

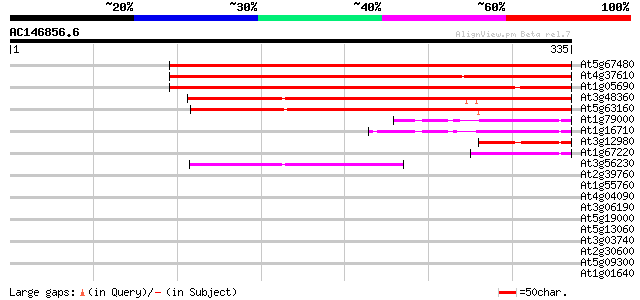
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146856.6 - phase: 0 /pseudo
(335 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g67480 unknown protein 372 e-103
At4g37610 unknown protein 326 1e-89
At1g05690 unknown protein 285 2e-77
At3g48360 unknown protein 216 2e-56
At5g63160 unknown protein 215 2e-56
At1g79000 hypothetical protein 67 1e-11
At1g16710 hypothetical protein 65 7e-11
At3g12980 putative CREB-binding protein 59 5e-09
At1g67220 hypothetical protein 45 7e-05
At3g56230 putative protein 41 8e-04
At2g39760 unknown protein 40 0.002
At1g55760 unknown protein 40 0.002
At4g04090 hypothetical protein 39 0.005
At3g06190 unknown protein 39 0.005
At5g19000 unknown protein 38 0.009
At5g13060 unknown protein 37 0.012
At3g03740 unknown protein 37 0.020
At2g30600 unknown protein 33 0.22
At5g09300 branched-chain alpha keto-acid dehydrogenase E1 alpha ... 33 0.29
At1g01640 hypothetical protein 32 0.37
>At5g67480 unknown protein
Length = 372
Score = 372 bits (954), Expect = e-103
Identities = 174/241 (72%), Positives = 201/241 (83%), Gaps = 1/241 (0%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
MLKQA R +W +ISI GVPHDAVRVFIR+LYSSCYEKEEM EF++HLL+LSH Y VP L
Sbjct: 89 MLKQAKRHGKWHTISIRGVPHDAVRVFIRFLYSSCYEKEEMNEFIMHLLLLSHAYVVPQL 148
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KR CE LE GLLT +N+VDVFQLALLCD PRLSLI HR I+K+F +S +E W AMK+S
Sbjct: 149 KRVCEWHLEHGLLTTENVVDVFQLALLCDFPRLSLISHRMIMKHFNELSATEAWTAMKKS 208
Query: 216 HPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDF 275
HP LEKE+ +S+I E N++KER+RK N++ +Y QLY+AMEALVHICRDGC+TIGPHDKDF
Sbjct: 209 HPFLEKEVRDSVIIEANTRKERMRKRNDQRIYSQLYEAMEALVHICRDGCKTIGPHDKDF 268
Query: 276 KANQ-PCRYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPL 334
K N C Y +CKGLE L+RHFAGCKLRVPGGC HCKRMWQLLELHSR+CA D CRVPL
Sbjct: 269 KPNHATCNYEACKGLESLIRHFAGCKLRVPGGCVHCKRMWQLLELHSRVCAGSDQCRVPL 328
Query: 335 C 335
C
Sbjct: 329 C 329
>At4g37610 unknown protein
Length = 368
Score = 326 bits (835), Expect = 1e-89
Identities = 154/241 (63%), Positives = 186/241 (76%), Gaps = 2/241 (0%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
M+KQ R + +SISI GVPH A+RVFIR+LYSSCYEK++M++F +HLLVLSHVY VPHL
Sbjct: 84 MMKQHKRKSHRKSISILGVPHHALRVFIRFLYSSCYEKQDMEDFAIHLLVLSHVYVVPHL 143
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KR CE + E LL +N++DVFQLALLCDAPRL L+CHR IL NF VS SEGW+AMK+S
Sbjct: 144 KRVCESEFESSLLNKENVIDVFQLALLCDAPRLGLLCHRMILNNFEEVSTSEGWQAMKES 203
Query: 216 HPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDF 275
HP L+KE+L S+ E NS K+R RK E + Y QLY+AMEA VHICRDGCR IGP K
Sbjct: 204 HPRLQKELLRSVAYELNSLKQRNRKQKEIQTYTQLYEAMEAFVHICRDGCREIGP-TKTE 262
Query: 276 KANQPCRYTSCKGLELLVRHFAGCKLR-VPGGCGHCKRMWQLLELHSRLCADPDFCRVPL 334
+ C + +C GLE L++H AGCKLR +PGGC CKRMWQLLELHSR+C D + C+VPL
Sbjct: 263 TPHMSCGFQACNGLEQLLKHLAGCKLRSIPGGCSRCKRMWQLLELHSRICVDSEQCKVPL 322
Query: 335 C 335
C
Sbjct: 323 C 323
>At1g05690 unknown protein
Length = 364
Score = 285 bits (730), Expect = 2e-77
Identities = 134/241 (55%), Positives = 181/241 (74%), Gaps = 3/241 (1%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
+L Q+ N + I GVP +AV +FIR+LYSSCYE+EEMK+FVLHLLVLSH Y+VP L
Sbjct: 83 LLNQSRDKNGNTYLKIHGVPCEAVYMFIRFLYSSCYEEEEMKKFVLHLLVLSHCYSVPSL 142
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KR C + L+ G + +N++DV QLA CD R+ +C ++K+F++VS +EGWK MK+S
Sbjct: 143 KRLCVEILDQGWINKENVIDVLQLARNCDVTRICFVCLSMVIKDFKSVSSTEGWKVMKRS 202
Query: 216 HPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDF 275
+P+LE+E++E++I+ ++ K+ER RK+ EREVYLQLY+AMEALVHICR+GC TIGP DK
Sbjct: 203 NPLLEQELIEAVIESDSRKQERRRKLEEREVYLQLYEAMEALVHICREGCGTIGPRDKAL 262
Query: 276 KANQP-CRYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPL 334
K + C++ +CKGLE +RHF GCK R C HCKRMWQLL+LHS +C D + C+V L
Sbjct: 263 KGSHTVCKFPACKGLEGALRHFLGCKSR--ASCSHCKRMWQLLQLHSCICDDSNSCKVSL 320
Query: 335 C 335
C
Sbjct: 321 C 321
>At3g48360 unknown protein
Length = 364
Score = 216 bits (549), Expect = 2e-56
Identities = 115/239 (48%), Positives = 157/239 (65%), Gaps = 11/239 (4%)
Query: 107 RSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKLELG 166
R I I GVP DAV VFI++LYSS ++EM+ + +HLL LSHVY V LK+ C + + +
Sbjct: 78 RVIKILGVPCDAVSVFIKFLYSSSLTEDEMERYGIHLLALSHVYMVTQLKQRCSKGV-VQ 136
Query: 167 LLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILES 226
LT +N+VDV QLA LCDAP + L R I F+TV ++EGWK +++ P LE +IL+
Sbjct: 137 RLTTENVVDVLQLARLCDAPDVCLRSMRLIHSQFKTVEQTEGWKFIQEHDPFLELDILQF 196
Query: 227 MIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPH---DKDFKA-----N 278
+ D E+ KK R R E+++Y+QL +AME + HIC GC +GP D + K+ +
Sbjct: 197 IDDAESRKKRRRRHRKEQDLYMQLSEAMECIEHICTQGCTLVGPSNVVDNNKKSMTAEKS 256
Query: 279 QPCR-YTSCKGLELLVRHFAGCKLR-VPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
+PC+ +++C GL+LL+RHFA CK R GC CKRM QL LHS +C PD CRVPLC
Sbjct: 257 EPCKAFSTCYGLQLLIRHFAVCKRRNNDKGCLRCKRMLQLFRLHSLICDQPDSCRVPLC 315
>At5g63160 unknown protein
Length = 365
Score = 215 bits (548), Expect = 2e-56
Identities = 115/235 (48%), Positives = 153/235 (64%), Gaps = 9/235 (3%)
Query: 109 ISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKLELGLL 168
I I GVP DAV VF+R+LYS + EM+++ +HLL LSHVY V LK+ C + + +
Sbjct: 70 IKILGVPCDAVSVFVRFLYSPSVTENEMEKYGIHLLALSHVYMVTQLKQRCTKGVG-ERV 128
Query: 169 TMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILESMI 228
T +N+VD+ QLA LCDAP L L C R I F+TV ++EGWK +++ P LE +IL+ +
Sbjct: 129 TAENVVDILQLARLCDAPDLCLKCMRFIHYKFKTVEQTEGWKFLQEHDPFLELDILQFID 188
Query: 229 DEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDK-DFKAN-----QPC- 281
D E+ KK R R E+ +YLQL +AME + HIC +GC +GP D K+ PC
Sbjct: 189 DAESRKKRRRRHRREQNLYLQLSEAMECIEHICTEGCTLVGPSSNLDNKSTCQAKPGPCS 248
Query: 282 RYTSCKGLELLVRHFAGCKLRVPG-GCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
+++C GL+LL+RHFA CK RV G GC CKRM QLL LHS +C + CRVPLC
Sbjct: 249 AFSTCYGLQLLIRHFAVCKKRVDGKGCVRCKRMIQLLRLHSSICDQSESCRVPLC 303
>At1g79000 hypothetical protein
Length = 1691
Score = 67.0 bits (162), Expect = 1e-11
Identities = 36/106 (33%), Positives = 58/106 (53%), Gaps = 18/106 (16%)
Query: 230 EENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDFKANQPCRYTSCKGL 289
++N++ + R++ LQL ++ LVH + CR+ C+Y +C+ +
Sbjct: 1566 DQNAQNKEARQLR----VLQLRKMLDLLVHASQ--CRSAH-----------CQYPNCRKV 1608
Query: 290 ELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
+ L RH CK+R GGC CK+MW LL+LH+R C + + C VP C
Sbjct: 1609 KGLFRHGINCKVRASGGCVLCKKMWYLLQLHARACKESE-CHVPRC 1653
>At1g16710 hypothetical protein
Length = 1706
Score = 64.7 bits (156), Expect = 7e-11
Identities = 39/121 (32%), Positives = 62/121 (51%), Gaps = 20/121 (16%)
Query: 215 SHPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKD 274
+HP K + ++N++ + R++ LQL ++ LVH + CR+
Sbjct: 1568 NHP--HKLTTHPSLADQNAQNKEARQLR----VLQLRKMLDLLVHASQ--CRS------- 1612
Query: 275 FKANQPCRYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPL 334
C Y +C+ ++ L RH CK+R GGC CK+MW LL+LH+R C + + C VP
Sbjct: 1613 ----PVCLYPNCRKVKGLFRHGLRCKVRASGGCVLCKKMWYLLQLHARACKESE-CDVPR 1667
Query: 335 C 335
C
Sbjct: 1668 C 1668
>At3g12980 putative CREB-binding protein
Length = 1670
Score = 58.5 bits (140), Expect = 5e-09
Identities = 27/55 (49%), Positives = 34/55 (61%), Gaps = 4/55 (7%)
Query: 281 CRYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
C+Y C+ ++ L+RH CK R GC CK+MW L LHSR C DP C+VP C
Sbjct: 1581 CQYPRCRVIKGLIRHGLVCKTR---GCIACKKMWSLFRLHSRNCRDPQ-CKVPKC 1631
>At1g67220 hypothetical protein
Length = 1357
Score = 44.7 bits (104), Expect = 7e-05
Identities = 21/61 (34%), Positives = 30/61 (48%), Gaps = 2/61 (3%)
Query: 276 KANQPCRYTSCKGLELLVRHFAGCKLRVPG-GCGHCKRMWQLLELHSRLCADPDFCRVPL 334
K + C Y C ++ L H CK+R G C C ++WQ + +H C D + C VP
Sbjct: 1297 KTTKSCSYPKCHEVKALFTHNVQCKIRKKGTRCNTCYKLWQTIRIHVYHCQDLN-CPVPQ 1355
Query: 335 C 335
C
Sbjct: 1356 C 1356
>At3g56230 putative protein
Length = 282
Score = 41.2 bits (95), Expect = 8e-04
Identities = 26/128 (20%), Positives = 65/128 (50%), Gaps = 1/128 (0%)
Query: 108 SISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKLELGL 167
+I++ + + ++ + +LY+ +++++ V L + + Y + +L+ CEQ + L
Sbjct: 154 AITLQELNSEQLQALLEFLYTGTLASDKLEKNVYALFIAADKYMIHYLQELCEQYM-LSS 212
Query: 168 LTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILESM 227
L + ++++V ++ L + L C +++N V S+ ++ Q + L EI +
Sbjct: 213 LDISSVLNVLDVSDLGSSKTLKEACVGFVVRNMDDVVFSDKYEPFSQKNQHLCVEITRAF 272
Query: 228 IDEENSKK 235
+ E SK+
Sbjct: 273 LMETRSKR 280
>At2g39760 unknown protein
Length = 408
Score = 39.7 bits (91), Expect = 0.002
Identities = 30/96 (31%), Positives = 45/96 (46%), Gaps = 3/96 (3%)
Query: 140 VLHLLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILK- 198
+ HLL + +Y + LK CE L L +DN+ LA +L C +
Sbjct: 276 IQHLLAAADLYDLARLKILCEVLL-CEKLDVDNVATTLALAEQHQFLQLKAFCLEFVASP 334
Query: 199 -NFRTVSESEGWKAMKQSHPVLEKEILESMIDEENS 233
N V +SEG+K +KQS P L E+L ++ + S
Sbjct: 335 ANLGAVMKSEGFKHLKQSCPTLLSELLNTVAAADKS 370
>At1g55760 unknown protein
Length = 329
Score = 39.7 bits (91), Expect = 0.002
Identities = 26/93 (27%), Positives = 46/93 (48%), Gaps = 8/93 (8%)
Query: 109 ISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVL---SHVYAVPHLKRECEQKLEL 165
I++ +P DA + F+ Y+Y + + ++F++H L L + Y + LK C L L
Sbjct: 205 INVLDMPLDACQAFLSYIYGNI----QNEDFLIHRLALLQAAEKYDIADLKEACHLSL-L 259
Query: 166 GLLTMDNLVDVFQLALLCDAPRLSLICHRKILK 198
+ N+++ Q A L P L C R ++K
Sbjct: 260 DDIDTKNVLERLQNAYLYQLPELKASCMRYLVK 292
>At4g04090 hypothetical protein
Length = 192
Score = 38.5 bits (88), Expect = 0.005
Identities = 29/116 (25%), Positives = 53/116 (45%), Gaps = 2/116 (1%)
Query: 103 SNRWRSISIFGVPHDAVRVFIRYLYSS-CYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQ 161
S++ +I++ + H+ + F+ + YS E+ K+ + L + Y +P L C +
Sbjct: 64 SSKLETITLSEMKHEVLEAFVDFTYSDGSMLSEKAKQHAMSLYSAAKDYEIPRLWCLCRK 123
Query: 162 KLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHP 217
+L + L M N + V QLA + LS+ I + +S S K +HP
Sbjct: 124 EL-IASLNMSNALRVLQLAQIPYDESLSVAAFTTIKNSKLKLSSSTKVKVFVVNHP 178
>At3g06190 unknown protein
Length = 406
Score = 38.5 bits (88), Expect = 0.005
Identities = 27/86 (31%), Positives = 45/86 (51%), Gaps = 3/86 (3%)
Query: 142 HLLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKIL--KN 199
HLL + YA+ L+ CE KL G+ +++ + LA +L C + I +N
Sbjct: 286 HLLAAADRYALERLRTICESKLCEGI-SINTVATTLALAEQHHCFQLKAACLKFIALPEN 344
Query: 200 FRTVSESEGWKAMKQSHPVLEKEILE 225
+ V E++G+ +K+S P L E+LE
Sbjct: 345 LKAVMETDGFDYLKESCPSLLSELLE 370
>At5g19000 unknown protein
Length = 407
Score = 37.7 bits (86), Expect = 0.009
Identities = 25/86 (29%), Positives = 45/86 (52%), Gaps = 3/86 (3%)
Query: 142 HLLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKIL--KN 199
HLL + YA+ LK CE KL G + ++ + LA +L +C + + +N
Sbjct: 287 HLLAAADRYALERLKAICESKLCEG-VAINTVATTLALAEQHHCLQLKAVCLKFVALPEN 345
Query: 200 FRTVSESEGWKAMKQSHPVLEKEILE 225
+ V +++G+ +K+S P L E+L+
Sbjct: 346 LKAVMQTDGFDYLKESCPSLLTELLQ 371
>At5g13060 unknown protein
Length = 709
Score = 37.4 bits (85), Expect = 0.012
Identities = 32/124 (25%), Positives = 60/124 (47%), Gaps = 10/124 (8%)
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
+ K+ N N + I + + + ++++YS K LLV + Y + L
Sbjct: 572 LYKERNAQN----VEIPNIRWEVFELMMKFIYSG--RINIAKHLAKDLLVAADQYLLEGL 625
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGW--KAMK 213
KR+CE + + +DN+ ++++LA +A L C +L++F +S S+ W K +K
Sbjct: 626 KRQCEYTIAQE-ICLDNIPEMYELADTFNASALRRACTLFVLEHFTKLS-SQLWFAKFVK 683
Query: 214 QSHP 217
Q P
Sbjct: 684 QIIP 687
>At3g03740 unknown protein
Length = 465
Score = 36.6 bits (83), Expect = 0.020
Identities = 22/85 (25%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Query: 143 LLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRT 202
LL + Y +P L CE L + ++D++ ++ LA +A L +C + +N
Sbjct: 304 LLGAADKYKLPRLSLMCESVLCKDI-SVDSVANILALADRYNASALKSVCLKFAAENLIA 362
Query: 203 VSESEGWKAMKQSHPVLEKEILESM 227
V S+G+ +++ P L+ E+L+++
Sbjct: 363 VMRSDGFDYLREHCPSLQSELLKTV 387
>At2g30600 unknown protein
Length = 586
Score = 33.1 bits (74), Expect = 0.22
Identities = 25/97 (25%), Positives = 48/97 (48%), Gaps = 5/97 (5%)
Query: 108 SISIFGVPHDAVRVFIRYLYSSCYEKEEMKEF---VLHLLVLSHVYAVPHLKRECEQKLE 164
+I + V +A + + ++YS E+ F ++HLL L+ + V L +EC K+
Sbjct: 393 TIYLTDVSPEAFKAMMNFMYSGELNMEDTVNFGTELIHLLFLADRFGVVPLHQEC-CKML 451
Query: 165 LGLLTMDNLVDVFQ-LALLCDAPRLSLICHRKILKNF 200
L L+ D++ V Q ++ + + +C RK +F
Sbjct: 452 LECLSEDSVCSVLQVVSSISSCKLIEEMCKRKFSMHF 488
>At5g09300 branched-chain alpha keto-acid dehydrogenase E1 alpha
subunit-like protein
Length = 472
Score = 32.7 bits (73), Expect = 0.29
Identities = 27/97 (27%), Positives = 45/97 (45%), Gaps = 21/97 (21%)
Query: 194 RKILKNFRTVSESEGWKAMKQSHPV---LEKEILESMIDEENSKKERIRKMNEREVYLQL 250
R L FRT ES GW + K + ++KE+LE++ E ++K ++ M + +
Sbjct: 386 RNPLSRFRTWIESNGWWSDKTESDLRSRIKKEMLEALRVAEKTEKPNLQNM-----FSDV 440
Query: 251 YDA-------MEALVHICRDGCRTIGPHDKDFKANQP 280
YD E LV +TI H +D+ ++ P
Sbjct: 441 YDVPPSNLREQELLVR------QTINSHPQDYPSDVP 471
>At1g01640 hypothetical protein
Length = 207
Score = 32.3 bits (72), Expect = 0.37
Identities = 28/127 (22%), Positives = 57/127 (44%), Gaps = 3/127 (2%)
Query: 108 SISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKLELGL 167
SI++ + HD ++ + +LYS + + L+L + Y + +L+ C +
Sbjct: 67 SITLPDLSHDELKSLLEFLYSGNLKAPYNQYRSLYL--AADKYDISYLQDVCRNHF-IAS 123
Query: 168 LTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILESM 227
L+ N++D+ +LA + L I+K+ V ++ Q +P L EI +
Sbjct: 124 LSSRNVLDILELASIPCDTILKDAAINHIVKHMEEVVVPMKYETFVQRNPDLSVEITRAY 183
Query: 228 IDEENSK 234
+ E +K
Sbjct: 184 LRETKAK 190
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.342 0.149 0.512
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,188,420
Number of Sequences: 26719
Number of extensions: 287919
Number of successful extensions: 1365
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 1331
Number of HSP's gapped (non-prelim): 40
length of query: 335
length of database: 11,318,596
effective HSP length: 100
effective length of query: 235
effective length of database: 8,646,696
effective search space: 2031973560
effective search space used: 2031973560
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146856.6


