

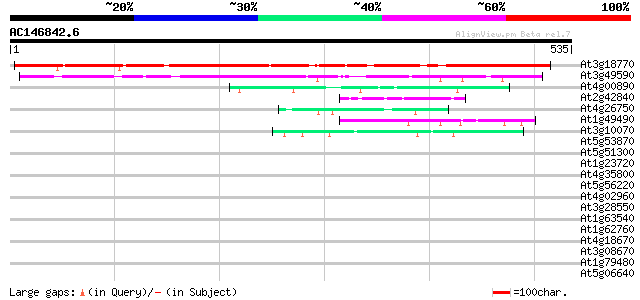
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146842.6 - phase: 0
(535 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g18770 unknown protein 451 e-127
At3g49590 unknown protein 232 4e-61
At4g00890 49 5e-06
At2g42840 En/Spm-like transposon protein 44 3e-04
At4g26750 unknown protein 43 4e-04
At1g49490 hypothetical protein 43 4e-04
At3g10070 unknown protein 43 5e-04
At5g53870 predicted GPI-anchored protein 42 0.001
At5g51300 unknown protein 41 0.002
At1g23720 unknown protein 40 0.002
At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain 40 0.003
At5g56220 unknown protein 39 0.006
At4g02960 putative polyprotein of LTR transposon 39 0.009
At3g28550 unknown protein 38 0.012
At1g63540 unknown protein 38 0.012
At1g62760 hypothetical protein 38 0.012
At4g18670 extensin-like protein 38 0.016
At3g08670 hypothetical protein 37 0.021
At1g79480 hypothetical protein 37 0.021
At5g06640 putative protein 37 0.027
>At3g18770 unknown protein
Length = 625
Score = 451 bits (1160), Expect = e-127
Identities = 272/529 (51%), Positives = 355/529 (66%), Gaps = 50/529 (9%)
Query: 5 SSHAQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFS---TLSSPSSTSSSSSSSVR 61
S++ +E AK EQII EFFAKSLHII+ESR SSRNFS + SPSS SSSSSSSVR
Sbjct: 8 SNNNNSEGAKAEQIIFEFFAKSLHIILESRTPFMSSRNFSGEQMICSPSS-SSSSSSSVR 66
Query: 62 PRDKWFNLALRECPTALENTDLWRHSNLQPIVVDVVLVHRNL------------SFSPKV 109
PRDKWFNLALRECP ALE+ D+ R S+L+P+VVDVVLV R L +FS K
Sbjct: 67 PRDKWFNLALRECPAALESFDIGRRSSLEPLVVDVVLVVRPLVGDQSGKRELIRNFSGK- 125
Query: 110 RSFVKERNPFEEFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLY 169
+ N ++ G +NE+I+ERWV+QY+++KI++ + ++RRSS+ LQ +Y
Sbjct: 126 -DYQSGWNSDQDELGCETKNEQIIERWVVQYDNRKIRE----SVTTSSRRSSSNKLQVMY 180
Query: 170 KKSTLLLRSLYATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFV 229
KK+TLLLRSL+ VRLLPA+KIF+EL+SS + F L RV + EPFT K+EAEM KF
Sbjct: 181 KKATLLLRSLFVMVRLLPAYKIFRELNSSGQIFKFKLVPRVPSIVEPFTRKEEAEMQKFS 240
Query: 230 FTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRM 289
FTPV+T G+LCLSV+YR +SDVS E +TP+SP ITDYVGSPL D L RFPSLP+
Sbjct: 241 FTPVETICGRLCLSVLYR-SLSDVSCEHSTPMSPTFITDYVGSPLADPLKRFPSLPL--- 296
Query: 290 PSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSL 349
S+G SP LPF RRHSWS++ +ASPP+V+ SPSPT S+SH LVS+ S+ L
Sbjct: 297 -SYG-SPPLLPFQRRHSWSFDRYKASPPSVS-CSPSPTRSDSHALVSHPCSR------HL 347
Query: 350 PPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIP 409
PPHPS++ +K +EY P +FSP PSPS+ G + R+ESAPV IP
Sbjct: 348 PPHPSDIPTGRRKESYPEEYSPCQDFSPPPSPSAPKHAVPRG------ITRTESAPVRIP 401
Query: 410 NAEVTYSPGHTSRQN-LPPSTPIRISRCTS-ETERSRNLMQSCTTPEKMFSIGKES-QKY 466
+P S++N + PS +++SR S + R+ +S +K+F G++ ++
Sbjct: 402 ------APTFQSKENVVAPSAHLKLSRHASLKPVRNLGPGESGAAIDKLFLYGRDDFRRP 455
Query: 467 SGGKIAPNSSPQISFSRSSSRSYQDEFDDTDFTCPFDVDDDDTTDPGSR 515
SG + + +SSP+ISFSRSSSRS+QD+FDD DF CPFDV+ DD TD SR
Sbjct: 456 SGVRPSSSSSPRISFSRSSSRSFQDDFDDPDFPCPFDVEYDDITDRNSR 504
>At3g49590 unknown protein
Length = 603
Score = 232 bits (591), Expect = 4e-61
Identities = 180/531 (33%), Positives = 259/531 (47%), Gaps = 80/531 (15%)
Query: 10 TEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNL 69
++ + EQI++ FF K+LHI++ SR S SR + S +VR DKWFNL
Sbjct: 9 SDIGRLEQIVSHFFPKALHIVLNSRIPSLQSRG-------RTRERLSGLNVRKSDKWFNL 61
Query: 70 ALRECPTALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKVRSFVKERNPFEEFGGGSEQN 129
+ + P ALE W + L +++D++LVH P + + + + S
Sbjct: 62 VMGDRPAALEKLHSWHRNILDSMIIDIILVH------PISNDNLDDDDDHSDSVVRSA-- 113
Query: 130 EKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLLPAF 189
E ++ERWV+QYE+ I SS++A T Q +YKKS +LLRSLYA RLLPA+
Sbjct: 114 ETVIERWVVQYENPLIMSPQSSDSA--------TRYQKVYKKSIILLRSLYAQTRLLPAY 165
Query: 190 KIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFVFTPVDTNSGKLCLSVMYRPC 249
++ ++LSSS S + L ++VS+F + F+ M +F F PV+ G+LC SV YR
Sbjct: 166 RVSRQLSSSLASSGYDLIYKVSSFSDIFSGPVTETMKEFRFAPVEVPPGRLCASVTYRSD 225
Query: 250 VSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSH------GSSP-SSLPFS 302
+SD + L P++ITDYVGSP TD + FPS P + H G P +
Sbjct: 226 LSDFNLGAHITLPPRIITDYVGSPATDPMRFFPS-PGRSVEGHSFTGRAGRPPLTGSSAE 284
Query: 303 RRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKK 362
R HSW+ R PPA + +P +Q + P S P +
Sbjct: 285 RPHSWTSGFHR--PPA-QFATP---------------NQSFSPAQSHQLSPGLHDFHWSR 326
Query: 363 NVNF-DEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIP-----NAEVTYS 416
F D + SP FSPS SP S+P Y G + + +R +APV IP N V+ +
Sbjct: 327 TDAFGDNHQLSPPFSPSGSP--STPRYISGGNSPRINVRPGTAPVTIPSSATLNRYVSSN 384
Query: 417 PGHTSRQNLPPSTP---------------IRISRCTSETERSRNLMQSCTTPEKMFSIGK 461
R LPP +P I + R + E LM T + + K
Sbjct: 385 FSEPGRNPLPPFSPKSTRRSPSSQDSLPGIALYRSSRSGESPSGLMNQYPTQK----LSK 440
Query: 462 ESQKYSG---GKIAPNSSPQISFSRSSSR-SYQDEFDDTDFTCPFDVDDDD 508
+S+ SG G ++ + SP+ +FSRS SR S QD+ DD D +CPFD DD D
Sbjct: 441 DSKYDSGRFSGVLSSSDSPRFAFSRSPSRLSSQDDLDDPDCSCPFDFDDVD 491
>At4g00890
Length = 431
Score = 49.3 bits (116), Expect = 5e-06
Identities = 63/276 (22%), Positives = 105/276 (37%), Gaps = 25/276 (9%)
Query: 210 VSTFFEPF--TTKQEAEMLKFVFTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVIT 267
V T+F P T+ + + + P M + S+P PL P
Sbjct: 152 VETWFRPSPPTSTETGDETQLPIPPPQEAKTPPSSPSMMLNATEEFESQPKPPLLPSKSI 211
Query: 268 DY--VGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPS 325
D + SPL + P LP + + + S P S PP + + S
Sbjct: 212 DETRLRSPLMSQASSPPPLPSKSIDENETRSQSPPIS-------------PPKSDKQARS 258
Query: 326 PTHSESHT--LVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSS 383
THS L+S +S+ + S +PP PS + + ++ PSP +P P
Sbjct: 259 QTHSSPSPPPLLSPKASENHQSKSPMPP-PSPTAQISLSSLKSP--IPSPATITAPPPPF 315
Query: 384 SSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTS-RQNL--PPSTPIRISRCTSET 440
SSP+ +L + E + P+ ++T + H S QNL P + + +E
Sbjct: 316 SSPLSQTTPSPKPSLPQIEPNQIKSPSPKLTNTESHASPEQNLVKPDANLMNKIPAKNEV 375
Query: 441 ERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSS 476
++R ++ T P M I Q ++ +SS
Sbjct: 376 PKNRGTLEKKTEPMIMMFINSNVQGFNTSLSLDSSS 411
>At2g42840 En/Spm-like transposon protein
Length = 306
Score = 43.5 bits (101), Expect = 3e-04
Identities = 36/120 (30%), Positives = 58/120 (48%), Gaps = 9/120 (7%)
Query: 315 SPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPN 374
SPP+ ++ +P P+H+ SN S Y P S P HPS S H + + P+P+
Sbjct: 39 SPPSGSHGTP-PSHTPPS---SNCGSPPYDPSPSTPSHPSPPS--HTPTPSTPSHTPTPH 92
Query: 375 FSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRIS 434
+PS +P+ +P + GS S S + + P S G+ S + PP TP+ ++
Sbjct: 93 -TPSHTPTPHTPPCNCGSPPSHPSTPSHPSTPSHPTPSHPPSGGYYS--SPPPRTPVVVT 149
Score = 29.6 bits (65), Expect = 4.4
Identities = 32/110 (29%), Positives = 43/110 (39%), Gaps = 11/110 (10%)
Query: 250 VSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSS-----LPFSRR 304
+S S TP S + GSP D PS P PSH +PS+ P +
Sbjct: 38 LSPPSGSHGTPPSHTPPSSNCGSPPYDPSPSTPSHP--SPPSHTPTPSTPSHTPTPHTPS 95
Query: 305 HSWSYEN--CRASPPAVNYYSPSPTHSESHTLVSNASSQGY--CPPSSLP 350
H+ + C P + +PS + SH S+ S GY PP P
Sbjct: 96 HTPTPHTPPCNCGSPPSHPSTPSHPSTPSHPTPSHPPSGGYYSSPPPRTP 145
>At4g26750 unknown protein
Length = 421
Score = 43.1 bits (100), Expect = 4e-04
Identities = 44/176 (25%), Positives = 69/176 (39%), Gaps = 25/176 (14%)
Query: 257 PTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHG-----SSPSSLPFSRRHS---WS 308
P PL P+ + +P D P P PS+ + PS P+ +S +
Sbjct: 211 PQHPLPPR----FYDNPTNDYPADVPPPPPSSYPSNDHLPPPTGPSDSPYPHPYSHQPYH 266
Query: 309 YENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDE 368
+ + PP NY S P+ + S S PS+ P +PS N +
Sbjct: 267 QDPPKHMPPPQNYSSHEPSPNSLPNFQSYPSFSESSLPSTSPHYPSHYQ-------NPEP 319
Query: 369 YYPSPNFSPSPSPSSSS------PIYSLGSLASKTLLRSESAPVNIPNAEVTYSPG 418
YY SP+ +P+PS +S S P S G + +L + + + +Y PG
Sbjct: 320 YYSSPHSAPAPSSTSFSSAPPPPPYSSNGRINIAPVLDPAPSSAQKYHYDSSYQPG 375
>At1g49490 hypothetical protein
Length = 847
Score = 43.1 bits (100), Expect = 4e-04
Identities = 56/208 (26%), Positives = 86/208 (40%), Gaps = 23/208 (11%)
Query: 315 SPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDE-YYPSP 373
SPP Y P PT S T +N G P+ P SE + + + D+ SP
Sbjct: 624 SPPPPVYSPPPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILSP 683
Query: 374 NFSPSP---SPSSSSPIYSLGSLASKTLLRSESAPVNIP--------NAEVTYSPGHTSR 422
+P+P S SS P +S++ +PV P ++E + P +S
Sbjct: 684 VQAPTPVQSSTPSSEPTQVPTPSSSESYQAPNLSPVQAPTPVQAPTTSSETSQVPTPSSE 743
Query: 423 QNLPPS---TPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYSGGK---IAPNSS 476
N PS TPI + + T S+ +QS T + S ++S++ + + P+S
Sbjct: 744 SNQSPSQAPTPI-LEPVHAPTPNSKP-VQSPTPSSEPVSSPEQSEEVEAPEPTPVNPSSV 801
Query: 477 PQISFSRSSS---RSYQDEFDDTDFTCP 501
P S S +S D+ DD DF P
Sbjct: 802 PSSSPSTDTSIPPPENNDDDDDGDFVLP 829
Score = 35.4 bits (80), Expect = 0.080
Identities = 64/263 (24%), Positives = 88/263 (33%), Gaps = 70/263 (26%)
Query: 256 EPTTPLSPQVITD-------YVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWS 308
EPT P+SP Y SP+ +R R P P V +P
Sbjct: 492 EPTKPVSPPNEAQGPTPDDPYDASPVKNR--RSPPPPKVEDTRVPPPQPPMP-------- 541
Query: 309 YENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDE 368
SPP+ Y P P HS + S+ PP PPH
Sbjct: 542 ----SPSPPSPIYSPPPPVHSPPPPVYSS-------PP---PPHV--------------- 572
Query: 369 YYPSPNFSPSPSPSSSSPIYS------------LGSLASKTLLRSESAPVNIPNAEV--- 413
Y P P + P PS P++S + S + + S P + P V
Sbjct: 573 YSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSP 632
Query: 414 ---TYSPGHTSRQNLPP---STPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYS 467
T+SP T N PP TP + +SET + + + + + S
Sbjct: 633 PPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILSPVQAPTPVQS 692
Query: 468 GGKIAPNSSPQISFSRSSSRSYQ 490
P+S P + SSS SYQ
Sbjct: 693 S---TPSSEPTQVPTPSSSESYQ 712
Score = 29.6 bits (65), Expect = 4.4
Identities = 47/180 (26%), Positives = 58/180 (32%), Gaps = 13/180 (7%)
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
SS+P TP +P+ + P + +P SP P S E +
Sbjct: 419 SSKPETPKTPEQPSPKPQPPKHESPKPEEPENKHELPKQKESPKPQPSKPEDSPKPEQPK 478
Query: 314 A--SPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYP 371
SP P PT S +QG P P + D P
Sbjct: 479 PEESPKPEQPQIPEPTKPVSPP----NEAQGPTPDDPYDASPVKNRRSPPPPKVEDTRVP 534
Query: 372 SPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPV-NIPNAEVTYSPGHTSRQNLPPSTP 430
P P PSPS SPIYS + S PV + P YSP PPS P
Sbjct: 535 PPQ-PPMPSPSPPSPIYS-----PPPPVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSPP 588
>At3g10070 unknown protein
Length = 539
Score = 42.7 bits (99), Expect = 5e-04
Identities = 65/269 (24%), Positives = 104/269 (38%), Gaps = 31/269 (11%)
Query: 251 SDVSSEPTTP-----LSPQVITDYVGSPLTDR-----LMRFPSLPVVRMPSHGSSPSSLP 300
S S P TP P +T +P T+ + PS P + PS +P+
Sbjct: 8 STASQPPETPPQPSDSKPSTLTQIQPTPSTNPSPSSVVSSIPSSPAPQSPSLNPNPNPPQ 67
Query: 301 FSR-------RHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHP 353
++R + PP Y P HS S+T S+ASS S+
Sbjct: 68 YTRPVTSPATQQQQHLSQPLVRPPPQAYSRPWQQHS-SYTHFSSASSPLLSSSSAPASSS 126
Query: 354 SEMSLLHKKNVNFDEYYP-SPNFSPSPSPSSSSPI-------YSLGSLASKTLLRSESAP 405
S + + ++ P SP SPSP+PS SP G L T+ SE A
Sbjct: 127 SSLPISGQQRGGMAIGVPASPIPSPSPTPSQHSPSAFPGSFGQQYGGLGRGTVGMSE-AT 185
Query: 406 VNIPNAEVTYSPGHTS---RQNLPPSTPIRISRCTSETER-SRNLMQSCTTPEKMFSIGK 461
N + +V G L + IR S T +R +++ ++ ++ + +
Sbjct: 186 SNTSSPQVRMMQGTQGIGMMGTLGSGSQIRPSGMTQHQQRPTQSSLRPASSTSTQSPVAQ 245
Query: 462 ESQKYSGGKIAPNSSPQISFSRSSSRSYQ 490
Q +S + +P SSP + + +S +S Q
Sbjct: 246 NFQGHSLMRPSPISSPNVQSTGASQQSLQ 274
>At5g53870 predicted GPI-anchored protein
Length = 370
Score = 41.6 bits (96), Expect = 0.001
Identities = 48/150 (32%), Positives = 59/150 (39%), Gaps = 27/150 (18%)
Query: 248 PCVSDVSSEP---TTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLP-FSR 303
P SS P T LSP T + SP T PV PSH SP+ P S
Sbjct: 178 PAQPPKSSSPISHTPALSPSHATSH--SPATPSPSPKSPSPVSHSPSH--SPAHTPSHSP 233
Query: 304 RHSWSYENCRA---SPPAVNYYSP------SPTHSESHTLVSNASSQGYCPPSSLPPHPS 354
H+ S+ A SP +SP SP HS SH+ + S P+ P PS
Sbjct: 234 AHTPSHSPAHAPSHSPAHAPSHSPAHAPSHSPAHSPSHSPATPKSPSPSSSPAQSPATPS 293
Query: 355 EMSLLHKKNVNFDEYYPSPNFSPSPSPSSS 384
M+ PSP SPSP S++
Sbjct: 294 PMT----------PQSPSPVSSPSPDQSAA 313
Score = 28.5 bits (62), Expect = 9.8
Identities = 25/88 (28%), Positives = 38/88 (42%), Gaps = 8/88 (9%)
Query: 346 PSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLA-SKTLLRSESA 404
P+ P S +S + + P + SP+ P SSSPI +L+ S S +
Sbjct: 147 PTQPPKSHSPVSPVAPASAPSKSQPPRSSVSPAQPPKSSSPISHTPALSPSHATSHSPAT 206
Query: 405 PVNIPN--AEVTYSPGHTSRQNLPPSTP 430
P P + V++SP H+ P TP
Sbjct: 207 PSPSPKSPSPVSHSPSHS-----PAHTP 229
>At5g51300 unknown protein
Length = 804
Score = 40.8 bits (94), Expect = 0.002
Identities = 54/222 (24%), Positives = 81/222 (36%), Gaps = 37/222 (16%)
Query: 282 PSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQ 341
P P + P+ G PS+ P S Y S V + P P+
Sbjct: 566 PGPPAPQPPTQGYPPSNQPPGAYPSQQYATGGYSTAPVPWGPPVPS-------------- 611
Query: 342 GYCPPSSLPPHPSEMSLLHKKNV-NFDEYYPSP----NFSPSP----SPSSSSPIYSL-G 391
Y P + PP P +H +++ + YP P +P P +PSSS P S
Sbjct: 612 -YSPYALPPPPPGSYHPVHGQHMPPYGMQYPPPPPHVTQAPPPGTTQNPSSSEPQQSFPP 670
Query: 392 SLASKTLLRSESAPVNIPNAEVTYSPGHTSRQN-------LPPSTPIRISRCTSETERSR 444
+ + + + S P N+ + VT PG + +PP TP S T+ S+
Sbjct: 671 GVQADSGAATSSIPPNVYGSSVTAMPGQPPYMSYPSYYNAVPPPTP---PAPASSTDHSQ 727
Query: 445 NLMQSCTTPEKMFSIGKESQKYSGGKIAPN--SSPQISFSRS 484
N+ S SQ APN P + +S+S
Sbjct: 728 NMGNMPWANNPSVSTPDHSQGLVNAPWAPNPPMPPTVGYSQS 769
>At1g23720 unknown protein
Length = 895
Score = 40.4 bits (93), Expect = 0.002
Identities = 39/122 (31%), Positives = 48/122 (38%), Gaps = 13/122 (10%)
Query: 315 SPPAVNYYS--PSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY-- 370
SPP YS P P +S S L +S Y S PP+ S + K+ Y
Sbjct: 431 SPPPPYVYSSPPPPYYSPSPKLTYKSSPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSS 490
Query: 371 -PSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTY-SPGHTSRQNLPPS 428
P P +SPSP PS SP + S P P+ +V Y SP H PP
Sbjct: 491 PPPPYYSPSPKPSYKSP-------PPPYVYNSPPPPYYSPSPKVIYKSPPHPHVCVCPPP 543
Query: 429 TP 430
P
Sbjct: 544 PP 545
Score = 35.0 bits (79), Expect = 0.10
Identities = 39/137 (28%), Positives = 52/137 (37%), Gaps = 21/137 (15%)
Query: 255 SEPTTPL--SPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENC 312
S P PL SP DY + P P V + S P L +S Y++
Sbjct: 12 SSPPPPLYDSPTPKVDY----------KSPPPPYV----YSSPPPPLSYSPSPKVDYKS- 56
Query: 313 RASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY-- 370
PP V P P +S S + + Y S PP+ S + K+ Y
Sbjct: 57 -PPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSS 115
Query: 371 -PSPNFSPSPSPSSSSP 386
P P +SPSP P+ SP
Sbjct: 116 PPPPYYSPSPKPTYKSP 132
Score = 33.5 bits (75), Expect = 0.30
Identities = 26/75 (34%), Positives = 32/75 (42%), Gaps = 9/75 (12%)
Query: 315 SPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---P 371
SPP YYSPSP + +S Y S PP+ S K+ Y P
Sbjct: 566 SPPPPPYYSPSPKPA------YKSSPPPYVYSSPPPPYYSPAPKPVYKSPPPPYVYNSPP 619
Query: 372 SPNFSPSPSPSSSSP 386
P +SPSP P+ SP
Sbjct: 620 PPYYSPSPKPTYKSP 634
Score = 33.1 bits (74), Expect = 0.40
Identities = 36/120 (30%), Positives = 43/120 (35%), Gaps = 14/120 (11%)
Query: 272 SPLTDRLMRFPSLPVVRM--PSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPT-- 327
SP + + P P V P SPS P + Y PP YYSPSP
Sbjct: 222 SPSPKPVYKSPPPPYVYSSPPPPYYSPSPKPAYKSPPPPYVYSSPPPP---YYSPSPKPI 278
Query: 328 -HSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSP 386
S V N+ Y PS P + S + P P +SPSP P SP
Sbjct: 279 YKSPPPPYVYNSPPPPYYSPSPKPAYKSPPPPY------VYSFPPPPYYSPSPKPVYKSP 332
Score = 32.7 bits (73), Expect = 0.52
Identities = 43/153 (28%), Positives = 55/153 (35%), Gaps = 29/153 (18%)
Query: 272 SPLTDRLMRFPSLPVV--RMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPT-- 327
SP + + P P V P SPS P + Y PP YYSPSP
Sbjct: 322 SPSPKPVYKSPPPPYVYNSPPPPYYSPSPKPAYKSPPPPYVYSSPPPP---YYSPSPKPT 378
Query: 328 -HSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSP 386
S V ++ Y PS P + S + P P +SPSP PS SP
Sbjct: 379 YKSPPPPYVYSSPPPPYYSPSPKPVYKSPPPPYIYNSP------PPPYYSPSPKPSYKSP 432
Query: 387 ----IYSLGSLASKTLLRSESAPVNIPNAEVTY 415
+YS S P P+ ++TY
Sbjct: 433 PPPYVYS-----------SPPPPYYSPSPKLTY 454
Score = 32.3 bits (72), Expect = 0.68
Identities = 37/130 (28%), Positives = 47/130 (35%), Gaps = 22/130 (16%)
Query: 314 ASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY--- 370
+SPP YYSPSP + + Y S PP+ S + K+ Y
Sbjct: 741 SSPPPPPYYSPSP------KVEYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSP 794
Query: 371 -PSPNFSPSPSPSSSSP----IYS------LGSLASKTLLRSESAP--VNIPNAEVTYSP 417
P P +SPSP SP +YS S + K +S P N P YSP
Sbjct: 795 PPPPYYSPSPKVEYKSPPPPYVYSSPPPPTYYSPSPKVEYKSPPPPYVYNSPPPPAYYSP 854
Query: 418 GHTSRQNLPP 427
PP
Sbjct: 855 SPKIEYKSPP 864
Score = 32.3 bits (72), Expect = 0.68
Identities = 24/74 (32%), Positives = 31/74 (41%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V + P P +S S V + Y S PP+ S K+ Y P
Sbjct: 309 PPYVYSFPPPPYYSPSPKPVYKSPPPPYVYNSPPPPYYSPSPKPAYKSPPPPYVYSSPPP 368
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP P+ SP
Sbjct: 369 PYYSPSPKPTYKSP 382
Score = 32.0 bits (71), Expect = 0.88
Identities = 43/145 (29%), Positives = 54/145 (36%), Gaps = 13/145 (8%)
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVR---MPSHGSSPS----SLPFSRRHS 306
SS P SP DY SP + P P P++ S P S P +S
Sbjct: 164 SSPPPPYYSPSPKVDYK-SPPPPYVYNSPPPPYYSPSPKPTYKSPPPPYIYSSPPPPYYS 222
Query: 307 WSYENCRASPPAVNYYS--PSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNV 364
S + SPP YS P P +S S + Y S PP+ S K+
Sbjct: 223 PSPKPVYKSPPPPYVYSSPPPPYYSPSPKPAYKSPPPPYVYSSPPPPYYSPSPKPIYKSP 282
Query: 365 NFDEYY---PSPNFSPSPSPSSSSP 386
Y P P +SPSP P+ SP
Sbjct: 283 PPPYVYNSPPPPYYSPSPKPAYKSP 307
Score = 31.2 bits (69), Expect = 1.5
Identities = 26/79 (32%), Positives = 33/79 (40%), Gaps = 14/79 (17%)
Query: 314 ASPPAVNYYSPSPT---HSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY 370
+SPP YYSPSP S V N+ PP+ P P + K+ Y
Sbjct: 818 SSPPPPTYYSPSPKVEYKSPPPPYVYNSPP----PPAYYSPSPK----IEYKSPPPPYVY 869
Query: 371 ---PSPNFSPSPSPSSSSP 386
P P++SPSP SP
Sbjct: 870 SSPPPPSYSPSPKAEYKSP 888
Score = 30.8 bits (68), Expect = 2.0
Identities = 63/227 (27%), Positives = 79/227 (34%), Gaps = 45/227 (19%)
Query: 210 VSTFFEPFTTKQEAEMLKFVFTP-VDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITD 268
V+ +EP+T L TP VD S P V S P PLS
Sbjct: 2 VAASYEPYTYSSPPPPLYDSPTPKVDYKS----------PPPPYVYSSPPPPLS------ 45
Query: 269 YVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTH 328
Y SP D + P P V SSP +S Y++ PP V P P +
Sbjct: 46 YSPSPKVD--YKSPPPPYVY-----SSPPPPYYSPSPKVEYKS--PPPPYVYSSPPPPYY 96
Query: 329 SESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PSPNFSPSPSPSSSS 385
S S + + Y S PP+ S K+ Y P P +SPSP S
Sbjct: 97 SPSPKVDYKSPPPPYVYSSPPPPYYSPSPKPTYKSPPPPYVYNSPPPPYYSPSPKVEYKS 156
Query: 386 P----IYSLGSLASKTLLRSESAPVNIPNAEVTY-SPGHTSRQNLPP 427
P +YS S P P+ +V Y SP N PP
Sbjct: 157 PPPPYVYS-----------SPPPPYYSPSPKVDYKSPPPPYVYNSPP 192
Score = 30.0 bits (66), Expect = 3.4
Identities = 38/146 (26%), Positives = 48/146 (32%), Gaps = 14/146 (9%)
Query: 246 YRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRM--PSHGSSPSSLPFSR 303
Y P V P P SP + P P V P SP+ P +
Sbjct: 598 YSPAPKPVYKSPPPPYVYNSPPPPYYSPSPKPTYKSPPPPYVYSSPPPPYYSPTPKPTYK 657
Query: 304 RHSWSYENCRASPPAVNYYSPSPT---HSESHTLVSNASSQGYCPPSSLPPHPSEMSLLH 360
Y PP YYSPSP S V ++ Y P+ P + S
Sbjct: 658 SPPPPYVYSSPPPP---YYSPSPKPTYKSPPPPYVYSSPPPPYYSPAPKPTYKSPPPPYV 714
Query: 361 KKNVNFDEYYPSPNFSPSPSPSSSSP 386
+ P P +SPSP P+ SP
Sbjct: 715 YSSP------PPPYYSPSPKPTYKSP 734
Score = 30.0 bits (66), Expect = 3.4
Identities = 30/106 (28%), Positives = 41/106 (38%), Gaps = 9/106 (8%)
Query: 290 PSHGSSPSSLPFSRRHSWSYENC-----RASPPAVNYYSPSPTH-SESHTLVSNASSQGY 343
P++ SSP +S Y ++ PP Y SP P + S S + Y
Sbjct: 579 PAYKSSPPPYVYSSPPPPYYSPAPKPVYKSPPPPYVYNSPPPPYYSPSPKPTYKSPPPPY 638
Query: 344 CPPSSLPPHPSEMSLLHKKNVNFDEYY---PSPNFSPSPSPSSSSP 386
S PP+ S K+ Y P P +SPSP P+ SP
Sbjct: 639 VYSSPPPPYYSPTPKPTYKSPPPPYVYSSPPPPYYSPSPKPTYKSP 684
>At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain
Length = 1840
Score = 40.0 bits (92), Expect = 0.003
Identities = 49/166 (29%), Positives = 66/166 (39%), Gaps = 19/166 (11%)
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
S PT+P Y SP + P+ P S G SP+S +S
Sbjct: 1670 SYSPTSPSYSPTSPSY--SPTSPSYS--PTSPAYSPTSPGYSPTSPSYSPTSPSYGPTSP 1725
Query: 314 ASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYP-S 372
+ P YSPS +S S+ +S AS P+ P PS + Y P S
Sbjct: 1726 SYNPQSAKYSPSIAYSPSNARLSPASPYSPTSPNYSPTSPS-------YSPTSPSYSPSS 1778
Query: 373 PNFSP-SPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSP 417
P +SP SP S +SP YS + S TL P P++ Y+P
Sbjct: 1779 PTYSPSSPYSSGASPDYSPSAGYSPTL------PGYSPSSTGQYTP 1818
Score = 38.1 bits (87), Expect = 0.012
Identities = 62/211 (29%), Positives = 90/211 (42%), Gaps = 30/211 (14%)
Query: 282 PSLPVVRMPSHGSSPSSLPFS-RRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASS 340
P+ P S G SP+S +S +S + SP + +Y SP++S + S +S
Sbjct: 1561 PTSPGYSPTSPGYSPTSPTYSPSSPGYSPTSPAYSPTSPSYSPTSPSYSPTSPSYS-PTS 1619
Query: 341 QGYCP--PSSLPPHPSEMSLLHKKNVNFDEYYP-SPNFSP-SPSPSSSSPIYSLGSLASK 396
Y P PS P PS + Y P SP +SP SPS S +SP YS S +
Sbjct: 1620 PSYSPTSPSYSPTSPSYSPTSPAYSPTSPAYSPTSPAYSPTSPSYSPTSPSYSPTSPS-- 1677
Query: 397 TLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKM 456
S ++P P + +YSP P +P TS + S T+P
Sbjct: 1678 ---YSPTSPSYSPTSP-SYSP------TSPAYSP------TSPGYSPTSPSYSPTSP--- 1718
Query: 457 FSIGKESQKYSGGKIAPNSSPQISFSRSSSR 487
S G S Y+ + SP I++S S++R
Sbjct: 1719 -SYGPTSPSYN--PQSAKYSPSIAYSPSNAR 1746
>At5g56220 unknown protein
Length = 973
Score = 39.3 bits (90), Expect = 0.006
Identities = 38/116 (32%), Positives = 50/116 (42%), Gaps = 6/116 (5%)
Query: 290 PSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSS- 348
PS S PSS S S Y + RA+ P++ + PSP S + + S Y PPS
Sbjct: 54 PSLVSPPSSAFVSALQS-PYISPRATTPSITTHKPSPPLSYKGSQSDDVPSSSYTPPSDQ 112
Query: 349 ---LPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRS 401
PS+ L + D P +FS P P S S+ S A+ T LRS
Sbjct: 113 YEFSDEQPSDRKLKLSASCTPDPAPPRISFS-FPVPRVSLAKVSVSSPATNTKLRS 167
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 38.5 bits (88), Expect = 0.009
Identities = 64/227 (28%), Positives = 89/227 (39%), Gaps = 49/227 (21%)
Query: 257 PTTPL---SPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
PTTPL +P + P D R PS P + SS S+LP S S S
Sbjct: 758 PTTPLVLPAPPCL-----GPHLDTSPRPPSSPSPLCTTQVSS-SNLPSSSISSPS----S 807
Query: 314 ASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSP 373
+ P A ++ P PT T SN++S P P+P+ PSP
Sbjct: 808 SEPTAPSHNGPQPTAQPHQTQNSNSNS-----PILNNPNPNS---------------PSP 847
Query: 374 NFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPI-R 432
N SP SPI S + + S ++ PN+ S TS LPP P
Sbjct: 848 NSPNQNSPLPQSPI-------SSPHIPTPSTSISEPNSP---SSSSTSTPPLPPVLPAPP 897
Query: 433 ISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYS-GGKIAPNSSPQ 478
I + ++ + + M + I K +QKYS +A NS P+
Sbjct: 898 IIQVNAQAPVNTHSMAT----RAKDGIRKPNQKYSYATSLAANSEPR 940
>At3g28550 unknown protein
Length = 1018
Score = 38.1 bits (87), Expect = 0.012
Identities = 38/128 (29%), Positives = 52/128 (39%), Gaps = 13/128 (10%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S +V + Y S PP+ S + K+ Y P
Sbjct: 515 PPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPP 574
Query: 373 PNFSPSPSPSSSSP----IYS-----LGSLASKTLLRSESAPVNIPNAEVTY-SPGHTSR 422
P +SPSP SP +YS S + K +S +P + P+ +V Y SP H
Sbjct: 575 PYYSPSPKVVYKSPPPPYVYSSPPPPYYSPSPKVYYKSPPSPYHAPSPKVLYKSPPHPHV 634
Query: 423 QNLPPSTP 430
PP P
Sbjct: 635 CVCPPPPP 642
Score = 35.4 bits (80), Expect = 0.080
Identities = 32/100 (32%), Positives = 39/100 (39%), Gaps = 13/100 (13%)
Query: 290 PSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSL 349
P H SP L S H + PP YSPSP +V +S Y S
Sbjct: 616 PYHAPSPKVLYKSPPHP----HVCVCPPPPPCYSPSPK------VVYKSSPPPYVYSSPP 665
Query: 350 PPHPSEMSLLHKKNVNFDEYY---PSPNFSPSPSPSSSSP 386
PP+ S +H K+ Y P P +SPSP SP
Sbjct: 666 PPYHSPSPKVHYKSPPPPYVYSSPPPPYYSPSPKVHYKSP 705
Score = 34.7 bits (78), Expect = 0.14
Identities = 26/78 (33%), Positives = 34/78 (43%), Gaps = 4/78 (5%)
Query: 313 RASPPAVNYYSPSPT-HSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY- 370
++SPP Y SP P HS S + + Y S PP+ S +H K+ Y
Sbjct: 653 KSSPPPYVYSSPPPPYHSPSPKVHYKSPPPPYVYSSPPPPYYSPSPKVHYKSPPPPYVYS 712
Query: 371 --PSPNFSPSPSPSSSSP 386
P P +SPSP SP
Sbjct: 713 SPPPPYYSPSPKVVYKSP 730
Score = 32.7 bits (73), Expect = 0.52
Identities = 52/189 (27%), Positives = 69/189 (35%), Gaps = 20/189 (10%)
Query: 248 PCVSDVSSEPTTPL--SPQVITDYVGSP----LTDRLMRFPSLPVVRMPSHGSSPSSLPF 301
P + DV S P PL SP DY P + + S P P +SP +
Sbjct: 46 PPLPDVYSSPPPPLEYSPAPKVDYKSPPPPYYSPSPKVEYKSPPP---PYVYNSPPPPYY 102
Query: 302 SRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHK 361
S Y++ PP V P P +S S + + Y S PP+ S +
Sbjct: 103 SPSPKVDYKS--PPPPYVYSSPPPPIYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVEY 160
Query: 362 KNVNFDEYYPSPNFSPSPSPS--SSSPIYSLGSLASKTLLRSESAPVNIPNAEVTY-SPG 418
K+ PSP SP PS S SP S + S P P+ +V Y SP
Sbjct: 161 KSP------PSPYVYNSPPPSYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVVYKSPP 214
Query: 419 HTSRQNLPP 427
+ PP
Sbjct: 215 PPYVYSSPP 223
Score = 30.4 bits (67), Expect = 2.6
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S +V + Y S PP+ S + K+ Y P
Sbjct: 832 PPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPP 891
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 892 PYYSPSPKVDYKSP 905
Score = 30.4 bits (67), Expect = 2.6
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S +V + Y S PP+ S + K+ Y P
Sbjct: 290 PPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSPTPKVDYKSPPPPYVYSSPPP 349
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 350 PYYSPSPKVDYKSP 363
Score = 30.0 bits (66), Expect = 3.4
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S +V + Y S PP+ S + K+ Y P
Sbjct: 240 PPYVYSSPPPPYYSPSPKIVYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPP 299
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 300 PYYSPSPKVVYKSP 313
Score = 29.6 bits (65), Expect = 4.4
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S +V + Y S PP+ S + K+ Y P
Sbjct: 807 PPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPP 866
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 867 PYYSPSPKVDYKSP 880
Score = 29.6 bits (65), Expect = 4.4
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S +V + Y S PP+ S + K+ Y P
Sbjct: 440 PPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPP 499
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 500 PYYSPSPKVLYKSP 513
Score = 29.3 bits (64), Expect = 5.7
Identities = 36/136 (26%), Positives = 47/136 (34%), Gaps = 20/136 (14%)
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
SS P SP DY P P SSP +S Y++
Sbjct: 320 SSPPPPYYSPTPKVDYKSPP---------------PPYVYSSPPPPYYSPSPKVDYKS-- 362
Query: 314 ASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY--- 370
PP V P P +S S +V + Y S PP+ + + K+ Y
Sbjct: 363 PPPPYVYSSPPPPYYSPSPKIVYKSPPPPYVYSSPPPPYYTPSPKVVYKSPPPPYVYSSP 422
Query: 371 PSPNFSPSPSPSSSSP 386
P P +SPSP SP
Sbjct: 423 PPPYYSPSPKVDYKSP 438
Score = 29.3 bits (64), Expect = 5.7
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S +V + Y S PP+ S + K+ Y P
Sbjct: 190 PPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPP 249
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 250 PYYSPSPKIVYKSP 263
Score = 28.9 bits (63), Expect = 7.5
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S +V + Y S PP+ S + K+ Y P
Sbjct: 782 PPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPP 841
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 842 PYYSPSPKVVYKSP 855
Score = 28.9 bits (63), Expect = 7.5
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S +V + Y S PP+ S + K+ Y P
Sbjct: 707 PPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPP 766
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 767 PYYSPSPKVVYKSP 780
Score = 28.9 bits (63), Expect = 7.5
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S +V + Y S PP+ S + K+ Y P
Sbjct: 732 PPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPP 791
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 792 PYYSPSPKVVYKSP 805
Score = 28.9 bits (63), Expect = 7.5
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S +V + Y S PP+ S + K+ Y P
Sbjct: 757 PPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPP 816
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 817 PYYSPSPKVVYKSP 830
Score = 28.5 bits (62), Expect = 9.8
Identities = 22/74 (29%), Positives = 30/74 (39%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P ++ S +V + Y S PP+ S + K+ Y P
Sbjct: 390 PPYVYSSPPPPYYTPSPKVVYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPP 449
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 450 PYYSPSPKVVYKSP 463
>At1g63540 unknown protein
Length = 635
Score = 38.1 bits (87), Expect = 0.012
Identities = 37/137 (27%), Positives = 60/137 (43%), Gaps = 14/137 (10%)
Query: 380 SPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGH---TSRQNLPPSTPI----R 432
S S+SSP+ +LG + T+ S S+PV I ++ ++ H T + P +P R
Sbjct: 59 STSTSSPVQALGHDSGATVSTSTSSPVQIFSSPFSFGSAHAAITPVSSGPAPSPTFGEPR 118
Query: 433 ISRCTSETERSRNLMQSCTTP---EKMFSIGKE----SQKYSGGKIAPNSSPQISFSRSS 485
I TS + S S ++P FS G + SG +P SSP++ R +
Sbjct: 119 ILLATSGSGASATSTSSTSSPLHSSSPFSFGSAPAAITSVSSGPAQSPASSPRLWIDRFA 178
Query: 486 SRSYQDEFDDTDFTCPF 502
+ S + + PF
Sbjct: 179 TSSSASATSSSSTSSPF 195
>At1g62760 hypothetical protein
Length = 312
Score = 38.1 bits (87), Expect = 0.012
Identities = 39/123 (31%), Positives = 54/123 (43%), Gaps = 13/123 (10%)
Query: 283 SLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQG 342
S P + SSPSS P S S P +++ SP P S L S + S
Sbjct: 32 SSPSLSPSPPSSSPSSAPPS-----SLSPSSPPPLSLSPSSPPPPPPSSSPLSSLSPSLS 86
Query: 343 YCPPSSLPPH--PSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLR 400
PPSS P PS +S ++ SP+ SP P P SSSP+ SL +S +
Sbjct: 87 PSPPSSSPSSAPPSSLSPSSPPPLSL-----SPS-SPPPPPPSSSPLSSLSPSSSSSTYS 140
Query: 401 SES 403
+++
Sbjct: 141 NQT 143
Score = 33.1 bits (74), Expect = 0.40
Identities = 36/114 (31%), Positives = 46/114 (39%), Gaps = 23/114 (20%)
Query: 323 SPSPTHSESHTLVSNASSQGYCPPSSL-----PPHPSEMSLLHKKNVNFDEYYPSPNFSP 377
SPSP S + ++ S PP SL PP P S L SP+ SP
Sbjct: 37 SPSPPSSSPSSAPPSSLSPSSPPPLSLSPSSPPPPPPSSSPLSSL---------SPSLSP 87
Query: 378 SP--SPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPST 429
SP S SS+P SL + L S S+P P P + +L PS+
Sbjct: 88 SPPSSSPSSAPPSSLSPSSPPPLSLSPSSPPPPP-------PSSSPLSSLSPSS 134
Score = 31.2 bits (69), Expect = 1.5
Identities = 38/129 (29%), Positives = 53/129 (40%), Gaps = 16/129 (12%)
Query: 372 SPNFSPSP--SPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPST 429
SP+ SPSP S SS+P SL + L S S+P P + SP + +L PS
Sbjct: 33 SPSLSPSPPSSSPSSAPPSSLSPSSPPPLSLSPSSP---PPPPPSSSPLSSLSPSLSPSP 89
Query: 430 PIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSY 489
P +S + + + + P S S P+SSP S S SSS S
Sbjct: 90 P-----SSSPSSAPPSSLSPSSPPPLSLS------PSSPPPPPPSSSPLSSLSPSSSSST 138
Query: 490 QDEFDDTDF 498
+ D+
Sbjct: 139 YSNQTNLDY 147
>At4g18670 extensin-like protein
Length = 839
Score = 37.7 bits (86), Expect = 0.016
Identities = 42/134 (31%), Positives = 52/134 (38%), Gaps = 5/134 (3%)
Query: 255 SEPTTPLSPQVITDYVGSPLTDRLMRFP-SLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
S PT+P +P SP T P S P P+ G SP S P + S +
Sbjct: 467 SPPTSPTTPTPGGSPPSSPTTPTPGGSPPSSPTT--PTPGGSPPSSPTTPSPGGSPPSPS 524
Query: 314 ASP-PAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPS 372
SP P + SP T + + S +S P S PP PS N PS
Sbjct: 525 ISPSPPITVPSPPSTPTSPGSPPSPSSPTPSSPIPS-PPTPSTPPTPISPGQNSPPIIPS 583
Query: 373 PNFSPSPSPSSSSP 386
P F+ PSS SP
Sbjct: 584 PPFTGPSPPSSPSP 597
Score = 36.6 bits (83), Expect = 0.036
Identities = 54/182 (29%), Positives = 69/182 (37%), Gaps = 13/182 (7%)
Query: 253 VSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENC 312
V S PTTP SP GSP + + P + V P+ SP P S S +
Sbjct: 410 VPSPPTTP-SPG------GSPPSPSISPSPPITVPSPPTT-PSPGGSPPSPSIVPSPPST 461
Query: 313 RASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSS-LPPHPSEMSLLHKKNVNFDEYYP 371
SP + +PT S + G PPSS P P + P
Sbjct: 462 TPSPGSPPTSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPSPGGSPP 521
Query: 372 SPNFSPSPS---PSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPS 428
SP+ SPSP PS S S GS S + + S+P+ P T + QN PP
Sbjct: 522 SPSISPSPPITVPSPPSTPTSPGSPPSPS-SPTPSSPIPSPPTPSTPPTPISPGQNSPPI 580
Query: 429 TP 430
P
Sbjct: 581 IP 582
Score = 36.2 bits (82), Expect = 0.047
Identities = 53/192 (27%), Positives = 72/192 (36%), Gaps = 46/192 (23%)
Query: 248 PCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSW 307
P S V S P+T SP GSP P+ P P+ G SP S P +
Sbjct: 450 PSPSIVPSPPSTTPSP-------GSP--------PTSPTT--PTPGGSPPSSPTTPTPGG 492
Query: 308 SYENCRASP-----PAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKK 362
S + +P P + +PSP S +S + PP ++P PS +
Sbjct: 493 SPPSSPTTPTPGGSPPSSPTTPSPGGSPPSPSISPS------PPITVPSPPSTPT----- 541
Query: 363 NVNFDEYYPSPNFSPSPS---PSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVT-YSPG 418
SP PSPS PSS P S + +++P IP+ T SP
Sbjct: 542 ---------SPGSPPSPSSPTPSSPIPSPPTPSTPPTPISPGQNSPPIIPSPPFTGPSPP 592
Query: 419 HTSRQNLPPSTP 430
+ LPP P
Sbjct: 593 SSPSPPLPPVIP 604
>At3g08670 hypothetical protein
Length = 567
Score = 37.4 bits (85), Expect = 0.021
Identities = 34/111 (30%), Positives = 48/111 (42%), Gaps = 10/111 (9%)
Query: 377 PSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTY-----SPGHTSRQNLPPSTPI 431
P+ S S + P S +S T RS S+ +N +A V+ SP S + PSTP
Sbjct: 151 PARSSSVTRPSISTSQYSSFTSGRSPSSILNTSSASVSSYIRPSSPSSRSSSSARPSTPT 210
Query: 432 RISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFS 482
R S + + SR S ++ S+ K S P S PQ+S S
Sbjct: 211 RTSSASRSSTPSRIRPGSSSS-----SMDKARPSLSSRPSTPTSRPQLSAS 256
Score = 32.0 bits (71), Expect = 0.88
Identities = 48/187 (25%), Positives = 67/187 (35%), Gaps = 43/187 (22%)
Query: 321 YYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY----PSPNFS 376
Y+S P S S T S ++SQ SS S S+L+ + + Y PS S
Sbjct: 146 YHSSRPARSSSVTRPSISTSQY----SSFTSGRSPSSILNTSSASVSSYIRPSSPSSRSS 201
Query: 377 PSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHT---------------- 420
S PS+ + S ++ + +R S+ ++ A + S +
Sbjct: 202 SSARPSTPTRTSSASRSSTPSRIRPGSSSSSMDKARPSLSSRPSTPTSRPQLSASSPNII 261
Query: 421 -SRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQI 479
SR N PSTP R R S T S SGG+ A N
Sbjct: 262 ASRPNSRPSTPTR--RSPSSTSLS----------------ATSGPTISGGRAASNGRTGP 303
Query: 480 SFSRSSS 486
S SR SS
Sbjct: 304 SLSRPSS 310
Score = 29.3 bits (64), Expect = 5.7
Identities = 16/30 (53%), Positives = 19/30 (63%)
Query: 33 SRALSASSRNFSTLSSPSSTSSSSSSSVRP 62
S L+ SS + S+ PSS SS SSSS RP
Sbjct: 177 SSILNTSSASVSSYIRPSSPSSRSSSSARP 206
>At1g79480 hypothetical protein
Length = 356
Score = 37.4 bits (85), Expect = 0.021
Identities = 47/183 (25%), Positives = 66/183 (35%), Gaps = 39/183 (21%)
Query: 267 TDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSP 326
T G T L + SLP + +P + P + S SY SPP P+P
Sbjct: 32 TQPYGVSTTLTLPPYVSLPPLSVPGNAPPFCINPPNTPPSSSYPGL--SPPPGPITLPNP 89
Query: 327 THSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSP------------- 373
S S N +S P SS P+P + S N N P+P
Sbjct: 90 PDSSS-----NPNSNPNPPESSSNPNPPDSSSNPNSNPNPPVTVPNPPESSSNPNPPDSS 144
Query: 374 ---NFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTP 430
N +P+P SSS+P + PV +PN + S + + P+ P
Sbjct: 145 SNPNSNPNPPESSSNP----------------NPPVTVPNPPESSSNPNPPESSSNPNPP 188
Query: 431 IRI 433
I I
Sbjct: 189 ITI 191
>At5g06640 putative protein
Length = 689
Score = 37.0 bits (84), Expect = 0.027
Identities = 45/155 (29%), Positives = 58/155 (37%), Gaps = 32/155 (20%)
Query: 282 PSLPVVRMPSHG-SSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASS 340
P P PSH SP P + + + SPP +YYSPSP + N +
Sbjct: 57 PQTPHYNSPSHEHKSPKYAPHPKPYVY------ISPPPPSYYSPSPKVNYKSPPPPNVYN 110
Query: 341 QGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PSPNFSPSPSPSSSSP----IYSLGSL 393
PP PP+ S + K+ Y P P +SPSP SP +YS
Sbjct: 111 S---PP---PPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYS---- 160
Query: 394 ASKTLLRSESAPVNIPNAEVTY-SPGHTSRQNLPP 427
S P P+ +V Y SP N PP
Sbjct: 161 -------SPPPPYYSPSPKVEYKSPPPPYVYNSPP 188
Score = 31.6 bits (70), Expect = 1.2
Identities = 25/77 (32%), Positives = 33/77 (42%), Gaps = 5/77 (6%)
Query: 315 SPPAVNYYS--PSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY-- 370
SPP YS P P HS S + + Y S PP+ S ++ K+ Y
Sbjct: 502 SPPPPYVYSSPPPPYHSPSPKVNYKSPPPPYVYSSHPPPYYSPSPKVNYKSPPPPYVYSS 561
Query: 371 -PSPNFSPSPSPSSSSP 386
P P +SPSP + SP
Sbjct: 562 PPPPYYSPSPKVNYKSP 578
Score = 31.2 bits (69), Expect = 1.5
Identities = 24/80 (30%), Positives = 34/80 (42%), Gaps = 4/80 (5%)
Query: 311 NCRASPPAVNYYSPSPT-HSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEY 369
N ++ PP Y SP P +S S + + Y S PP+ S ++ K+
Sbjct: 549 NYKSPPPPYVYSSPPPPYYSPSPKVNYKSPPPPYVYSSPPPPYYSPSPMVDYKSTPPPYV 608
Query: 370 Y---PSPNFSPSPSPSSSSP 386
Y P P +SPSP SP
Sbjct: 609 YSFPPLPYYSPSPKVDYKSP 628
Score = 30.4 bits (67), Expect = 2.6
Identities = 48/182 (26%), Positives = 61/182 (33%), Gaps = 36/182 (19%)
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
SS P SP DY P P SSP +S Y++
Sbjct: 135 SSPPPPYYSPSPKVDYKSPP---------------PPYVYSSPPPPYYSPSPKVEYKS-- 177
Query: 314 ASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY--- 370
PP V P P +S S + + Y S PP+ S + K+ Y
Sbjct: 178 PPPPYVYNSPPPPYYSPSPKIEYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYNSP 237
Query: 371 PSPNFSPSPSPSSSSP----IYSLGSLASKTLLRSESAPVNIPNAEVTY-SPGHTSRQNL 425
P P +SPSP SP +YS S P P+ +V Y SP N
Sbjct: 238 PPPYYSPSPKVDYKSPPPPYVYS-----------SPPPPYFSPSPKVEYKSPPPPYVYNS 286
Query: 426 PP 427
PP
Sbjct: 287 PP 288
Score = 30.4 bits (67), Expect = 2.6
Identities = 36/136 (26%), Positives = 47/136 (34%), Gaps = 20/136 (14%)
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
SS P SP DY P P SSP +S +Y++
Sbjct: 360 SSPPPPYYSPSPKVDYKSPP---------------PPYVYSSPPPQYYSPSPKVAYKS-- 402
Query: 314 ASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY--- 370
PP V P P +S S + + Y S PP+ S + K+ Y
Sbjct: 403 PPPPYVYSSPPPPYYSPSPKVAYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSP 462
Query: 371 PSPNFSPSPSPSSSSP 386
P P +SPSP SP
Sbjct: 463 PPPYYSPSPKVEYKSP 478
Score = 30.4 bits (67), Expect = 2.6
Identities = 28/96 (29%), Positives = 38/96 (39%), Gaps = 5/96 (5%)
Query: 294 SSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHP 353
SSP FS Y++ PP V P P +S S + + Y S PP+
Sbjct: 260 SSPPPPYFSPSPKVEYKS--PPPPYVYNSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPYY 317
Query: 354 SEMSLLHKKNVNFDEYY---PSPNFSPSPSPSSSSP 386
S ++ K+ Y P P +SPSP SP
Sbjct: 318 SPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVDYKSP 353
Score = 30.0 bits (66), Expect = 3.4
Identities = 34/125 (27%), Positives = 47/125 (37%), Gaps = 23/125 (18%)
Query: 315 SPPAVNYYS--PSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY-- 370
SPP YS P P +S S + + Y S PP+ S + K+ Y
Sbjct: 427 SPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSS 486
Query: 371 -PSPNFSPSPSPSSSSP----IYSLGSLASKTLLRSESAPVNIPNAEVTYS---PGHTSR 422
P P +SPSP SP +YS S P + P+ +V Y P +
Sbjct: 487 PPPPYYSPSPKVEYKSPPPPYVYS-----------SPPPPYHSPSPKVNYKSPPPPYVYS 535
Query: 423 QNLPP 427
+ PP
Sbjct: 536 SHPPP 540
Score = 30.0 bits (66), Expect = 3.4
Identities = 40/134 (29%), Positives = 49/134 (35%), Gaps = 41/134 (30%)
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
SS P SP + DY +P + FP LP PS S P +S
Sbjct: 585 SSPPPPYYSPSPMVDYKSTP-PPYVYSFPPLPYYS-PSPKVDYKSPPLPYVYS------- 635
Query: 314 ASPPAVNYYSPSP-THSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPS 372
SPP + YYSPSP H +S PP PP+ P
Sbjct: 636 -SPPPL-YYSPSPKVHYKS-------------PP---PPYVYNSP-------------PP 664
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP + SP
Sbjct: 665 PYYSPSPKVTYKSP 678
Score = 30.0 bits (66), Expect = 3.4
Identities = 25/84 (29%), Positives = 35/84 (40%), Gaps = 5/84 (5%)
Query: 306 SWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYC---PPSSLPPHPSEMSLLHKK 362
S+ Y + +SP +Y SPS H +S + Y PPS P P +++
Sbjct: 47 SYPYSSPYSSPQTPHYNSPSHEH-KSPKYAPHPKPYVYISPPPPSYYSPSP-KVNYKSPP 104
Query: 363 NVNFDEYYPSPNFSPSPSPSSSSP 386
N P P +SPSP SP
Sbjct: 105 PPNVYNSPPPPYYSPSPKVDYKSP 128
Score = 28.5 bits (62), Expect = 9.8
Identities = 22/74 (29%), Positives = 29/74 (38%), Gaps = 3/74 (4%)
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY---PS 372
PP V P P +S S + + Y S PP+ S + K+ Y P
Sbjct: 305 PPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPP 364
Query: 373 PNFSPSPSPSSSSP 386
P +SPSP SP
Sbjct: 365 PYYSPSPKVDYKSP 378
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.127 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,798,855
Number of Sequences: 26719
Number of extensions: 626947
Number of successful extensions: 3439
Number of sequences better than 10.0: 170
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 143
Number of HSP's that attempted gapping in prelim test: 2775
Number of HSP's gapped (non-prelim): 466
length of query: 535
length of database: 11,318,596
effective HSP length: 104
effective length of query: 431
effective length of database: 8,539,820
effective search space: 3680662420
effective search space used: 3680662420
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146842.6


