

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.6 - phase: 0 /pseudo
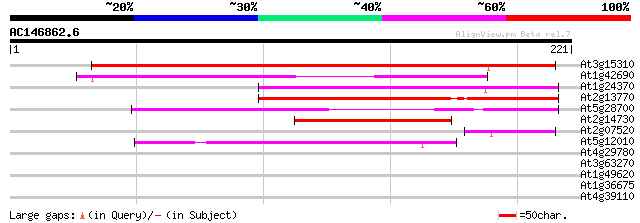
(221 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g15310 unknown protein 203 7e-53
At1g42690 unknown protein 150 7e-37
At1g24370 hypothetical protein 131 2e-31
At2g13770 hypothetical protein 100 5e-22
At5g28700 putative protein 100 6e-22
At2g14730 hypothetical protein 66 1e-11
At2g07520 pseudogene 53 1e-07
At5g12010 putative protein 40 7e-04
At4g29780 unknown protein 35 0.031
At3g63270 unknown protein 34 0.070
At1g49620 hypothetical protein 30 0.77
At1g36675 putative protein 29 1.7
At4g39110 putative receptor-like protein kinase 27 8.5
>At3g15310 unknown protein
Length = 415
Score = 203 bits (516), Expect = 7e-53
Identities = 99/215 (46%), Positives = 135/215 (62%), Gaps = 32/215 (14%)
Query: 33 EAANQRLIDDYFANEPTYDDAMFRRRYRMQKHVFLRIVGDLSITDNYFTQRVDAANKEGI 92
E + L +DYF++ + FRRR+RM+K +FLRIV LS +F R DA + G
Sbjct: 53 EEGHVLLWNDYFSDNAIFPLQTFRRRFRMKKPLFLRIVDRLSSELMFFQHRRDATGRFGH 112
Query: 93 SPLAKCTTTMRMLAYGVAADAVDEYIKIGGTTTLECLRRFCKGIIRLYEEVYLRAPNQDD 152
SP+ KCT +R+LAYG A+DAVDEY+++G TT + CL F KGII + + YLRAP +
Sbjct: 113 SPIQKCTAAIRLLAYGYASDAVDEYLRMGETTAMSCLENFTKGIISFFGDEYLRAPTATN 172
Query: 153 LQRILHVSEMQGFPGMIGSIDCMHWEWKNCPKAWE------------------------- 187
L+R+L++ +++GFPGMIGS+DCMHWEWKNCP AW+
Sbjct: 173 LRRLLNIGKIRGFPGMIGSLDCMHWEWKNCPTAWKGQYTRGSGKPTIVLEAIASQDLWIW 232
Query: 188 -------GTLNDINVLDRSPVFDDVNQGKTPRVEF 215
GTLNDIN+LDRSP+FDD+ QG+ P V++
Sbjct: 233 HVFFGPPGTLNDINILDRSPIFDDILQGRAPNVKY 267
>At1g42690 unknown protein
Length = 333
Score = 150 bits (378), Expect = 7e-37
Identities = 79/167 (47%), Positives = 96/167 (57%), Gaps = 35/167 (20%)
Query: 27 RRKKL-----EEAANQRLIDDYFANEPTYDDAMFRRRYRMQKHVFLRIVGDLSITDNYFT 81
R+KKL E + +L++DYF PTY +FRRR+RM K +F+RIV S YF
Sbjct: 38 RKKKLYIERNREEGHIQLVNDYFTENPTYPPHIFRRRFRMNKSLFMRIVERFSNEVPYFK 97
Query: 82 QRVDAANKEGISPLAKCTTTMRMLAYGVAADAVDEYIKIGGTTTLECLRRFCKGIIRLYE 141
QR DA + G S L K T +RMLAYG+AADA
Sbjct: 98 QRRDATRRLGFSALQKSTAAIRMLAYGIAADA---------------------------- 129
Query: 142 EVYLRAPNQDDLQRILHVSEMQGFPGMIGSIDCMHWEWKNCPKAWEG 188
YLR P +DDL R+LH+ E +GFPGMIGSIDCMHWEWKNCP AW+G
Sbjct: 130 --YLRRPTRDDLIRLLHIGEQRGFPGMIGSIDCMHWEWKNCPTAWKG 174
>At1g24370 hypothetical protein
Length = 413
Score = 131 bits (330), Expect = 2e-31
Identities = 63/150 (42%), Positives = 88/150 (58%), Gaps = 32/150 (21%)
Query: 99 TTTMRMLAYGVAADAVDEYIKIGGTTTLECLRRFCKGIIRLYEEVYLRAPNQDDLQRILH 158
T RMLAYGV AD+ DEYIKIG +T LE L+RFC+ I+ ++ YLR+P+ +D+ R+LH
Sbjct: 2 TAAFRMLAYGVPADSTDEYIKIGESTALESLKRFCRAIVEVFACRYLRSPDANDVARLLH 61
Query: 159 VSEMQGFPGMIGSIDCMHWEWKNCPKAW-------------------------------- 186
+ E +GFP M+GS+DCMHW+WKNCP AW
Sbjct: 62 IGESRGFPRMLGSLDCMHWKWKNCPTAWGGQYAGRSRSPTIILEAVADYDLWIWHAYFGL 121
Query: 187 EGTLNDINVLDRSPVFDDVNQGKTPRVEFL 216
G+ NDINVL+ S +F ++ +G P ++
Sbjct: 122 PGSNNDINVLEASHLFANLAEGTAPPASYV 151
>At2g13770 hypothetical protein
Length = 244
Score = 100 bits (250), Expect = 5e-22
Identities = 50/118 (42%), Positives = 74/118 (62%), Gaps = 3/118 (2%)
Query: 99 TTTMRMLAYGVAADAVDEYIKIGGTTTLECLRRFCKGIIRLYEEVYLRAPNQDDLQRILH 158
T RMLAYGV AD+ DEYIKIG +T LE L+RFC+ I+ ++ YLR+P+ +D+ R+LH
Sbjct: 2 TAAFRMLAYGVPADSTDEYIKIGESTALESLKRFCRAIVEVFACCYLRSPDANDVARLLH 61
Query: 159 VSEMQGFPGMIGSIDCMHWEWKNCPKAWEGTLNDINVLDRSPVFDDVNQGKTPRVEFL 216
+ E +GFP + D W W + G+ N INVL+ S +F ++ +G P ++
Sbjct: 62 IGESRGFPEAVADYDL--WIW-HAYFGLPGSNNGINVLEASHLFANLAEGTAPPASYV 116
>At5g28700 putative protein
Length = 292
Score = 100 bits (249), Expect = 6e-22
Identities = 65/168 (38%), Positives = 83/168 (48%), Gaps = 44/168 (26%)
Query: 49 TYDDAMFRRRYRMQKHVFLRIVGDLSITDNYFTQRVDAANKEGISPLAKCTTTMRMLAYG 108
TY +FR R+RM K +F+RIV S YF QR DA + S L K T +RMLAYG
Sbjct: 33 TYPPHIFRHRFRMNKPLFMRIVERFSNEVPYFKQRRDATGRLDFSALQKSTAAIRMLAYG 92
Query: 109 VAADAVDEYIKIGGTTTLECLRRFCKGIIRLYEEVYLRAPNQDDLQRILHVSEMQGFPGM 168
+AADAVDEY++IG +T L +
Sbjct: 93 IAADAVDEYLRIGESTLL-----------------------------------------V 111
Query: 169 IGSIDCMHWEWKNCPKAWEGTLNDINVLDRSPVFDDVNQGKTPRVEFL 216
+ S D W P GTLNDINVLDRSPVFDD+ QG+ +V+++
Sbjct: 112 VASQDLWIWHAIFGP---PGTLNDINVLDRSPVFDDILQGRASKVKYV 156
>At2g14730 hypothetical protein
Length = 117
Score = 66.2 bits (160), Expect = 1e-11
Identities = 29/62 (46%), Positives = 43/62 (68%)
Query: 113 AVDEYIKIGGTTTLECLRRFCKGIIRLYEEVYLRAPNQDDLQRILHVSEMQGFPGMIGSI 172
AVD+Y++IG T + C+ + II L+ + YLR P + DL+R+L + E++GF GM GSI
Sbjct: 19 AVDKYLRIGENTLMSCMIHSVEAIIYLFGKEYLRRPTRQDLKRLLRIGELRGFLGMTGSI 78
Query: 173 DC 174
DC
Sbjct: 79 DC 80
>At2g07520 pseudogene
Length = 222
Score = 52.8 bits (125), Expect = 1e-07
Identities = 30/69 (43%), Positives = 31/69 (44%), Gaps = 33/69 (47%)
Query: 180 KNCPKAWEG---------------------------------TLNDINVLDRSPVFDDVN 206
KNCP AWEG TLNDIN LDRSPVFDD+
Sbjct: 16 KNCPIAWEGQFTRGDKGTSTISFEAVASHDLWIWHTFFGCASTLNDINFLDRSPVFDDIE 75
Query: 207 QGKTPRVEF 215
QG TPRV F
Sbjct: 76 QGNTPRVNF 84
>At5g12010 putative protein
Length = 502
Score = 40.4 bits (93), Expect = 7e-04
Identities = 32/128 (25%), Positives = 55/128 (42%), Gaps = 5/128 (3%)
Query: 50 YDDAMFRRRYRMQKHVFLRIVGDLSITDNYFTQRVDAANKEGISPLAKCTTTMRMLAYGV 109
Y + F++ +RM K F I +L N + D A + I + + LA G
Sbjct: 170 YPEEDFKKAFRMSKSTFELICDEL----NSAVAKEDTALRNAIPVRQRVAVCIWRLATGE 225
Query: 110 AADAVDEYIKIGGTTTLECLRRFCKGIIRLYEEVYLRAPNQDDLQRILHVSE-MQGFPGM 168
V + +G +T + + CK I + YL+ P+ + L+ I E + G P +
Sbjct: 226 PLRLVSKKFGLGISTCHKLVLEVCKAIKDVLMPKYLQWPDDESLRNIRERFESVSGIPNV 285
Query: 169 IGSIDCMH 176
+GS+ H
Sbjct: 286 VGSMYTTH 293
>At4g29780 unknown protein
Length = 540
Score = 35.0 bits (79), Expect = 0.031
Identities = 29/132 (21%), Positives = 52/132 (38%), Gaps = 5/132 (3%)
Query: 46 NEPTYDDAMFRRRYRMQKHVFLRIVGDLSITDNYFTQRVDAANKEGISPLAKCTTTMRML 105
+ P + + FRR +RM K F I +L T + + ++ I + + L
Sbjct: 204 SRPDFPEDEFRREFRMSKSTFNLICEELDTT----VTKKNTMLRDAIPAPKRVGVCVWRL 259
Query: 106 AYGVAADAVDEYIKIGGTTTLECLRRFCKGIIRLYEEVYLRAPNQDDLQRI-LHVSEMQG 164
A G V E +G +T + + C+ I + YL P+ ++ +
Sbjct: 260 ATGAPLRHVSERFGLGISTCHKLVIEVCRAIYDVLMPKYLLWPSDSEINSTKAKFESVHK 319
Query: 165 FPGMIGSIDCMH 176
P ++GSI H
Sbjct: 320 IPNVVGSIYTTH 331
>At3g63270 unknown protein
Length = 396
Score = 33.9 bits (76), Expect = 0.070
Identities = 35/132 (26%), Positives = 55/132 (41%), Gaps = 5/132 (3%)
Query: 48 PTYDDAMFRRRYRMQKHVFLRIVGDLSITDNYFTQRVDAANKEG--ISPLAKCTTTMRML 105
P+ +D F+ +R K F I L D N EG +S + +R L
Sbjct: 58 PSDEDYAFKHFFRASKTTFSYICS-LVREDLISRPPSGLINIEGRLLSVEKQVAIALRRL 116
Query: 106 AYGVAADAVDEYIKIGGTTTLECLRRFCKGIIRLYEEVYLRAPNQDDLQRILH-VSEMQG 164
A G + +V +G +T + RF + + + +LR P+ D ++ I EM G
Sbjct: 117 ASGDSQVSVGAAFGVGQSTVSQVTWRFIEALEERAKH-HLRWPDSDRIEEIKSKFEEMYG 175
Query: 165 FPGMIGSIDCMH 176
P G+ID H
Sbjct: 176 LPNCCGAIDTTH 187
>At1g49620 hypothetical protein
Length = 195
Score = 30.4 bits (67), Expect = 0.77
Identities = 15/57 (26%), Positives = 27/57 (47%), Gaps = 6/57 (10%)
Query: 23 RFIQRRKKLEEAANQRLIDDYFANEPTYDDAMFRRRYRMQKHVFLRIVGDLSITDNY 79
R +++KK+E++ Q +DD+F+ Y+ F +Y IV D + Y
Sbjct: 138 RKTEKKKKMEKSPTQAELDDFFSAAERYEQKRFTEKYNYD------IVNDTPLEGRY 188
>At1g36675 putative protein
Length = 268
Score = 29.3 bits (64), Expect = 1.7
Identities = 12/19 (63%), Positives = 15/19 (78%)
Query: 72 DLSITDNYFTQRVDAANKE 90
DLS +DNYFT+R +A KE
Sbjct: 175 DLSSSDNYFTKRFEATKKE 193
>At4g39110 putative receptor-like protein kinase
Length = 878
Score = 26.9 bits (58), Expect = 8.5
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 1/34 (2%)
Query: 29 KKLEEAANQRLIDDYFANEPTYDDAMFRRRYRMQ 62
KK EAA ++ ++DY + PT D ++ Y +Q
Sbjct: 768 KKFAEAA-EKCLEDYGVDRPTMGDVLWNLEYALQ 800
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.139 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,113,374
Number of Sequences: 26719
Number of extensions: 212451
Number of successful extensions: 471
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 456
Number of HSP's gapped (non-prelim): 19
length of query: 221
length of database: 11,318,596
effective HSP length: 95
effective length of query: 126
effective length of database: 8,780,291
effective search space: 1106316666
effective search space used: 1106316666
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146862.6


