

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.25 + phase: 0
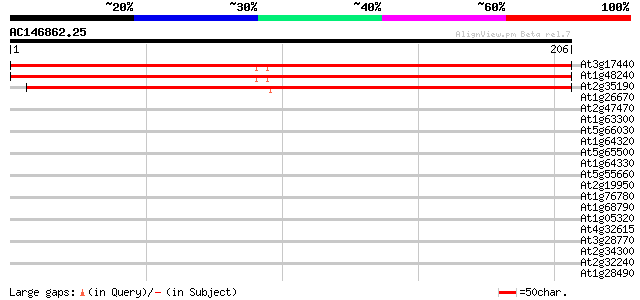
(206 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g17440 unknown protein 315 8e-87
At1g48240 NPSN12 like protein 313 5e-86
At2g35190 unknown protein 253 4e-68
At1g26670 v-SNARE AtVTI1b (VTI1b) 40 0.001
At2g47470 putative protein disulfide-isomerase 39 0.002
At1g63300 hypothetical protein 39 0.002
At5g66030 Golgi-localized protein GRIP 37 0.006
At1g64320 bZIP transcription factor, putative 37 0.007
At5g65500 putative protein 35 0.036
At1g64330 unknown protein 35 0.036
At5g55660 putative protein 34 0.062
At2g19950 hypothetical protein 33 0.080
At1g76780 putative heat shock protein 33 0.080
At1g68790 putative nuclear matrix constituent protein 1 (NMCP1) 33 0.10
At1g05320 unknown protein 33 0.10
At4g32615 putative protein 33 0.14
At3g28770 hypothetical protein 33 0.14
At2g34300 unknown protein 33 0.14
At2g32240 putative myosin heavy chain 33 0.14
At1g28490 unknown protein (At1g28490) 33 0.14
>At3g17440 unknown protein
Length = 269
Score = 315 bits (808), Expect = 8e-87
Identities = 172/237 (72%), Positives = 188/237 (78%), Gaps = 31/237 (13%)
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MASNL M+PQLEQIHGEIRD FRALANGFQ+LDKIKDS RQS QLEELT KMR+CKRL+K
Sbjct: 1 MASNLPMSPQLEQIHGEIRDHFRALANGFQRLDKIKDSTRQSKQLEELTDKMRECKRLVK 60
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSMT-EKNS-------------------------- 93
+FDRE+KDE A N EVNKQLNDEKQSM E NS
Sbjct: 61 EFDRELKDEEARNSPEVNKQLNDEKQSMIKELNSYVALRKTYMSTLGNKKVELFDMGAGV 120
Query: 94 ----TAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQ 149
TAE NVQ+AS MSNQELV+AGMK MDETDQAIERSKQVV QT+EVGTQTA+ LKGQ
Sbjct: 121 SGEPTAEENVQVASSMSNQELVDAGMKRMDETDQAIERSKQVVEQTLEVGTQTAANLKGQ 180
Query: 150 TEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
T+QMGR+VN LD+IQFSIKKASQLVKEIGRQVATDKCIM FLFLIVCGV+AII+VK+
Sbjct: 181 TDQMGRVVNHLDTIQFSIKKASQLVKEIGRQVATDKCIMGFLFLIVCGVVAIIIVKI 237
>At1g48240 NPSN12 like protein
Length = 265
Score = 313 bits (801), Expect = 5e-86
Identities = 170/237 (71%), Positives = 187/237 (78%), Gaps = 31/237 (13%)
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MAS L M+P LEQIHGEIRD FRALANGFQ+LDKIKDS+RQS QLEEL KMRDCKRL+K
Sbjct: 1 MASELPMSPHLEQIHGEIRDHFRALANGFQRLDKIKDSSRQSKQLEELAEKMRDCKRLVK 60
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSMT-EKNS-------------------------- 93
+FDRE+KD A N +VNKQLNDEKQSM E NS
Sbjct: 61 EFDRELKDGEARNSPQVNKQLNDEKQSMIKELNSYVALRKTYLNTLGNKKVELFDTGAGV 120
Query: 94 ----TAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQ 149
TAE NVQ+AS MSNQELV+AGMK MDETDQAIERSKQVVHQT+EVGTQTAS LKGQ
Sbjct: 121 SGEPTAEENVQMASTMSNQELVDAGMKRMDETDQAIERSKQVVHQTLEVGTQTASNLKGQ 180
Query: 150 TEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
T+QMGR+VN+LD+IQFS+KKASQLVKEIGRQVATDKCIM FLFLIVCGVIAII+VK+
Sbjct: 181 TDQMGRVVNDLDTIQFSLKKASQLVKEIGRQVATDKCIMAFLFLIVCGVIAIIIVKI 237
>At2g35190 unknown protein
Length = 265
Score = 253 bits (647), Expect = 4e-68
Identities = 136/229 (59%), Positives = 164/229 (71%), Gaps = 29/229 (12%)
Query: 7 MTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREI 66
++ +L +I G+I DIFRAL+NGFQKL+KIKD+NRQS QLEELT KMRDCK LIKDFDREI
Sbjct: 7 VSEELAEIEGQINDIFRALSNGFQKLEKIKDANRQSRQLEELTDKMRDCKSLIKDFDREI 66
Query: 67 KDEGAGNPEEVNKQLNDEKQSMTEKNST-----------------------------AEG 97
K +GN N+ LND +QSM ++ ++ E
Sbjct: 67 KSLESGNDASTNRMLNDRRQSMVKELNSYVALKKKYSSNLASNNKRVDLFDGPGEEHMEE 126
Query: 98 NVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIV 157
NV LAS MSNQEL++ G MD+TDQAIER K++V +TI VGT T++ LK QTEQM R+V
Sbjct: 127 NVLLASNMSNQELMDKGNSMMDDTDQAIERGKKIVQETINVGTDTSAALKAQTEQMSRVV 186
Query: 158 NELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
NELDSI FS+KKAS+LVKEIGRQVATDKCIM FLFLIV GVIAII+VK+
Sbjct: 187 NELDSIHFSLKKASKLVKEIGRQVATDKCIMAFLFLIVIGVIAIIIVKI 235
>At1g26670 v-SNARE AtVTI1b (VTI1b)
Length = 222
Score = 39.7 bits (91), Expect = 0.001
Identities = 43/198 (21%), Positives = 84/198 (41%), Gaps = 29/198 (14%)
Query: 25 LANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDE 84
L+NG +K KI + + + L KM R ++ + + E LN
Sbjct: 29 LSNGEEKKGKIAEIKSGIDEADVLIRKMDLEARSLQPSAKAVC---LSKLREYKSDLNQL 85
Query: 85 KQSMTEKNSTAEGNVQLASEMSNQELVNAGM------------------KTMDETDQAIE 126
K+ +S A S +EL+ +GM + +D++ I
Sbjct: 86 KKEFKRVSSAD------AKPSSREELMESGMADLHAVSADQRGRLAMSVERLDQSSDRIR 139
Query: 127 RSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKC 186
S++++ +T EVG L Q + + N+L + +I K+ +++ + R++ +K
Sbjct: 140 ESRRLMLETEEVGISIVQDLSQQRQTLLHAHNKLHGVDDAIDKSKKVLTAMSRRMTRNKW 199
Query: 187 IMLFLFLIVCGVIAIIVV 204
I+ +IV V+AII++
Sbjct: 200 II--TSVIVALVLAIILI 215
>At2g47470 putative protein disulfide-isomerase
Length = 361
Score = 38.5 bits (88), Expect = 0.002
Identities = 37/132 (28%), Positives = 64/132 (48%), Gaps = 13/132 (9%)
Query: 67 KDEGAGNPEEVNKQLNDEKQSMTEKNSTA-EGNVQLASEMSNQELVNAGMKTMDETDQAI 125
KD AG+ + + L+D + EK+ T+ + QL S+ E ++A +K E A
Sbjct: 224 KDNKAGHDYDGGRDLDDFVSFINEKSGTSRDSKGQLTSKAGIVESLDALVK---ELVAAS 280
Query: 126 ERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNEL------DSIQFSIKKASQLVKEIGR 179
E K+ V IE + ASTLKG T + G++ +L ++ K+ +L + +G+
Sbjct: 281 EDEKKAVLSRIE---EEASTLKGSTTRYGKLYLKLAKSYIEKGSDYASKETERLGRVLGK 337
Query: 180 QVATDKCIMLFL 191
++ K L L
Sbjct: 338 SISPVKADELTL 349
>At1g63300 hypothetical protein
Length = 1029
Score = 38.5 bits (88), Expect = 0.002
Identities = 35/179 (19%), Positives = 90/179 (49%), Gaps = 9/179 (5%)
Query: 14 IHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELT--GKMRDCKRLIKDFDREIKDEGA 71
+ G+++D F+ L+ +++D + SN + ++ +T ++R KR +++ ++ DE
Sbjct: 619 VAGKLQDEFKRLS---EQMDSMFTSNEKMA-MKAMTEANELRMQKRQLEEMIKDANDELR 674
Query: 72 GNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQV 131
N E +L++ + ++ K S E ++ E SN+ ++ + ++ + + ++
Sbjct: 675 ANQAEYEAKLHELSEKLSFKTSQMERMLENLDEKSNE--IDNQKRHEEDVTANLNQEIKI 732
Query: 132 VHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKA-SQLVKEIGRQVATDKCIML 189
+ + IE + +L Q EQ + +L+ + S+ +A + L +E +++ + I L
Sbjct: 733 LKEEIENLKKNQDSLMLQAEQAENLRVDLEKTKKSVMEAEASLQRENMKKIELESKISL 791
>At5g66030 Golgi-localized protein GRIP
Length = 788
Score = 37.4 bits (85), Expect = 0.006
Identities = 37/149 (24%), Positives = 69/149 (45%), Gaps = 12/149 (8%)
Query: 30 QKLDK-IKDSNRQSTQLE-ELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQS 87
QKLD+ IK+ + + L+ + T + K+ I++ +E KD+ EVN+
Sbjct: 138 QKLDQEIKERDEKYADLDAKFTRLHKRAKQRIQEIQKE-KDDLDARFREVNET------- 189
Query: 88 MTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLK 147
E+ S+ ++Q E + Q+ N +K MD Q + + + TIE + +
Sbjct: 190 -AERASSQHSSMQQELERTRQQ-ANEALKAMDAERQQLRSANNKLRDTIEELRGSLQPKE 247
Query: 148 GQTEQMGRIVNELDSIQFSIKKASQLVKE 176
+ E + + + + D I +KK Q V+E
Sbjct: 248 NKIETLQQSLLDKDQILEDLKKQLQAVEE 276
>At1g64320 bZIP transcription factor, putative
Length = 476
Score = 37.0 bits (84), Expect = 0.007
Identities = 32/146 (21%), Positives = 68/146 (45%), Gaps = 9/146 (6%)
Query: 48 LTGKMRDCKRLIKDFDREI-----KDEGAGNPEEVNKQLNDE--KQSMTEKNSTAEGNVQ 100
LT ++ D K +K+ ++EI ++ G EV K E K M + N
Sbjct: 48 LTSRIEDLKCQLKNLEQEIGFLRARNAGLAGNLEVTKVEEKERVKGLMDQVNGMKHELES 107
Query: 101 LASEMSNQEL-VNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNE 159
L S+ E + ++ + ET ++ K+ + ++ LKG+ + + R ++E
Sbjct: 108 LRSQKDESEAKLEKKVEEVTETKMQLKSLKEETEEERNRLSEEIDQLKGENQMLHRRISE 167
Query: 160 LDSIQFSIK-KASQLVKEIGRQVATD 184
LDS+ +K K++ +++ +++ T+
Sbjct: 168 LDSLHMEMKTKSAHEMEDASKKLDTE 193
>At5g65500 putative protein
Length = 765
Score = 34.7 bits (78), Expect = 0.036
Identities = 31/103 (30%), Positives = 53/103 (51%), Gaps = 4/103 (3%)
Query: 55 CKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTE-KNSTAEGNVQLASEMSNQELVNA 113
C I +F+ IK+E E++ L+ +K+ + E KN +G +L S QE++++
Sbjct: 283 CNSRIGEFEAWIKEESERR-EKLQATLDSDKECIEEAKNYVEKGKTKLHSLAELQEVLSS 341
Query: 114 GMKTMDET-DQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGR 155
+KTM E QA ++VV Q E+ T+ L+ Q + R
Sbjct: 342 KVKTMMEAKSQAEVELERVVLQRGEMITE-IEKLRSQRDVFNR 383
>At1g64330 unknown protein
Length = 555
Score = 34.7 bits (78), Expect = 0.036
Identities = 32/143 (22%), Positives = 60/143 (41%), Gaps = 22/143 (15%)
Query: 41 QSTQLEELTGKMRDCKRLIKDFDREI----------------KDEGAGNPEEVNKQLND- 83
+S +EE K L+KDF +E K G G + + +D
Sbjct: 45 ESGDIEEDESKRLVVAELVKDFYKEYESLYHQYDDLTGEIRKKVHGKGENDSSSSSSSDS 104
Query: 84 EKQSMTEKNSTAEGNVQLAS---EMSNQELVNAGMK--TMDETDQAIERSKQVVHQTIEV 138
+ +++N E ++L E +N E+ + MK T DE +A+E Q + + ++
Sbjct: 105 DSDKKSKRNGRGENEIELLKKQMEDANLEIADLKMKLATTDEHKEAVESEHQEILKKLKE 164
Query: 139 GTQTASTLKGQTEQMGRIVNELD 161
+ L+ +TE++ EL+
Sbjct: 165 SDEICGNLRVETEKLTSENKELN 187
>At5g55660 putative protein
Length = 759
Score = 33.9 bits (76), Expect = 0.062
Identities = 30/158 (18%), Positives = 73/158 (45%), Gaps = 12/158 (7%)
Query: 31 KLDKIKDSNRQS-------TQLEELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLND 83
K +++K++N++ E+ K + K +D + E +DE EE N D
Sbjct: 211 KGEEVKEANKEDDVEADTKVAEPEVEDKKTESKDENEDKEEEKEDEKEDEKEESNDDKED 270
Query: 84 EKQSMTEKNSTAEGNVQ--LASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQ 141
+K+ + + N +G + S++E + KT +D+ + R ++ V + + V +
Sbjct: 271 KKEDIKKSNKRGKGKTEKTRGKTKSDEEKKDIEPKTPFFSDRPV-RERKSVERLVAVVDK 329
Query: 142 TASTLKGQTEQMGRIVNELDSIQFSI--KKASQLVKEI 177
+S + G + ++ ++ + + KK+ ++ K++
Sbjct: 330 DSSREFHVEKGKGTPLKDIPNVAYKVSRKKSDEVFKQL 367
>At2g19950 hypothetical protein
Length = 713
Score = 33.5 bits (75), Expect = 0.080
Identities = 29/123 (23%), Positives = 53/123 (42%), Gaps = 14/123 (11%)
Query: 3 SNLKMTPQLEQIHGEIRD-----------IFRALANGFQKLDKIKDSNRQSTQLEELTGK 51
+++K+ QLE+ G ++ + R A +L +IK N QLEEL
Sbjct: 267 TSMKIQDQLEEAQGLLKATVSTGQSKEARLARVCAGLSSRLQEIKAEN---AQLEELLTA 323
Query: 52 MRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELV 111
++ + + R ++ + + EV K + +++ KNS E V + NQ +
Sbjct: 324 EQELTKSYEASIRHLQKDLSAAKSEVTKVESSMVEALAAKNSEIETLVSAMDALKNQAAL 383
Query: 112 NAG 114
N G
Sbjct: 384 NEG 386
>At1g76780 putative heat shock protein
Length = 1871
Score = 33.5 bits (75), Expect = 0.080
Identities = 23/86 (26%), Positives = 39/86 (44%), Gaps = 11/86 (12%)
Query: 64 REIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQ 123
++ +D+G G ++ Q D QS EK+++ + +E +N T ET +
Sbjct: 1672 KKSRDDGFGKVRKIEVQRKDNDQSFVEKDTSGKA----------KENLNDEEPTKTET-K 1720
Query: 124 AIERSKQVVHQTIEVGTQTASTLKGQ 149
A + + +HQ E GT LK Q
Sbjct: 1721 ATDNESRKIHQIKEQGTSEQERLKEQ 1746
Score = 26.9 bits (58), Expect = 7.5
Identities = 19/76 (25%), Positives = 32/76 (42%), Gaps = 12/76 (15%)
Query: 36 KDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTE--KNS 93
K++ ++EL GK +CK D+G G EE KQ + + + K
Sbjct: 1286 KETKEVEAHVQELEGKTENCK----------DDDGEGRREERGKQGMTAENMLRQRFKTK 1335
Query: 94 TAEGNVQLASEMSNQE 109
+ +G V+ E +E
Sbjct: 1336 SDDGIVRKIQETKEEE 1351
>At1g68790 putative nuclear matrix constituent protein 1 (NMCP1)
Length = 1085
Score = 33.1 bits (74), Expect = 0.10
Identities = 36/161 (22%), Positives = 70/161 (43%), Gaps = 24/161 (14%)
Query: 10 QLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDE 69
Q+ + H R+ + N ++LD +K Q E M D + ++ D E + +
Sbjct: 564 QISEKHRLKREEMTSRDNLKRELDGVK------MQKESFEADMEDLEMQKRNLDMEFQRQ 617
Query: 70 GAGNPEEVNKQLNDEKQSMTEKNSTAE-GNVQLASEMSNQELVNAGMKTMDETDQAIERS 128
EE ++ +E+ EK S E N+ +++ +E M+ M A+ER
Sbjct: 618 -----EEAGERDFNERARTYEKRSQEELDNINYTKKLAQRE-----MEEMQYEKLALERE 667
Query: 129 KQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKK 169
++ + + LK Q +M + + ELD ++ S+K+
Sbjct: 668 REQI-------SVRKKLLKEQEAEMHKDITELDVLRSSLKE 701
>At1g05320 unknown protein
Length = 841
Score = 33.1 bits (74), Expect = 0.10
Identities = 31/145 (21%), Positives = 64/145 (43%), Gaps = 15/145 (10%)
Query: 30 QKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREI------KDEGAGNPEEVNKQLND 83
+K I+ N++ T+ ++L K++ + +I++ R++ D EE +LN
Sbjct: 426 EKETAIEKLNQKDTEAKDLITKLKSHENVIEEHKRQVLEASGVADTRKVEVEEALLKLNT 485
Query: 84 EKQSMTE----KNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVG 139
+ ++ E AE N++L NQ+L N G +T D + + Q E+
Sbjct: 486 LESTIEELEKENGDLAEVNIKL-----NQKLANQGSETDDFQAKLSVLEAEKYQQAKELQ 540
Query: 140 TQTASTLKGQTEQMGRIVNELDSIQ 164
K T + R+ +++ S++
Sbjct: 541 ITIEDLTKQLTSERERLRSQISSLE 565
>At4g32615 putative protein
Length = 315
Score = 32.7 bits (73), Expect = 0.14
Identities = 28/134 (20%), Positives = 55/134 (40%), Gaps = 26/134 (19%)
Query: 36 KDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDEGA-------GNPEEVN---------- 78
K++ RQ ++ E ++ + + L+ DF KDE G +EVN
Sbjct: 155 KEAERQLSKKERKKKELAELEALLADFGVATKDENGQQDSQDKGEKKEVNDEGEKKENTT 214
Query: 79 --------KQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQ 130
K+ D+++ + E S + N ASE + QE ++ + + + K+
Sbjct: 215 GESKASKKKKKKDKQKELKESQSEVKSNSDAASESAEQEESSSSIDVKERLKKIASMKKK 274
Query: 131 VVHQTIEVGTQTAS 144
+ ++ G TA+
Sbjct: 275 KSSKEVD-GASTAA 287
>At3g28770 hypothetical protein
Length = 2081
Score = 32.7 bits (73), Expect = 0.14
Identities = 27/137 (19%), Positives = 59/137 (42%), Gaps = 14/137 (10%)
Query: 33 DKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDEGAG-----------NPEEVNKQL 81
+K++D + +E K + + + + ++E++ +G + E+V ++
Sbjct: 244 NKVEDLKEGNNVVENGETKENNGENVESNNEKEVEGQGESIGDSAIEKNLESKEDVKSEV 303
Query: 82 NDEKQ---SMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEV 138
K SMTE A+GN +++ + +E+ G D + SK+ V +E
Sbjct: 304 EAAKNDGSSMTENLGEAQGNNGVSTIDNEKEVEGQGESIEDSDIEKNLESKEDVKSEVEA 363
Query: 139 GTQTASTLKGQTEQMGR 155
S++ G+ E+ R
Sbjct: 364 AKNAGSSMTGKLEEAQR 380
Score = 29.3 bits (64), Expect = 1.5
Identities = 28/126 (22%), Positives = 56/126 (44%), Gaps = 19/126 (15%)
Query: 30 QKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMT 89
+K ++ + +N +S + E L K + +E+KD+ + + N+ +EK+ T
Sbjct: 584 EKREETQGNNGESVKNENLENK---------EDKKELKDDESVGAKTNNETSLEEKREQT 634
Query: 90 EK------NSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTA 143
+K NS N ++ + ++ V+ G D T+ SK+ +EV
Sbjct: 635 QKGHDNSINSKIVDNKGGNADSNKEKEVHVG----DSTNDNNMESKEDTKSEVEVKKNDG 690
Query: 144 STLKGQ 149
S+ KG+
Sbjct: 691 SSEKGE 696
>At2g34300 unknown protein
Length = 770
Score = 32.7 bits (73), Expect = 0.14
Identities = 24/100 (24%), Positives = 45/100 (45%), Gaps = 10/100 (10%)
Query: 67 KDEGAGNPEEVNKQLNDEKQSMTEKN-------STAEGNVQLASEMSNQELVNAGMKTMD 119
K+EG +P+ + + N+E ++ TE N ++AEGN S+ E AG + +
Sbjct: 66 KEEGDRDPKNFSDEKNEENEAATENNQVKTDSENSAEGN---QVNESSGEKTEAGEERKE 122
Query: 120 ETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNE 159
D + + EVG+++ T + + Q+ E
Sbjct: 123 SDDNNGDGDGEKEKNVKEVGSESDETTQKEKTQLEESTEE 162
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 32.7 bits (73), Expect = 0.14
Identities = 29/158 (18%), Positives = 66/158 (41%), Gaps = 30/158 (18%)
Query: 35 IKDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDEGAGNP----------------EEVN 78
+++ + ++ LT K+RD + IK ++ ++ + + E VN
Sbjct: 812 MEEFTSRDSEASSLTEKLRDLEGKIKSYEEQLAEASGKSSSLKEKLEQTLGRLAAAESVN 871
Query: 79 KQLNDEKQSMTEKNSTAEGNVQLASEMSNQ---------ELVNAGM----KTMDETDQAI 125
++L E EK+ + +L +E +NQ L+ +G + ++AI
Sbjct: 872 EKLKQEFDQAQEKSLQSSSESELLAETNNQLKIKIQELEGLIGSGSVEKETALKRLEEAI 931
Query: 126 ERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSI 163
ER Q ++ ++ + T + Q E+ ++ +E +
Sbjct: 932 ERFNQKETESSDL-VEKLKTHENQIEEYKKLAHEASGV 968
Score = 28.1 bits (61), Expect = 3.4
Identities = 21/111 (18%), Positives = 46/111 (40%), Gaps = 4/111 (3%)
Query: 46 EELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEM 105
E+ ++ K I+D +++ EG +++ + Q ST E + +++
Sbjct: 1040 EQTANELEASKTTIEDLTKQLTSEGEKLQSQISSHTEENNQVNAMFQSTKEELQSVIAKL 1099
Query: 106 SNQELVNAG----MKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQ 152
Q V + + + E +A+ K V+ E +T S +K Q ++
Sbjct: 1100 EEQLTVESSKADTLVSEIEKLRAVAAEKSVLESHFEELEKTLSEVKAQLKE 1150
>At1g28490 unknown protein (At1g28490)
Length = 245
Score = 32.7 bits (73), Expect = 0.14
Identities = 32/143 (22%), Positives = 64/143 (44%), Gaps = 18/143 (12%)
Query: 70 GAGNPEEVNKQL----NDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAI 125
GAG+ EV ++L N + S ++ + + + SE Q L+ + + D+ +
Sbjct: 110 GAGHASEVRRELMRMPNSGEASRYDQYGGRDDDGFVQSESDRQMLL------IKQQDEEL 163
Query: 126 ERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDS----IQFSIKKASQLVKEIGRQV 181
+ + V + VG L Q + + E+DS ++F KK ++K+ G +
Sbjct: 164 DELSKSVQRIGGVGLTIHDELVAQERIIDELDTEMDSTKNRLEFVQKKVGMVMKKAGAKG 223
Query: 182 ATDKCIMLFLFLIVCGVIAIIVV 204
+M+ FL+V +I ++V
Sbjct: 224 Q----MMMICFLLVLFIILFVLV 242
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.130 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,799,772
Number of Sequences: 26719
Number of extensions: 145307
Number of successful extensions: 763
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 86
Number of HSP's that attempted gapping in prelim test: 703
Number of HSP's gapped (non-prelim): 138
length of query: 206
length of database: 11,318,596
effective HSP length: 95
effective length of query: 111
effective length of database: 8,780,291
effective search space: 974612301
effective search space used: 974612301
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146862.25


