

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.17 + phase: 0
(310 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
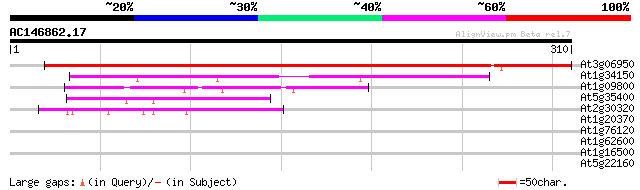
At3g06950 putative tRNA pseudouridine synthase 408 e-114
At1g34150 pseudouridine synthase-like protein 75 5e-14
At1g09800 hypothetical protein 68 6e-12
At5g35400 unknown protein 61 9e-10
At2g30320 putative pseudouridine synthase 47 1e-05
At1g20370 putative tRNA pseudouridine synthase 36 0.023
At1g76120 unknown protein 35 0.040
At1g62600 unknown protein 28 6.3
At1g16500 unknown protein 28 6.3
At5g22160 putative protein 28 8.3
>At3g06950 putative tRNA pseudouridine synthase
Length = 298
Score = 408 bits (1049), Expect = e-114
Identities = 199/293 (67%), Positives = 234/293 (78%), Gaps = 3/293 (1%)
Query: 20 TSLLSPNQDGYKWRLLLSYDGTRYAGWQYQESPPTVQCAVEKALIRATKLQRKDLQLVGA 79
+S+ + YKWRL+++YDGTR+AGWQYQESPPT+Q +EKALI+ T+L RK+LQL+GA
Sbjct: 6 SSVKVSDSGAYKWRLVIAYDGTRFAGWQYQESPPTIQSMLEKALIQITELGRKELQLIGA 65
Query: 80 SRTDAGVHAWGQVAHFLTPFNYDNLDSVHAALNGLLPSDIRVREISPASAEFHARFSVKS 139
RTDAGVHAWGQVAHF+TPFNY +LDS HAALNGLLP DIRVRE+S A EFHARFS S
Sbjct: 66 GRTDAGVHAWGQVAHFVTPFNYTSLDSFHAALNGLLPKDIRVRELSAAVPEFHARFSASS 125
Query: 140 KIYHYKIYSDTIMDPFQRHFAYHNMYKLNSAAMREAARYFIGKHDFTAFENASHNDRIPD 199
K+Y Y+IY+DT MDPFQRH+AYH YKLN++ MREAA F+GKHDF+AF NA+ D +PD
Sbjct: 126 KVYRYQIYNDTFMDPFQRHWAYHCAYKLNASKMREAANLFVGKHDFSAFANATREDGVPD 185
Query: 200 PVKHIFPIDVKEMGALLQLEVEGSGFLYRQVRNMVALLLQIGKEATPPDIVPHILASRDR 259
P+K I DV +MG+LLQLEVEGSGFLYRQVRNMVALL+QIGKEA DIVP IL ++DR
Sbjct: 186 PLKTISRFDVIQMGSLLQLEVEGSGFLYRQVRNMVALLIQIGKEALDSDIVPMILETKDR 245
Query: 260 KELAKYCYYLP--PHGLCLVSINYNESHLLPPPGCPAKSFGMHHTIRKCKVVF 310
+ LAKY LP PHGLCLVS+ Y E HL P CP S G HHTI KCK+ F
Sbjct: 246 RVLAKYT-SLPASPHGLCLVSVKYKEDHLKLPLDCPVTSSGRHHTITKCKLPF 297
>At1g34150 pseudouridine synthase-like protein
Length = 446
Score = 75.1 bits (183), Expect = 5e-14
Identities = 76/257 (29%), Positives = 106/257 (40%), Gaps = 41/257 (15%)
Query: 34 LLLSYDGTRYAGWQYQ-ESPPTVQCAVEKALIRATKL--QRKDLQLVGASRTDAGVHAWG 90
L + Y G R+ G+ + + P+++ V KAL R L +KD RTD GV + G
Sbjct: 94 LKIMYFGKRFYGFSAEAQMEPSIESEVFKALERTRLLVGDKKDSCYSRCGRTDKGVSSTG 153
Query: 91 QV-AHFLTPFNYDNLDSVHAALNG----------------LLPSDIRVREISPASAEFHA 133
QV A FL A +NG LP DIRV SPA +FHA
Sbjct: 154 QVIALFLRSRLKSPPGDSKAQVNGRTGERPEYDYVRVLNRALPDDIRVIGWSPAPIDFHA 213
Query: 134 RFSVKSKIYHYKIYSDTIMDPFQRHFAYHNMYKLNSAAMREAARYFIGKHDFTAFENAS- 192
RFS ++ Y Y + LN +AM A + FIG+HDF F
Sbjct: 214 RFSCYAREYKYFFWRQ----------------NLNLSAMDFAGKKFIGEHDFRNFCKMDV 257
Query: 193 ---HNDRIPDPVKHIFPIDVKEMG-ALLQLEVEGSGFLYRQVRNMVALLLQIGKEATPPD 248
H + P G L + GS FL+ Q+R MVA+L IG+ D
Sbjct: 258 ANVHCYTRRVTFFEVSPCQNSHEGDQLCTFTMRGSAFLWHQIRCMVAVLFMIGQGVESVD 317
Query: 249 IVPHILASRDRKELAKY 265
++ +L ++ +Y
Sbjct: 318 VIDTLLDTKKTPRKPQY 334
>At1g09800 hypothetical protein
Length = 372
Score = 68.2 bits (165), Expect = 6e-12
Identities = 51/186 (27%), Positives = 89/186 (47%), Gaps = 25/186 (13%)
Query: 31 KWRLLLSYDGTRYAGWQYQESPPTVQCAVEKALIRATKLQRKDLQLVGASRTDAGVHAWG 90
++ + + Y GTR++G Q Q TV +++A K + +++ +SRTDAGVHA
Sbjct: 56 RYLVAIEYIGTRFSGSQQQAKDRTVVGVLQEAF---HKFIGQPVKIFCSSRTDAGVHALS 112
Query: 91 QVAHF------------LTPFNYDNLDSVHAALNGLLP---SDIRVREISPASAEFHARF 135
V H + P + + V A+N L D+ V ++ + +HAR+
Sbjct: 113 NVCHVDVERISKRKPGEVLPPHEPGV--VQRAVNHFLQRKDGDVMVIDVRCVPSNYHARY 170
Query: 136 SVKSKIYHYKIYSDTIMDPF---QRHFAYHNMYKLNSAAMREAARYFIGKHDFTAFENAS 192
+ + Y Y++ S + DP ++ +H +LN M+EA R +G HDF++F A
Sbjct: 171 KAQERTYFYRLLSGS--DPLSILEKDRCWHVPEELNLRFMQEACRVLVGSHDFSSFRAAG 228
Query: 193 HNDRIP 198
+ P
Sbjct: 229 CQAKSP 234
>At5g35400 unknown protein
Length = 420
Score = 60.8 bits (146), Expect = 9e-10
Identities = 45/123 (36%), Positives = 60/123 (48%), Gaps = 10/123 (8%)
Query: 32 WRLLLSYDGTRYAGWQYQESPPTVQCAVEKAL------IRATKLQRKDLQLVG----ASR 81
++++LSY+G + GWQ Q TVQ VEK+L +A L++K L G A R
Sbjct: 95 FKIVLSYNGASFDGWQKQPDLHTVQGVVEKSLGGFVDERKAQLLKKKCKPLEGRVLVAGR 154
Query: 82 TDAGVHAWGQVAHFLTPFNYDNLDSVHAALNGLLPSDIRVREISPASAEFHARFSVKSKI 141
TD GV A QV F T ++ A+N +RV IS S FH FS K +
Sbjct: 155 TDKGVSALNQVCSFYTWRKDIEPIAIEDAINKDASGKLRVVSISKVSRSFHPNFSAKWRR 214
Query: 142 YHY 144
Y Y
Sbjct: 215 YLY 217
>At2g30320 putative pseudouridine synthase
Length = 510
Score = 47.0 bits (110), Expect = 1e-05
Identities = 46/161 (28%), Positives = 71/161 (43%), Gaps = 26/161 (16%)
Query: 17 PSPTSLLSPNQDGY---KWR--------LLLSYDGTRYAGWQYQESPP---TVQCAVEKA 62
PS TSL P+ D + KW + + Y GT Y G Q Q P T++ +E A
Sbjct: 39 PSSTSLSPPSSDNFLADKWESYRKKKVVIRIGYVGTDYRGLQIQRDDPSIKTIEGELEVA 98
Query: 63 LIRATKLQRK---DLQLVG---ASRTDAGVHAWGQVAHFL-----TPFNYDNLDSVHA-A 110
+ +A ++ DL +G +SRTD GVH+ T + D +V A
Sbjct: 99 IYKAGGIRDSNYGDLHKIGWARSSRTDKGVHSLATSISLKMEIPETAWKDDPQGTVLAKC 158
Query: 111 LNGLLPSDIRVREISPASAEFHARFSVKSKIYHYKIYSDTI 151
++ LP +IRV + P++ F R + Y Y + D +
Sbjct: 159 ISKHLPENIRVFSVLPSNRRFDPRRECTLRKYSYLLPVDVL 199
Score = 28.1 bits (61), Expect = 6.3
Identities = 9/35 (25%), Positives = 20/35 (56%)
Query: 216 LQLEVEGSGFLYRQVRNMVALLLQIGKEATPPDIV 250
+++ + G F+ Q+R M+ + + +E P DI+
Sbjct: 344 VEISIWGESFMLHQIRKMIGTAVAVKRELLPRDII 378
>At1g20370 putative tRNA pseudouridine synthase
Length = 549
Score = 36.2 bits (82), Expect = 0.023
Identities = 36/144 (25%), Positives = 59/144 (40%), Gaps = 9/144 (6%)
Query: 9 TSSVVTPIPSPTSLLSPNQDGYKWRLLLSYDGTRYAGWQYQESPPTVQCAVEKALIRATK 68
+S+V T + +P K ++ ++ G Y G Q T++ +E+AL A
Sbjct: 37 SSAVTTGGSNVAERRAPKYRRRKVAIVFAFCGVGYQGMQKNPGAKTIEGELEEALFHAGA 96
Query: 69 LQRKD------LQLVGASRTDAGVHAWGQVAHFLTPFNYDNLDSVHAALNGLLPSDIRVR 122
+ D + ++RTD GV A GQV ++ Y + LN LP IRV
Sbjct: 97 VPDADRNKPRNYEWARSARTDKGVSAVGQV---VSGRFYVDPPGFVERLNSKLPDQIRVF 153
Query: 123 EISPASAEFHARFSVKSKIYHYKI 146
+ F ++ + Y Y I
Sbjct: 154 GYKRVAPSFSSKKFCDRRRYVYLI 177
>At1g76120 unknown protein
Length = 463
Score = 35.4 bits (80), Expect = 0.040
Identities = 33/120 (27%), Positives = 51/120 (42%), Gaps = 9/120 (7%)
Query: 31 KWRLLLSYDGTRYAGWQYQESPPTVQCAVEKALIRATKLQR------KDLQLVGASRTDA 84
K ++ ++ G Y G Q T++ +E+AL A + K ++RTD
Sbjct: 23 KVAIIFAFCGVGYQGMQKNPGAKTIEGELEEALFHAGAVPESIRGKPKLYDFARSARTDK 82
Query: 85 GVHAWGQVAHFLTPFNYDNLDSVHAALNGLLPSDIRVREISPASAEFHARFSVKSKIYHY 144
GV A GQV F D L V+ LN LP+ IR+ + F ++ + Y Y
Sbjct: 83 GVSAVGQVVS--GRFIVDPLGFVN-RLNSNLPNQIRIFGYKHVTPSFSSKKFCDRRRYVY 139
>At1g62600 unknown protein
Length = 452
Score = 28.1 bits (61), Expect = 6.3
Identities = 28/126 (22%), Positives = 49/126 (38%), Gaps = 15/126 (11%)
Query: 77 VGASRTDAGVHAWGQVAHFLTPFNYDNLDSVHAALNGLLPSDIRVREISPASAEFHARFS 136
V SR + G+V +L F + A+ ++ D V +++PA+ E ++
Sbjct: 99 VSESRDPRRFPSHGEVLAYLQDF------AKEFAIEEMIRFDTAVVKVAPAAEEGSGKWR 152
Query: 137 VKSKIYHYKIYSDTIMDPFQRHFAYHNMYKLNSAAMREAARYFIGKHDFTAFENASHNDR 196
++S K+ D I D + N + G + E SHN R
Sbjct: 153 IESTEKEKKVLRDEIYDA---------VVVCNGHYIEPRHAEIPGISSWPGKEMHSHNYR 203
Query: 197 IPDPVK 202
IP+P +
Sbjct: 204 IPEPFR 209
>At1g16500 unknown protein
Length = 259
Score = 28.1 bits (61), Expect = 6.3
Identities = 13/26 (50%), Positives = 15/26 (57%)
Query: 2 SLALAAATSSVVTPIPSPTSLLSPNQ 27
SL L SS+ P+P P S SPNQ
Sbjct: 141 SLELKLVPSSISRPLPPPLSSFSPNQ 166
>At5g22160 putative protein
Length = 353
Score = 27.7 bits (60), Expect = 8.3
Identities = 19/56 (33%), Positives = 27/56 (47%), Gaps = 6/56 (10%)
Query: 92 VAHFLTPFNYDNLDSVHAALNGLLPSDIRVREISPASAEFHARFSVKSKIYHYKIY 147
VA +L F+ N HA + I+ +SP+SA +H F VK+ Y IY
Sbjct: 52 VAVYLLIFHVVNNAHCHAKFS------IQSIAVSPSSATWHVDFLVKNPSSRYSIY 101
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,070,529
Number of Sequences: 26719
Number of extensions: 294908
Number of successful extensions: 637
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 621
Number of HSP's gapped (non-prelim): 12
length of query: 310
length of database: 11,318,596
effective HSP length: 99
effective length of query: 211
effective length of database: 8,673,415
effective search space: 1830090565
effective search space used: 1830090565
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146862.17


