

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.5 + phase: 0 /pseudo
(624 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
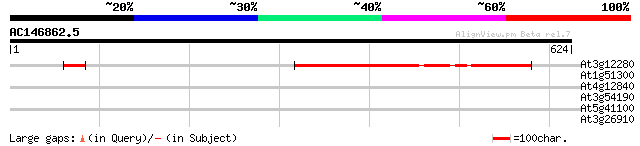
At3g12280 putative retinoblastoma-associated protein 402 e-112
At1g51300 hypothetical protein 33 0.62
At4g12840 unknown protein 32 1.4
At3g54190 unknown protein 30 3.1
At5g41100 unknown protein 29 6.9
At3g26910 unknown protein 29 9.0
>At3g12280 putative retinoblastoma-associated protein
Length = 1013
Score = 402 bits (1032), Expect = e-112
Identities = 201/263 (76%), Positives = 227/263 (85%), Gaps = 11/263 (4%)
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
+KSK+LPPPLQSAFASPT+PNPGGGGETCAETGI++FF+KI KL AVRI+GMVERLQLSQ
Sbjct: 697 VKSKMLPPPLQSAFASPTRPNPGGGGETCAETGINIFFTKINKLAAVRINGMVERLQLSQ 756
Query: 378 QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC 437
QIRE+VY FQ +L Q TSL F+RHIDQIILCCFYGVAKISQ++LTFREIIYNYRKQPQC
Sbjct: 757 QIRESVYCFFQHVLAQRTSLLFSRHIDQIILCCFYGVAKISQMSLTFREIIYNYRKQPQC 816
Query: 438 KPQVFRSVFVDWSSARRNGGSRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRS 497
KP VFRSV+VD RR G R G +H+DII+FYNE+FIP+VKPLLVELGP VR+
Sbjct: 817 KPLVFRSVYVDALQCRRQG----RIGPDHVDIITFYNEIFIPAVKPLLVELGP----VRN 868
Query: 498 DQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKVSAAHNVYVSPLRSSKMDALISHS 557
D+ EANNK +G Q PGSP++S FPS+PDMSPKKVSA HNVYVSPLR SKMDALISHS
Sbjct: 869 DRAVEANNKPEG---QCPGSPKVSVFPSVPDMSPKKVSAVHNVYVSPLRGSKMDALISHS 925
Query: 558 SKSYYACXGESTHAYQSPXKDLT 580
+KSYYAC GESTHAYQSP KDL+
Sbjct: 926 TKSYYACVGESTHAYQSPSKDLS 948
Score = 42.7 bits (99), Expect = 6e-04
Identities = 20/24 (83%), Positives = 21/24 (87%)
Query: 61 RLLPPPVSTAMTTAKWLRTVISPL 84
+L PVSTAMTTAKWLRTVISPL
Sbjct: 402 KLAATPVSTAMTTAKWLRTVISPL 425
>At1g51300 hypothetical protein
Length = 212
Score = 32.7 bits (73), Expect = 0.62
Identities = 24/78 (30%), Positives = 37/78 (46%), Gaps = 4/78 (5%)
Query: 465 EHIDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANNKNDGHLVQNPGSPRISPFP 524
E + S YN +I PL+ +G GGA R+ ++ N + H + +++ F
Sbjct: 55 ELVKSFSLYNVKWICPSSPLISNVGFGGAPARACKISLLQNFKEEHAISIHRGFKVNEFS 114
Query: 525 S-LPD---MSPKKVSAAH 538
S +PD M K SAAH
Sbjct: 115 SRMPDPYEMEGLKNSAAH 132
>At4g12840 unknown protein
Length = 413
Score = 31.6 bits (70), Expect = 1.4
Identities = 18/54 (33%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query: 385 SLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQCK 438
++F +L S+F+ R + Q+IL FYG+ KIS L + E ++ K+ K
Sbjct: 172 NMFVTLLGGLQSVFYTRILSQLILTFFYGM-KISVLTILMLEGMFQLLKKKSLK 224
>At3g54190 unknown protein
Length = 467
Score = 30.4 bits (67), Expect = 3.1
Identities = 17/54 (31%), Positives = 23/54 (42%)
Query: 396 SLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSVFVDW 449
SLF+N++ D +I Y S L I Y R QP +F S + W
Sbjct: 132 SLFYNKNNDSLITVSVYASDNFSSLKCRSTRIEYILRGQPDAGFALFESESLKW 185
>At5g41100 unknown protein
Length = 582
Score = 29.3 bits (64), Expect = 6.9
Identities = 17/47 (36%), Positives = 24/47 (50%), Gaps = 5/47 (10%)
Query: 517 SPRISPFPSLPDMSPKKVSAAHNV-----YVSPLRSSKMDALISHSS 558
SPRISP S P S +++ H + +P R SK L+ HS+
Sbjct: 438 SPRISPTASPPLASSPRINELHELPRPPGQFAPPRRSKSPGLVGHSA 484
>At3g26910 unknown protein
Length = 621
Score = 28.9 bits (63), Expect = 9.0
Identities = 15/48 (31%), Positives = 24/48 (49%), Gaps = 6/48 (12%)
Query: 517 SPRISPFPSLPDMSPKKVSAAHNV------YVSPLRSSKMDALISHSS 558
SPR+SP S P S +++ H + + P R +K L+ HS+
Sbjct: 472 SPRVSPTASPPPASSPRLNELHELPRPPGHFAPPPRRAKSPGLVGHSA 519
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.342 0.148 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,994,259
Number of Sequences: 26719
Number of extensions: 465148
Number of successful extensions: 1622
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1615
Number of HSP's gapped (non-prelim): 8
length of query: 624
length of database: 11,318,596
effective HSP length: 105
effective length of query: 519
effective length of database: 8,513,101
effective search space: 4418299419
effective search space used: 4418299419
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146862.5


