

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146864.1 + phase: 0
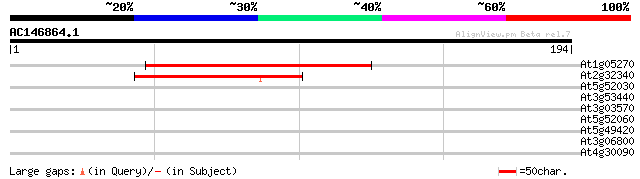
(194 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g05270 unknown protein 87 4e-18
At2g32340 hypothetical protein 61 3e-10
At5g52030 unknown protein 39 0.002
At3g53440 unknown protein 33 0.095
At3g03570 unknown protein 31 0.36
At5g52060 putative protein 30 0.80
At5g49420 MADS-box protein-like 27 5.2
At3g06800 hypothetical protein 27 6.8
At4g30090 putative protein 27 8.9
>At1g05270 unknown protein
Length = 371
Score = 87.4 bits (215), Expect = 4e-18
Identities = 40/78 (51%), Positives = 57/78 (72%)
Query: 48 RLELPKELTKNVIQLSCDSMEKGGVCDVYLVGTFHGNKESSKQVEEIVKFLKPEIVFLEL 107
++ELP+EL K+V+ L+C+S GG CDVYLVGT H +K+S +VE I+ LKPE+VF+EL
Sbjct: 84 KVELPEELAKSVVILTCESTADGGSCDVYLVGTAHVSKQSCLEVEAIISILKPEVVFVEL 143
Query: 108 CSDRQEVLLHDNMEVSTI 125
CS R VL +++ T+
Sbjct: 144 CSSRLSVLKPQTLKIPTM 161
>At2g32340 hypothetical protein
Length = 302
Score = 61.2 bits (147), Expect = 3e-10
Identities = 29/61 (47%), Positives = 43/61 (69%), Gaps = 3/61 (4%)
Query: 44 ERQRRLELPKELTKNVIQLSCDSMEKGGVCDVYLVGTFHGNK---ESSKQVEEIVKFLKP 100
E +LELP+E K+V+ L+C+S +GG CDVYLVGT H ++ ES ++VE ++ LKP
Sbjct: 66 ETTEKLELPEEFAKSVMVLTCESTAEGGSCDVYLVGTAHVSQVILESCREVEAVISALKP 125
Query: 101 E 101
+
Sbjct: 126 Q 126
>At5g52030 unknown protein
Length = 391
Score = 38.5 bits (88), Expect = 0.002
Identities = 17/41 (41%), Positives = 28/41 (67%)
Query: 75 VYLVGTFHGNKESSKQVEEIVKFLKPEIVFLELCSDRQEVL 115
++LVGT H + ES+ VE +V+ +KP+ V +ELC R ++
Sbjct: 94 IWLVGTSHISPESASIVERVVRTVKPDNVAVELCRSRAGIM 134
>At3g53440 unknown protein
Length = 512
Score = 33.1 bits (74), Expect = 0.095
Identities = 19/65 (29%), Positives = 40/65 (61%), Gaps = 2/65 (3%)
Query: 15 RKVIQTTFTTLPCFSTTSAAFCRDQSPE-KERQRRLELPKELTKNVIQLSCDSMEKGGVC 73
R+ +T +T+P FS +++ + SP+ K RQRR L K++ +++++ + M++G
Sbjct: 315 RRATKTILSTIPQFSLSASKLLKPNSPKTKIRQRRNNLSKKVVRHLLEKQ-NHMDQGLGF 373
Query: 74 DVYLV 78
D++ V
Sbjct: 374 DLFSV 378
>At3g03570 unknown protein
Length = 607
Score = 31.2 bits (69), Expect = 0.36
Identities = 20/78 (25%), Positives = 35/78 (44%)
Query: 85 KESSKQVEEIVKFLKPEIVFLELCSDRQEVLLHDNMEVSTILVHISDNAIMTIRCSFSCI 144
KE +EIV+FLK + F+ + R ++L N++ T S I S+ C
Sbjct: 175 KEWKVVAQEIVRFLKSDTAFMNIRPLRYSLVLDPNLDAGTPRASRSLRLTDAILSSYYCN 234
Query: 145 DNQHYHSIINFFNFSQAL 162
+ ++ ++ F Q L
Sbjct: 235 EVKYSELTLDSFRMLQCL 252
>At5g52060 putative protein
Length = 326
Score = 30.0 bits (66), Expect = 0.80
Identities = 21/109 (19%), Positives = 49/109 (44%), Gaps = 5/109 (4%)
Query: 39 QSPEKERQRRLELPK-----ELTKNVIQLSCDSMEKGGVCDVYLVGTFHGNKESSKQVEE 93
+ P + +R LE+ K + +K + +S + GG + + T G K + K +
Sbjct: 120 EDPLSQEKRFLEMRKIAKTEKASKAISDISLEVDRLGGRVSAFEMVTKKGGKIAEKDLVT 179
Query: 94 IVKFLKPEIVFLELCSDRQEVLLHDNMEVSTILVHISDNAIMTIRCSFS 142
+++ L E++ L+ +V L M+V + ++ + ++ S +
Sbjct: 180 VIELLMNELIKLDAIVAEGDVKLQRKMQVKRVQNYVETLDALKVKNSMA 228
>At5g49420 MADS-box protein-like
Length = 368
Score = 27.3 bits (59), Expect = 5.2
Identities = 15/36 (41%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Query: 8 CCLSTRLRKVIQTTFTTLPCFSTTSAAFC-RDQSPE 42
CC+ L ++ Q TL S+TSA C D SPE
Sbjct: 189 CCIPENLNEITQEPNQTLNIQSSTSAICCVPDNSPE 224
>At3g06800 hypothetical protein
Length = 314
Score = 26.9 bits (58), Expect = 6.8
Identities = 20/73 (27%), Positives = 33/73 (44%), Gaps = 2/73 (2%)
Query: 76 YLVGTFHGNKESSKQVEEIVKFLKPEIVFLELCSDRQEVLLHDNMEVSTILVHIS-DNAI 134
YL T G E + ++ E+V +L+ I E + L+H + + ++ H S D I
Sbjct: 189 YLASTSEGKPERNPKMFELVDWLRKNIP-AEDSTGATSGLVHGDFRIDNLVFHPSEDRVI 247
Query: 135 MTIRCSFSCIDNQ 147
I S + NQ
Sbjct: 248 GIIDWELSTLGNQ 260
>At4g30090 putative protein
Length = 312
Score = 26.6 bits (57), Expect = 8.9
Identities = 20/62 (32%), Positives = 33/62 (52%), Gaps = 2/62 (3%)
Query: 42 EKERQRRLELPKELT-KNVIQLSCDSMEKGGVCDVYLVGTFHGNKESSKQVEEIVKFLKP 100
E +R+RRLE+ EL K V QLS +E + +G + S ++E++ K+ +P
Sbjct: 124 ETQRKRRLEVEAELADKKVAQLS-SKLENIDGWFLSKLGLNPTESQVSMKIEQVQKWSEP 182
Query: 101 EI 102
I
Sbjct: 183 HI 184
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,109,576
Number of Sequences: 26719
Number of extensions: 157808
Number of successful extensions: 554
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 548
Number of HSP's gapped (non-prelim): 9
length of query: 194
length of database: 11,318,596
effective HSP length: 94
effective length of query: 100
effective length of database: 8,807,010
effective search space: 880701000
effective search space used: 880701000
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146864.1


