P+A L F L GLWIG+LA +L+ +V T+W Q + A+E
Sbjct: 414 AFYLWGIPIAASLAFWVHLKGVGLWIGILAGAVLQTLLLALVTGCTNWKTQAREARERMA 473
Query: 421 SSTTSDDVDAKLPI 434
+ S+ +++LPI
Sbjct: 474 VAHESELTESELPI 487
>At1g71140 hypothetical protein
Length = 485
Score = 272 bits (696), Expect = 3e-73
Identities = 147/421 (34%), Positives = 247/421 (57%), Gaps = 8/421 (1%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+IS++ +G+LGE+ L+ ++++ F ++TG+SV+ GLA +E +CGQA GAKQ++ LG+
Sbjct: 49 VISIMMVGHLGELFLSSTAIAVSFCSVTGFSVVFGLASALETLCGQANGAKQYEKLGVHT 108
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
++ L IP+S +W + IL GQD ++ A F +L+P LF + L PL
Sbjct: 109 YTGIVSLFLVCIPLSLLWTYIGDILSLIGQDAMVAQEAGKFATWLIPALFGYATLQPLVR 168
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
+ + Q + LPL S S+ +HI L + LV F +G G +IA+ ++ V +L ++
Sbjct: 169 FFQAQSLILPLVMSSVSSLCIHIVLCWSLVFKFGLGSLGAAIAIGVSYWLNVTVLGLYMT 228
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIAS 240
FSS KS + S+ +G IP+ +CLEWW +EF++++ G+L NPK +
Sbjct: 229 FSSSCSKSRATISMSLFEGMGEFFRFGIPSASMICLEWWSFEFLVLLSGILPNPKLEASV 288
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMV--LGLGAML 298
+ + + T S +Y P SL STR+ NELGA P++AR+++ ++ + V + +GA++
Sbjct: 289 LSVCLSTQSSLYQIPESLGAAASTRVANELGAGNPKQARMAVYTAMVITGVESIMVGAIV 348
Query: 299 FTTLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANIN 358
F RN +G F+++ E+++ + P++ L + + GV RGS R IGA +N
Sbjct: 349 FGA--RNVFGYLFSSETEVVDYVKSMAPLLSLSVIFDALHAALSGVARGSGRQDIGAYVN 406
Query: 359 LGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSC--AMLMLVVLCRTDWNLQVQRAK 416
L ++YL G+P AI L F K+ GLWIG+ GSC A+L+ +++ T+W Q ++A+
Sbjct: 407 LAAYYLFGIPTAILLAFGFKMRGRGLWIGITV--GSCVQAVLLGLIVILTNWKKQARKAR 464
Query: 417 E 417
E
Sbjct: 465 E 465
>At2g04050 hypothetical protein
Length = 476
Score = 270 bits (691), Expect = 1e-72
Identities = 148/425 (34%), Positives = 248/425 (57%), Gaps = 6/425 (1%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+IS++ G+ GE++L+G +L+ F N++G+S++ GLA +E +CGQAYGAKQ++ +G
Sbjct: 50 VISVMVAGHNGELQLSGVALATSFTNVSGFSILFGLAGALETLCGQAYGAKQYEKIGTYT 109
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
+ + IS +WI ++++L+ GQD +IS +A S+ L+L+P LF + PL
Sbjct: 110 YSATASNIPICVLISVLWIYIEKLLISLGQDPDISRVAGSYALWLIPALFAHAFFIPLTR 169
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
+L QG+ LPL YC+ ++L HIP+ + V F +G G ++A+ ++ V++LS ++
Sbjct: 170 FLLAQGLVLPLLYCTLTTLLFHIPVCWAFVYAFGLGSNGAAMAISVSFWFYVVILSCYVR 229
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIAS 240
+SS K+ + S D + +P+ VCLEWW +E +I+ GLL NPK +
Sbjct: 230 YSSSCDKTRVFVSSDFVSCIKQFFHFGVPSAAMVCLEWWLFELLILCSGLLPNPKLETSV 289
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFT 300
+ I + T SL YV P ++ VSTR+ N+LGA PQ AR+S++ L L +V + F+
Sbjct: 290 LSICLTTASLHYVIPGGVAAAVSTRVSNKLGAGIPQVARVSVLAGLCLWLV---ESAFFS 346
Query: 301 TLM---RNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANI 357
TL+ RN G F+N +E+++ + + P++ L + + GV RGS IGA
Sbjct: 347 TLLFTCRNIIGYAFSNSKEVVDYVANLTPLLCLSFILDGFTAVLNGVARGSGWQHIGALN 406
Query: 358 NLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKE 417
N+ ++YLVG PV ++L F +L GLW G++ A+++ V +W Q ++A++
Sbjct: 407 NVVAYYLVGAPVGVYLAFNRELNGKGLWCGVVVGSAVQAIILAFVTASINWKEQAEKARK 466
Query: 418 LTKSS 422
SS
Sbjct: 467 RMVSS 471
>At1g15160 hypothetical protein
Length = 487
Score = 270 bits (691), Expect = 1e-72
Identities = 148/435 (34%), Positives = 246/435 (56%), Gaps = 4/435 (0%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+I+M+ +G+LG + LA S +I F N+TG+S I GL+ ++ + GQAYGAK ++ LG+
Sbjct: 54 IITMVIVGHLGRLSLASASFAISFCNVTGFSFIMGLSCALDTLSGQAYGAKLYRKLGVQA 113
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
+ L +P+S +W NM ++L+ GQD I+ A F +L+P LF ++L PL
Sbjct: 114 YTAMFCLTLVCLPLSLLWFNMGKLLVILGQDPSIAHEAGRFAAWLIPGLFAYAVLQPLTR 173
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
Y + Q + PL S V LH+PL +LLV + G ++A+ L+ I L SF+Y
Sbjct: 174 YFKNQSLITPLLITSCVVFCLHVPLCWLLVYKSGLDHIGGALALSLSYWLYAIFLGSFMY 233
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIAS 240
FSS ++ +++ +G + A+P+ +CLEWW YE +I++ GLL NP+ +
Sbjct: 234 FSSACSETRAPLTMEIFEGVREFIKYALPSAAMLCLEWWSYELIILLSGLLPNPQLETSV 293
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFT 300
+ + +QT S+ Y P +++ STRI NELGA + A I + ++ LA+V L M+ T
Sbjct: 294 LSVCLQTLSMTYSIPLAIAAAASTRISNELGAGNSRAAHIVVYAAMSLAVVDAL--MVGT 351
Query: 301 TLM--RNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANIN 358
+L+ +N G+ F++D+ ++ + + P+V + + + Q GV G IGA IN
Sbjct: 352 SLLAGKNLLGQVFSSDKNTIDYVAKMAPLVSISLILDSLQGVLSGVASGCGWQHIGAYIN 411
Query: 359 LGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKEL 418
G+FYL G+P+A L F L GLWIG++A +L+ +V +W Q + A++
Sbjct: 412 FGAFYLWGIPIAASLAFWVHLKGVGLWIGIIAGAVLQTLLLALVTGCINWENQAREARKR 471
Query: 419 TKSSTTSDDVDAKLP 433
+ S+ +++LP
Sbjct: 472 MAVAHESELTESELP 486
>At1g15180 unknown protein
Length = 482
Score = 267 bits (683), Expect = 8e-72
Identities = 148/418 (35%), Positives = 238/418 (56%), Gaps = 4/418 (0%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+ISM+ +G+LG + LA SL+ F N+TG+S I GL+ ++ + GQAYGAK ++ +G+
Sbjct: 58 IISMVMVGHLGNLSLASASLASSFCNVTGFSFIVGLSCALDTLSGQAYGAKLYRKVGVQT 117
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
+ L +P++ IW+NM+ +L+F GQD I+ A + L+P LF ++L PL
Sbjct: 118 YTAMFCLALVCLPLTLIWLNMETLLVFLGQDPSIAHEAGRYAACLIPGLFAYAVLQPLTR 177
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
Y + Q + PL S LH+PL +LLV +G G ++A+ +N I+L S +
Sbjct: 178 YFQNQSMITPLLITSCFVFCLHVPLCWLLVYKSGLGNLGGALALSFSNCLYTIILGSLMC 237
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIAS 240
FSS ++ S++ G A+P+ +CLEWW YE +I++ GLL NP+ +
Sbjct: 238 FSSACSETRAPLSMEIFDGIGEFFRYALPSAAMICLEWWSYELIILLSGLLPNPQLETSV 297
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFT 300
+ + +QTT+ VY +++ STRI NELGA + A I + ++ LA+V L +L T
Sbjct: 298 LSVCLQTTATVYSIHLAIAAAASTRISNELGAGNSRAANIVVYAAMSLAVVEIL--ILST 355
Query: 301 TLM--RNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANIN 358
+L+ RN +G F++D+E ++ + + P+V + + + Q G+ RG IGA IN
Sbjct: 356 SLLVGRNVFGHVFSSDKETIDYVAKMAPLVSISLILDGLQGVLSGIARGCGWQHIGAYIN 415
Query: 359 LGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAK 416
LG+FYL G+P+A L F L GLWIG+ A +L+ +V T+W Q +A+
Sbjct: 416 LGAFYLWGIPIAASLAFWIHLKGVGLWIGIQAGAVLQTLLLTLVTGCTNWESQADKAR 473
>At1g73700 putative integral membrane protein
Length = 476
Score = 266 bits (680), Expect = 2e-71
Identities = 144/429 (33%), Positives = 244/429 (56%), Gaps = 3/429 (0%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+IS++F+G+LG + L+ S++ FA++TG++ + G A +E +CGQAYGAK + LG+ +
Sbjct: 47 VISVMFVGHLGSLPLSAASIATSFASVTGFTFLLGTASALETLCGQAYGAKLYGKLGIQM 106
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
QR + +LL S+P+S IW N ++IL+ QD I+S+A S+ +++P LF +L +
Sbjct: 107 QRAMFVLLILSVPLSIIWANTEQILVLVHQDKSIASVAGSYAKYMIPSLFAYGLLQCINR 166
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
+L+ Q P+ CS ++ LH+ L +L V+ +G G ++A+ ++ VILLS ++
Sbjct: 167 FLQAQNNVFPVFVCSGITTCLHLLLCWLFVLKTGLGYRGAALAISVSYWFNVILLSCYVK 226
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIAS 240
FS SW S + + +A P+ V VCLE W +E +++ GLL NP +
Sbjct: 227 FSPSCSHSWTGFSKEAFQELYDFSKIAFPSAVMVCLELWSFELLVLASGLLPNPVLETSV 286
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFT 300
+ I + T+ ++ L S R+ NELGA PQ A++++ V + +A+ G+ +
Sbjct: 287 LSICLNTSLTIWQISVGLGGAASIRVSNELGAGNPQVAKLAVYVIVGIAVAEGIVVVTVL 346
Query: 301 TLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLG 360
+R G F++D +I+ + ++PIV + Q GV RG IGA +NLG
Sbjct: 347 LSIRKILGHAFSSDPKIIAYAASMIPIVACGNFLDGLQCVLSGVARGCGWQKIGACVNLG 406
Query: 361 SFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTK 420
S+YLVG+P+ + LGF +G GLW+G++ A + + +V T+W+ + AK+ T
Sbjct: 407 SYYLVGVPLGLLLGFHFHIGGRGLWLGIVTALSVQVLCLSLVTIFTNWD---KEAKKATN 463
Query: 421 SSTTSDDVD 429
+SDD D
Sbjct: 464 RVGSSDDKD 472
>At1g64820 hypothetical protein
Length = 502
Score = 259 bits (661), Expect = 3e-69
Identities = 146/440 (33%), Positives = 247/440 (55%), Gaps = 6/440 (1%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+ISM+ G+L E+ L+ +++ N+TG+S+I G A ++ +CGQA+GA+Q+ +G
Sbjct: 52 VISMVMAGHLDELSLSAVAIATSLTNVTGFSLIVGFAGALDTLCGQAFGAEQFGKIGAYT 111
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
++L LL IS +W M ++L QD IS +A + ++L+P LF ++L P+
Sbjct: 112 YSSMLCLLVFCFSISIVWFFMDKLLEIFHQDPLISQLACRYSIWLIPALFGFTLLQPMTR 171
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
Y ++QGITLPL S ++ HIP +LLV + GI G ++++ + V LL F+
Sbjct: 172 YFQSQGITLPLFVSSLGALCFHIPFCWLLVYKLKFGIVGAALSIGFSYWLNVFLLWIFMR 231
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIAS 240
+S+++++ + I ++LAIP+ + +CLEWW +E +++M GLL N K +
Sbjct: 232 YSALHREMKNLGLQELISSMKQFIALAIPSAMMICLEWWSFEILLLMSGLLPNSKLETSV 291
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFT 300
+ I + T+++ +V +++ ST + NELGA + AR ++ ++FL G+GA++ T
Sbjct: 292 ISICLTTSAVHFVLVNAIGASASTHVSNELGAGNHRAARAAVNSAIFLG---GVGALITT 348
Query: 301 TLM---RNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANI 357
+ R WG F+N+RE++ + + PI+ L N GV RGS IG
Sbjct: 349 ITLYSYRKSWGYVFSNEREVVRYATQITPILCLSIFVNSFLAVLSGVARGSGWQRIGGYA 408
Query: 358 NLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKE 417
+LGS+YLVG+P+ FL FV KL GLWIG+L A ++ +V T+W + +A++
Sbjct: 409 SLGSYYLVGIPLGWFLCFVMKLRGKGLWIGILIASTIQLIVFALVTFFTNWEQEATKARD 468
Query: 418 LTKSSTTSDDVDAKLPIFME 437
T + K I +E
Sbjct: 469 RVFEMTPQVKGNQKTQIIVE 488
>At2g34360 hypothetical protein
Length = 466
Score = 258 bits (659), Expect = 5e-69
Identities = 142/419 (33%), Positives = 243/419 (57%), Gaps = 18/419 (4%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+IS++F+G+LG + L+ S++ FA++TG++ + G A M+ +CGQ+YGAK + +LG+ +
Sbjct: 50 IISVMFVGHLGSLPLSAASIATSFASVTGFTFLMGTASAMDTVCGQSYGAKMYGMLGIQM 109
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
QR +L+L S+P+S +W N + L+F GQD I+ ++ S+ F++P +F +L L
Sbjct: 110 QRAMLVLTLLSVPLSIVWANTEHFLVFFGQDKSIAHLSGSYARFMIPSIFAYGLLQCLNR 169
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
+L+ Q +P+ CS V+ LH+ + ++LV+ +G G ++A ++ VILLS ++
Sbjct: 170 FLQAQNNVIPVVICSGVTTSLHVIICWVLVLKSGLGFRGAAVANAISYWLNVILLSCYVK 229
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVC-LEWWWYEFMIMMCGLLVNPKATIA 239
FS +W S + + + L IP+ VC LE W +E +++ GLL NP
Sbjct: 230 FSPSCSLTWTGFSKEARRDIIPFMKLVIPSAFMVCSLEMWSFELLVLSSGLLPNP----- 284
Query: 240 SMGILIQTT--SLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAM 297
+++T+ V++ P LS STR+ NELG+ P+ A++++ V L ++V +
Sbjct: 285 ----VLETSCPRTVWMIPFGLSGAASTRVSNELGSGNPKGAKLAVRVVLSFSIVESILVG 340
Query: 298 LFTTLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANI 357
L+R WG +++D E++ + +LPI+ L + QT GV RG IGA +
Sbjct: 341 TVLILIRKIWGFAYSSDPEVVSHVASMLPILALGHSLDSFQTVLSGVARGCGWQKIGAFV 400
Query: 358 NLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAA---QGSCAMLMLVVLCRTDWNLQVQ 413
NLGS+YLVG+P + LGF +G GLW+G++ A QG C L+ T+W+ +V+
Sbjct: 401 NLGSYYLVGVPFGLLLGFHFHVGGRGLWLGIICALIVQGVCLSLITFF---TNWDEEVK 456
>At2g04100 unknown protein
Length = 483
Score = 256 bits (653), Expect = 3e-68
Identities = 139/425 (32%), Positives = 240/425 (55%)
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+IS++ G+ E++L+G +L+ F N++G+SV+ GLA +E +CGQAYGAKQ+ +G
Sbjct: 53 VISVMVAGHRSELQLSGVALATSFTNVSGFSVMFGLAGALETLCGQAYGAKQYAKIGTYT 112
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
++ + + IS +W M ++ + GQD +IS +A S+ + L+P L ++ PL
Sbjct: 113 FSAIVSNVPIVVLISILWFYMDKLFVSLGQDPDISKVAGSYAVCLIPALLAQAVQQPLTR 172
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
+L+TQG+ LPL YC+ ++L HIP+ +LV F +G G ++A+ L+ V++L+ ++
Sbjct: 173 FLQTQGLVLPLLYCAITTLLFHIPVCLILVYAFGLGSNGAALAIGLSYWFNVLILALYVR 232
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIAS 240
FSS +K+ S D + IP+ +EW +EF+I+ GLL NPK +
Sbjct: 233 FSSSCEKTRGFVSDDFVLSVKQFFQYGIPSAAMTTIEWSLFEFLILSSGLLPNPKLETSV 292
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFT 300
+ I + T+SL YV P + S R+ NELGA P+ AR+++ +FL +
Sbjct: 293 LSICLTTSSLHYVIPMGIGAAGSIRVSNELGAGNPEVARLAVFAGIFLWFLEATICSTLL 352
Query: 301 TLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLG 360
+ R+ +G F+N +E+++ + + P++ + L + GV RGS IGA N+
Sbjct: 353 FICRDIFGYAFSNSKEVVDYVTELSPLLCISFLVDGFSAVLGGVARGSGWQHIGAWANVV 412
Query: 361 SFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTK 420
++YL+G PV +FLGF + GLWIG++ + +++ +V WN Q +A++
Sbjct: 413 AYYLLGAPVGLFLGFWCHMNGKGLWIGVVVGSTAQGIILAIVTACMSWNEQAAKARQRIV 472
Query: 421 SSTTS 425
T+S
Sbjct: 473 VRTSS 477
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,945,754
Number of Sequences: 26719
Number of extensions: 395943
Number of successful extensions: 1277
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1145
Number of HSP's gapped (non-prelim): 70
length of query: 482
length of database: 11,318,596
effective HSP length: 103
effective length of query: 379
effective length of database: 8,566,539
effective search space: 3246718281
effective search space used: 3246718281
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146866.3


