

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146819.11 - phase: 0
(316 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
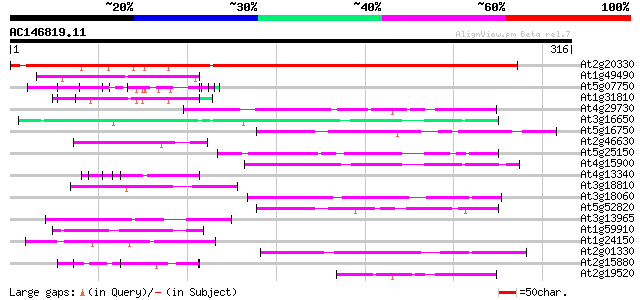
Score E
Sequences producing significant alignments: (bits) Value
At2g20330 putative WD-40 repeat protein 376 e-105
At1g49490 hypothetical protein 59 5e-09
At5g07750 putative protein 53 2e-07
At1g31810 hypothetical protein 49 5e-06
At4g29730 WD-40 repeat - like protein 48 6e-06
At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2 47 1e-05
At5g16750 WD40-repeat protein 47 2e-05
At2g46630 putative extensin 46 2e-05
At5g25150 transcription initiation factor IID-associated factor-... 46 3e-05
At4g15900 PRL1 protein 46 3e-05
At4g13340 extensin-like protein 46 3e-05
At3g18810 protein kinase, putative 45 4e-05
At3g18060 WD40-repeat protein 45 4e-05
At5g52820 Notchless protein homolog 45 5e-05
At3g13965 unknown protein 45 5e-05
At1g59910 hypothetical protein 45 5e-05
At1g24150 unknown protein 45 5e-05
At2g01330 putative WD-40-repeat protein 44 9e-05
At2g15880 unknown protein 44 1e-04
At2g19520 WD-40 repeat protein MSI4 (MSI4) 44 1e-04
>At2g20330 putative WD-40 repeat protein
Length = 648
Score = 376 bits (966), Expect = e-105
Identities = 198/323 (61%), Positives = 232/323 (71%), Gaps = 41/323 (12%)
Query: 1 MEDSGEIYDGVRAEFPLSFGKQSKSQTPLETIHKATRRS-----------VPTSNPKTTS 49
MED E G+RA FP+SFGK SK E IH ATRR+ + T P +
Sbjct: 1 MEDELE---GLRAHFPVSFGKTSKVSASTEAIHSATRRTDVANDAGVSSDITTKKPDSDK 57
Query: 50 DDFPS-----KECLHSIRPP-KNPN--------PPSPPL---------QDDSPLVGPPPP 86
FPS + L S+R P +NPN PP PP +D+ ++GPPPP
Sbjct: 58 SGFPSLSSSSQTWLRSVRGPNRNPNSSGDVAMGPPPPPSSKRTQEEEEEDEEVMMGPPPP 117
Query: 87 PP---HHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFRIPLSNEIVLKGH 143
P + DD+G+MIGPPPPPP + +D ED D+D D D NR++IPLSNEI LKGH
Sbjct: 118 PKGLGNINDSDDEGDMIGPPPPPPAARDSD-EDSDDDDDNDNEENRYQIPLSNEIQLKGH 176
Query: 144 TKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWSPTA 203
TK+VS+LAVD G+RVLSGSYDY VRMYDFQGMNSRLQSFRQIEPSEGHQ+R++SWSPT+
Sbjct: 177 TKIVSSLAVDSAGARVLSGSYDYTVRMYDFQGMNSRLQSFRQIEPSEGHQVRSVSWSPTS 236
Query: 204 DRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILT 263
+FLCVTGSAQAKI+DRDGLTLGEF+KGDMYIRDLKNTKGHI GLTCGEWHP+TKET+LT
Sbjct: 237 GQFLCVTGSAQAKIFDRDGLTLGEFMKGDMYIRDLKNTKGHICGLTCGEWHPRTKETVLT 296
Query: 264 SSEDGSLRIWDVNDFTTQKQVWK 286
SSEDGSLRIWDVN+F +Q QV K
Sbjct: 297 SSEDGSLRIWDVNNFLSQTQVIK 319
Score = 33.9 bits (76), Expect = 0.12
Identities = 31/126 (24%), Positives = 60/126 (47%), Gaps = 6/126 (4%)
Query: 141 KGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWS 200
K HT ++++ G +LS S+D ++++D + M L+ F + P+ Q N+++S
Sbjct: 370 KAHTDDITSVKFSSDGRILLSRSFDGSLKVWDLRQMKEALKVFEGL-PNYYPQ-TNVAFS 427
Query: 201 PTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKET 260
P D + +TG++ K GL L + + + I + + C WHP+ +
Sbjct: 428 P--DEQIILTGTSVEKDSTTGGL-LCFYDRTKLEIVQKVGISPTSSVVQCA-WHPRLNQI 483
Query: 261 ILTSSE 266
TS +
Sbjct: 484 FATSGD 489
Score = 33.1 bits (74), Expect = 0.20
Identities = 32/135 (23%), Positives = 56/135 (40%), Gaps = 13/135 (9%)
Query: 155 TGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGH-QIRNLSWSPTADRFLCVTGSA 213
T VL+ S D +R++D S+ Q + G + +W R G
Sbjct: 290 TKETVLTSSEDGSLRIWDVNNFLSQTQVIKPKLARPGRVPVTTCAWDRDGKRIAGGVGDG 349
Query: 214 QAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILTSSEDGSLRIW 273
+I+ L G + D+Y+ K H +T ++ + +L+ S DGSL++W
Sbjct: 350 SIQIWS---LKPGWGSRPDIYVG-----KAHTDDITSVKFSSDGR-ILLSRSFDGSLKVW 400
Query: 274 DVNDFTTQKQVWKIF 288
D+ K+ K+F
Sbjct: 401 DLRQ---MKEALKVF 412
>At1g49490 hypothetical protein
Length = 847
Score = 58.5 bits (140), Expect = 5e-09
Identities = 37/104 (35%), Positives = 48/104 (45%), Gaps = 13/104 (12%)
Query: 16 PLSFGKQSKSQTP---LETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSP 72
P S QS SQ P LE +H T S P +P +S+ S E + P+ P P +P
Sbjct: 740 PSSESNQSPSQAPTPILEPVHAPTPNSKPVQSPTPSSEPVSSPEQSEEVEAPE-PTPVNP 798
Query: 73 P-LQDDSPLVGPPPPPPHHEADDDDGEMIGPP--------PPPP 107
+ SP PPP + DDDDG+ + PP PPPP
Sbjct: 799 SSVPSSSPSTDTSIPPPENNDDDDDGDFVLPPHIGFQYASPPPP 842
Score = 38.1 bits (87), Expect = 0.006
Identities = 21/66 (31%), Positives = 27/66 (40%), Gaps = 6/66 (9%)
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMI 100
P +P S + +HS PP +PP P + P V PPPP +
Sbjct: 539 PMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASPPPP------SPPPPVH 592
Query: 101 GPPPPP 106
PPPPP
Sbjct: 593 SPPPPP 598
Score = 35.8 bits (81), Expect = 0.031
Identities = 30/112 (26%), Positives = 47/112 (41%), Gaps = 19/112 (16%)
Query: 24 KSQTPLETIHKATRRS-VPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVG 82
++ TP++ ++ S VPT P + S+ PS+ + P P P S P+Q +P
Sbjct: 720 QAPTPVQAPTTSSETSQVPT--PSSESNQSPSQAPTPILEPVHAPTPNSKPVQSPTPSSE 777
Query: 83 PPPPPPHHEADDDDGEMIGPPPPP----------PGSNFNDSEDEDNDSDQD 124
P P E E+ P P P P ++ + E+ND D D
Sbjct: 778 PVSSPEQSE------EVEAPEPTPVNPSSVPSSSPSTDTSIPPPENNDDDDD 823
Score = 35.4 bits (80), Expect = 0.041
Identities = 25/76 (32%), Positives = 30/76 (38%), Gaps = 11/76 (14%)
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMI 100
P ++P S P +HS PP +PP P P PPPP H +
Sbjct: 578 PVASPPPPSPPPP----VHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPP---PVY 630
Query: 101 GPPP----PPPGSNFN 112
PPP PPP N N
Sbjct: 631 SPPPPTFSPPPTHNTN 646
Score = 34.7 bits (78), Expect = 0.070
Identities = 21/78 (26%), Positives = 31/78 (38%), Gaps = 7/78 (8%)
Query: 63 PPKNP--NPPSPPLQDDSPLVGPPPP-----PPHHEADDDDGEMIGPPPPPPGSNFNDSE 115
PP +P +PP P P+ PPPP P H+ G P P S
Sbjct: 610 PPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSETTQVP 669
Query: 116 DEDNDSDQDELGNRFRIP 133
++SDQ ++ + + P
Sbjct: 670 TPSSESDQSQILSPVQAP 687
Score = 34.3 bits (77), Expect = 0.091
Identities = 22/68 (32%), Positives = 26/68 (37%), Gaps = 2/68 (2%)
Query: 40 VPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEM 99
VP P S PS ++S PP + PP P V PPPP
Sbjct: 533 VPPPQPPMPSPSPPSP--IYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSPPPP 590
Query: 100 IGPPPPPP 107
+ PPPPP
Sbjct: 591 VHSPPPPP 598
Score = 33.9 bits (76), Expect = 0.12
Identities = 20/55 (36%), Positives = 27/55 (48%), Gaps = 5/55 (9%)
Query: 39 SVPTSNPKTTSDDFPSKECLHSIRP-PKNPNPPSPPLQDDSPLVGPPPPPPHHEA 92
S P+ NP TS+ PSK + P P + + P P + P P P PP HE+
Sbjct: 392 SSPSPNPPRTSEPKPSKP--EPVMPKPSDSSKPETPKTPEQP--SPKPQPPKHES 442
Score = 33.1 bits (74), Expect = 0.20
Identities = 19/65 (29%), Positives = 27/65 (41%), Gaps = 5/65 (7%)
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMI 100
P S P P+ + + P KN P PP +D+ + PPP P + +
Sbjct: 496 PVSPPNEAQG--PTPDDPYDASPVKNRRSPPPPKVEDTRV---PPPQPPMPSPSPPSPIY 550
Query: 101 GPPPP 105
PPPP
Sbjct: 551 SPPPP 555
Score = 32.7 bits (73), Expect = 0.26
Identities = 27/96 (28%), Positives = 38/96 (39%), Gaps = 5/96 (5%)
Query: 16 PLSFGKQSKSQTPLETIHKA---TRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSP 72
P+S +++ TP + + RRS P + T P + + S PP P P
Sbjct: 496 PVSPPNEAQGPTPDDPYDASPVKNRRSPPPPKVEDTRVP-PPQPPMPSPSPPSPIYSPPP 554
Query: 73 PLQDDSPLV-GPPPPPPHHEADDDDGEMIGPPPPPP 107
P+ P V PPPP + P PPPP
Sbjct: 555 PVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSPPPP 590
Score = 27.7 bits (60), Expect = 8.5
Identities = 28/104 (26%), Positives = 35/104 (32%), Gaps = 5/104 (4%)
Query: 21 KQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPL 80
K S S P ET + S PK S E H + K P P +DSP
Sbjct: 415 KPSDSSKP-ETPKTPEQPSPKPQPPKHESPKPEEPENKHELPKQKESPKPQPSKPEDSP- 472
Query: 81 VGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQD 124
P P E + P P P S N+++ D D
Sbjct: 473 ---KPEQPKPEESPKPEQPQIPEPTKPVSPPNEAQGPTPDDPYD 513
>At5g07750 putative protein
Length = 1289
Score = 52.8 bits (125), Expect = 2e-07
Identities = 32/109 (29%), Positives = 48/109 (43%), Gaps = 10/109 (9%)
Query: 11 VRAEFPLSFGKQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPP 70
+R+ P+ S + ++ A+ P P TS+ +K LHS + +P PP
Sbjct: 594 MRSRAPICSSPDSSPKETPSSLPPASPHQAPPPLPSLTSE---AKTVLHSSQAVASPPPP 650
Query: 71 SPPLQ-------DDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFN 112
PP S L PPPPPP ++ + + PPPPPP F+
Sbjct: 651 PPPPPLPTYSHYQTSQLPPPPPPPPPFSSERPNSGTVLPPPPPPPPPFS 699
Score = 48.5 bits (114), Expect = 5e-06
Identities = 30/81 (37%), Positives = 34/81 (41%), Gaps = 4/81 (4%)
Query: 28 PLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPP 87
PL I A V T+ P F + + S PP +PP PP S PPPPP
Sbjct: 902 PLHGISSAPSPPVKTAPPPPPPPPFSNAHSVLSPPPPSYGSPPPPPPPPPSYGSPPPPPP 961
Query: 88 PHHEADDDDGEMIGPPPPPPG 108
P G PPPPPPG
Sbjct: 962 P----PPSYGSPPPPPPPPPG 978
Score = 43.9 bits (102), Expect = 1e-04
Identities = 29/94 (30%), Positives = 36/94 (37%), Gaps = 15/94 (15%)
Query: 40 VPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPL----QDDSPLVGPPPPPPH------ 89
+P+ P F S P P PP PP ++ L+ PPPPPP
Sbjct: 786 LPSPPPPPPPPPFASVRRNSETLLPPPPPPPPPPFASVRRNSETLLPPPPPPPPWKSLYA 845
Query: 90 -----HEADDDDGEMIGPPPPPPGSNFNDSEDED 118
HEA PPPPPP S N ++ D
Sbjct: 846 STFETHEACSTSSSPPPPPPPPPFSPLNTTKAND 879
Score = 43.1 bits (100), Expect = 2e-04
Identities = 20/41 (48%), Positives = 21/41 (50%), Gaps = 6/41 (14%)
Query: 67 PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P PP PP S + PPPPPP H G PPPPPP
Sbjct: 996 PPPPPPPFSHVSSIPPPPPPPPMH------GGAPPPPPPPP 1030
Score = 42.0 bits (97), Expect = 4e-04
Identities = 24/64 (37%), Positives = 28/64 (43%), Gaps = 13/64 (20%)
Query: 57 CLHSIRPPKNPNPPSPP------------LQDDS-PLVGPPPPPPHHEADDDDGEMIGPP 103
C S P +P PP PP LQ P PPPPPP + + E + PP
Sbjct: 752 CSTSQAPTSSPTPPPPPPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFASVRRNSETLLPP 811
Query: 104 PPPP 107
PPPP
Sbjct: 812 PPPP 815
Score = 41.6 bits (96), Expect = 6e-04
Identities = 20/41 (48%), Positives = 22/41 (52%), Gaps = 8/41 (19%)
Query: 67 PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P PP PP+ +P PPPPPP H G PPPPPP
Sbjct: 1011 PPPPPPPMHGGAP--PPPPPPPMH------GGAPPPPPPPP 1043
Score = 41.6 bits (96), Expect = 6e-04
Identities = 20/41 (48%), Positives = 22/41 (52%), Gaps = 8/41 (19%)
Query: 67 PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P PP PP+ +P PPPPPP H G PPPPPP
Sbjct: 1024 PPPPPPPMHGGAP--PPPPPPPMH------GGAPPPPPPPP 1056
Score = 41.6 bits (96), Expect = 6e-04
Identities = 26/86 (30%), Positives = 39/86 (45%), Gaps = 11/86 (12%)
Query: 28 PLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVG----- 82
P + ++ ++ P P S + P+ ++ PP P PP PP + P G
Sbjct: 657 PTYSHYQTSQLPPPPPPPPPFSSERPNSG---TVLPP--PPPPPPPFSSERPNSGTVLPP 711
Query: 83 PPPPP-PHHEADDDDGEMIGPPPPPP 107
PPPPP P + G ++ PPP PP
Sbjct: 712 PPPPPLPFSSERPNSGTVLPPPPSPP 737
Score = 41.6 bits (96), Expect = 6e-04
Identities = 19/41 (46%), Positives = 22/41 (53%), Gaps = 11/41 (26%)
Query: 67 PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P PP PP++ +P PPPPPP H PPPPPP
Sbjct: 1098 PPPPPPPMRGGAP---PPPPPPMHGG--------APPPPPP 1127
Score = 41.2 bits (95), Expect = 7e-04
Identities = 25/71 (35%), Positives = 30/71 (42%), Gaps = 17/71 (23%)
Query: 53 PSKECLHSIRPPKNPNPP----SPPLQDDSPLVG----PPPPPPHHEADDDDGEMIGPPP 104
P +H PP P PP +PP P+ G PPPPPP H G PP
Sbjct: 1013 PPPPPMHGGAPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPPMHG---------GAPP 1063
Query: 105 PPPGSNFNDSE 115
PPP F ++
Sbjct: 1064 PPPPPMFGGAQ 1074
Score = 38.9 bits (89), Expect = 0.004
Identities = 20/41 (48%), Positives = 20/41 (48%), Gaps = 7/41 (17%)
Query: 67 PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P PP PP SP PPPPPP I PPPPPP
Sbjct: 983 PPPPPPPPSYGSP--PPPPPPPFSHVSS-----IPPPPPPP 1016
Score = 38.5 bits (88), Expect = 0.005
Identities = 26/73 (35%), Positives = 31/73 (41%), Gaps = 6/73 (8%)
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPL-----QDDSPLVGPPPPPPHHEAD-D 94
P P S S + S P P PP PP ++ L PPPPPP A
Sbjct: 765 PPPPPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFASVRRNSETLLPPPPPPPPPPFASVR 824
Query: 95 DDGEMIGPPPPPP 107
+ E + PPPPPP
Sbjct: 825 RNSETLLPPPPPP 837
Score = 38.5 bits (88), Expect = 0.005
Identities = 18/41 (43%), Positives = 21/41 (50%), Gaps = 11/41 (26%)
Query: 67 PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P PP PP++ +P PPPPPP PPPPPP
Sbjct: 1086 PPPPPPPMRGGAP---PPPPPPMRGG--------APPPPPP 1115
Score = 37.7 bits (86), Expect = 0.008
Identities = 22/63 (34%), Positives = 26/63 (40%), Gaps = 19/63 (30%)
Query: 53 PSKECLHSIRPPKNPNP--------PSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPP 104
P +H PP P P P PP++ +P PPPPPP PPP
Sbjct: 1052 PPPPPMHGGAPPPPPPPMFGGAQPPPPPPMRGGAP---PPPPPPMRGG--------APPP 1100
Query: 105 PPP 107
PPP
Sbjct: 1101 PPP 1103
Score = 37.7 bits (86), Expect = 0.008
Identities = 20/44 (45%), Positives = 22/44 (49%), Gaps = 16/44 (36%)
Query: 67 PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIG---PPPPPP 107
P PP PP+ +P PPPPPP M G PPPPPP
Sbjct: 1050 PPPPPPPMHGGAP---PPPPPP----------MFGGAQPPPPPP 1080
Score = 37.0 bits (84), Expect = 0.014
Identities = 19/44 (43%), Positives = 21/44 (47%), Gaps = 8/44 (18%)
Query: 67 PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSN 110
P PP PP SP PPPPP + PPPPPP S+
Sbjct: 970 PPPPPPPPGYGSPPPPPPPPPSYGSPP--------PPPPPPFSH 1005
Score = 37.0 bits (84), Expect = 0.014
Identities = 29/85 (34%), Positives = 37/85 (43%), Gaps = 14/85 (16%)
Query: 27 TPLETIHKATRRSVPTSNPKTTSDDFPSKECL--HSIRPPKNPNPPSPPLQDDSPLVGPP 84
+PL T P P T+ PS + L H I + PSPP++ P PP
Sbjct: 870 SPLNTTKANDYILPPPPLPYTSIAPSPSVKILPLHGIS-----SAPSPPVKTAPP---PP 921
Query: 85 PPPPHHEADDDDGEMIGPPPPPPGS 109
PPPP A ++ PPPP GS
Sbjct: 922 PPPPFSNAHS----VLSPPPPSYGS 942
Score = 36.6 bits (83), Expect = 0.018
Identities = 22/60 (36%), Positives = 23/60 (37%), Gaps = 12/60 (20%)
Query: 53 PSKECLHSIRPPKNPNP------PSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPP 106
P +H PP P P P PP PPPPPP G GPPPPP
Sbjct: 1112 PPPPPMHGGAPPPPPPPMRGGAPPPPPPPGGRGPGAPPPPPP------PGGRAPGPPPPP 1165
Score = 36.6 bits (83), Expect = 0.018
Identities = 23/55 (41%), Positives = 25/55 (44%), Gaps = 12/55 (21%)
Query: 63 PPKNPNPPS----PPLQDDSPLVG-PPPPPPHHEADDDDGEMIGPPPPPPGSNFN 112
PP P PPS PP P G PPPPPP + G PPPPP F+
Sbjct: 957 PPPPPPPPSYGSPPPPPPPPPGYGSPPPPPPPPPS-------YGSPPPPPPPPFS 1004
Score = 35.8 bits (81), Expect = 0.031
Identities = 19/48 (39%), Positives = 22/48 (45%), Gaps = 8/48 (16%)
Query: 67 PNPPSPPLQDDSP------LVGPPPPPPHHEADDDDGEMIGPPPPPPG 108
P PP PP+ +P + G PPPP G PPPPPPG
Sbjct: 1110 PPPPPPPMHGGAPPPPPPPMRGGAPPPPPPPGGRGPG--APPPPPPPG 1155
Score = 34.7 bits (78), Expect = 0.070
Identities = 20/48 (41%), Positives = 22/48 (45%), Gaps = 12/48 (25%)
Query: 63 PPKNPNPPSPPLQDDSPLVG--PPPPPPHHEADDDDGEMIGPPPPPPG 108
P + PP PP P+ G PPPPPP PPPPPPG
Sbjct: 1104 PMRGGAPPPPP----PPMHGGAPPPPPPPMRGG------APPPPPPPG 1141
Score = 33.5 bits (75), Expect = 0.16
Identities = 18/46 (39%), Positives = 19/46 (41%), Gaps = 8/46 (17%)
Query: 63 PP--KNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPP 106
PP + P P PP GPPPPP GPPPPP
Sbjct: 1139 PPGGRGPGAPPPPPPPGGRAPGPPPPPGPRPPGG------GPPPPP 1178
Score = 30.4 bits (67), Expect = 1.3
Identities = 21/68 (30%), Positives = 24/68 (34%), Gaps = 20/68 (29%)
Query: 63 PPKNPNPPSPPLQDD--SPLVGPPPPPPHHEADDDDGEMI------------------GP 102
PP P PP PL + + PPPP P+ I P
Sbjct: 861 PPPPPPPPFSPLNTTKANDYILPPPPLPYTSIAPSPSVKILPLHGISSAPSPPVKTAPPP 920
Query: 103 PPPPPGSN 110
PPPPP SN
Sbjct: 921 PPPPPFSN 928
Score = 28.1 bits (61), Expect = 6.5
Identities = 19/50 (38%), Positives = 19/50 (38%), Gaps = 4/50 (8%)
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPP----HHEADDDDGEMIGPPPPPPG 108
PP P PP P G PPPPP A D G G P PG
Sbjct: 1153 PPGGRAPGPPPPPGPRPPGGGPPPPPMLGARGAAVDPRGAGRGRGLPRPG 1202
>At1g31810 hypothetical protein
Length = 1201
Score = 48.5 bits (114), Expect = 5e-06
Identities = 27/91 (29%), Positives = 39/91 (42%), Gaps = 10/91 (10%)
Query: 25 SQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPP--------LQD 76
++ PL T+H+ ++ P P PS+ + P P PP PP +
Sbjct: 557 NRDPLTTLHQPINKTPPP--PPPPPPPLPSRSIPPPLAQPPPPRPPPPPPPPPSSRSIPS 614
Query: 77 DSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
S PPPPPP + + + PPPPPP
Sbjct: 615 PSAPPPPPPPPPSFGSTGNKRQAQPPPPPPP 645
Score = 43.5 bits (101), Expect = 1e-04
Identities = 27/77 (35%), Positives = 33/77 (42%), Gaps = 8/77 (10%)
Query: 38 RSVPTSN----PKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPP---HH 90
RS+P+ + P F S +PP P PP PP + + PPPPPP H
Sbjct: 610 RSIPSPSAPPPPPPPPPSFGSTGNKRQAQPPPPP-PPPPPTRIPAAKCAPPPPPPPPTSH 668
Query: 91 EADDDDGEMIGPPPPPP 107
G PPPPPP
Sbjct: 669 SGSIRVGPPSTPPPPPP 685
Score = 43.5 bits (101), Expect = 1e-04
Identities = 30/101 (29%), Positives = 39/101 (37%), Gaps = 16/101 (15%)
Query: 28 PLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPP--------------SPP 73
PL T + S P P S F +++ L ++ P N PP PP
Sbjct: 533 PLFTSTTSFSPSQPPPPPPLPS--FSNRDPLTTLHQPINKTPPPPPPPPPPLPSRSIPPP 590
Query: 74 LQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDS 114
L P PPPPPP + PPPPPP +F +
Sbjct: 591 LAQPPPPRPPPPPPPPPSSRSIPSPSAPPPPPPPPPSFGST 631
Score = 40.4 bits (93), Expect = 0.001
Identities = 26/77 (33%), Positives = 31/77 (39%), Gaps = 7/77 (9%)
Query: 44 NPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVG------PPPPPPHHEADDDDG 97
NP D PS ++ PP P PP PPL + PPPPPP +
Sbjct: 463 NPLNLPSDPPSSGDHVTLLPPPPP-PPPPPLFTSTTSFSPSQPPPPPPPPPLFMSTTSFS 521
Query: 98 EMIGPPPPPPGSNFNDS 114
PPPPPP F +
Sbjct: 522 PSQPPPPPPPPPLFTST 538
Score = 37.0 bits (84), Expect = 0.014
Identities = 24/73 (32%), Positives = 32/73 (42%), Gaps = 8/73 (10%)
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPP---SPPLQDDSPLVGPPPPPPHHEADDDDG 97
P P P+ +C PP P PP S ++ P PPPPPP +A+ +
Sbjct: 640 PPPPPPPPPTRIPAAKCA----PPPPPPPPTSHSGSIRVGPPSTPPPPPPPPPKANISNA 695
Query: 98 -EMIGPPPPPPGS 109
+ PPP PP S
Sbjct: 696 PKPPAPPPLPPSS 708
Score = 36.2 bits (82), Expect = 0.024
Identities = 20/49 (40%), Positives = 23/49 (46%), Gaps = 5/49 (10%)
Query: 61 IRPPKNPNPPSPP--LQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
+ PP P PP PP + S PP PPP + G PPPPPP
Sbjct: 674 VGPPSTPPPPPPPPPKANISNAPKPPAPPPLPPSSTRLG---APPPPPP 719
Score = 35.8 bits (81), Expect = 0.031
Identities = 24/80 (30%), Positives = 30/80 (37%), Gaps = 7/80 (8%)
Query: 28 PLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPP 87
P T A + + P P TS + S PP P PP + + PPP P
Sbjct: 646 PPPTRIPAAKCAPPPPPPPPTSHSGSIRVGPPSTPPPPPPPPPKANISNAPKPPAPPPLP 705
Query: 88 PHHEADDDDGEMIGPPPPPP 107
P +G PPPPP
Sbjct: 706 P-------SSTRLGAPPPPP 718
Score = 35.8 bits (81), Expect = 0.031
Identities = 25/82 (30%), Positives = 31/82 (37%), Gaps = 1/82 (1%)
Query: 28 PLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPP 87
P T H + R P S P P ++ +PP P P + +P PPPP
Sbjct: 663 PPPTSHSGSIRVGPPSTPPPPPPPPPKANISNAPKPPAPPPLPPSSTRLGAPPPPPPPPL 722
Query: 88 PHHEADDDDGEMIGP-PPPPPG 108
A P PPPPPG
Sbjct: 723 SKTPAPPPPPLSKTPVPPPPPG 744
Score = 35.0 bits (79), Expect = 0.053
Identities = 25/81 (30%), Positives = 32/81 (38%), Gaps = 7/81 (8%)
Query: 40 VPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGP--PPPPPHHE---ADD 94
+P P F S +PP P PP PPL + P PPPPP
Sbjct: 481 LPPPPPPPPPPLFTSTTSFSPSQPP--PPPPPPPLFMSTTSFSPSQPPPPPPPPPLFTST 538
Query: 95 DDGEMIGPPPPPPGSNFNDSE 115
PPPPPP +F++ +
Sbjct: 539 TSFSPSQPPPPPPLPSFSNRD 559
Score = 32.7 bits (73), Expect = 0.26
Identities = 27/109 (24%), Positives = 40/109 (35%), Gaps = 8/109 (7%)
Query: 2 EDSGEIYDGV-RAEFPLSFGKQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHS 60
+ S E ++ + R + F L H+++ P + P+K+ S
Sbjct: 406 DSSDEGFEAIQRPRIHIPFDNDDTDDITLSVAHESSEE--PHEFSHHHHHEIPAKD---S 460
Query: 61 IRPPKNPNPPSPPLQDDSPLVGPPPPPPHHE--ADDDDGEMIGPPPPPP 107
+ P N P D L+ PPPPPP PPPPPP
Sbjct: 461 VDNPLNLPSDPPSSGDHVTLLPPPPPPPPPPLFTSTTSFSPSQPPPPPP 509
Score = 30.0 bits (66), Expect = 1.7
Identities = 19/48 (39%), Positives = 19/48 (39%), Gaps = 9/48 (18%)
Query: 63 PP--KNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPG 108
PP K P PP PP GPPP G PPPPP G
Sbjct: 731 PPLSKTPVPPPPPGLGRGTSSGPPPL-------GAKGSNAPPPPPPAG 771
>At4g29730 WD-40 repeat - like protein
Length = 496
Score = 48.1 bits (113), Expect = 6e-06
Identities = 44/179 (24%), Positives = 73/179 (40%), Gaps = 21/179 (11%)
Query: 99 MIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFRIPLSNEIVLKGHTKVVSALAV-DHTGS 157
M G PGS+F + + + + +G R + GH V +A +
Sbjct: 244 MAGSDSKSPGSSFKQTGEGSDKTGGPSVGPRG--------IYNGHKDTVEDVAFCPSSAQ 295
Query: 158 RVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWSPTADRFLCVTGSAQ--A 215
S D + ++D + S ++E + + + W+P D L +TGSA
Sbjct: 296 EFCSVGDDSCLMLWDARTGTSPAM---KVEKAHDADLHCVDWNPH-DNNLILTGSADNTV 351
Query: 216 KIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILTSSEDGSLRIWD 274
+++DR LT V +Y +GH + C +W P +S+EDG L IWD
Sbjct: 352 RVFDRRNLT-SNGVGSPVY-----KFEGHRAAVLCVQWSPDKSSVFGSSAEDGLLNIWD 404
Score = 37.7 bits (86), Expect = 0.008
Identities = 45/170 (26%), Positives = 74/170 (43%), Gaps = 35/170 (20%)
Query: 137 EIVLKGHTKVVS-ALAVDHTGSRVLSGSYDYMVRMYDFQ------GMNSRL--QSFRQIE 187
+++L GH ALA+ T VLSG D V +++ Q G +S+ SF+Q
Sbjct: 201 DLLLIGHQDDAEFALAMCPTEPFVLSGGKDKSVILWNIQDHITMAGSDSKSPGSSFKQTG 260
Query: 188 --------PS-------EGHQ--IRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVK 230
PS GH+ + ++++ P++ + C G D L L +
Sbjct: 261 EGSDKTGGPSVGPRGIYNGHKDTVEDVAFCPSSAQEFCSVG-------DDSCLMLWDART 313
Query: 231 GDMYIRDLKNTKGHITGLTCGEWHPKTKETILTSSEDGSLRIWDVNDFTT 280
G +K K H L C +W+P ILT S D ++R++D + T+
Sbjct: 314 GTSPA--MKVEKAHDADLHCVDWNPHDNNLILTGSADNTVRVFDRRNLTS 361
>At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2
Length = 479
Score = 47.4 bits (111), Expect = 1e-05
Identities = 61/278 (21%), Positives = 105/278 (36%), Gaps = 30/278 (10%)
Query: 6 EIYDGVRAEFPLSFGKQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKEC---LHSIR 62
EI+ V +FP +SK I A P S P +D K ++
Sbjct: 28 EIFSPVHGQFPQP-DPESKRIRLCHKIQVAFGGVEPASKPTRIADHNSEKTAPLKALALP 86
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSD 122
PK + + +VGP PP + G+ P P +F++ + +
Sbjct: 87 GPKGSKELRKSATEKALVVGPTLPPRDLNNTGNPGKSTAILPAP--GSFSE-RNLSTAAL 143
Query: 123 QDELGNRF-----RIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMN 177
+ + +R+ P N VL+GH V ++A D + +GS D ++++D
Sbjct: 144 MERMPSRWPRPEWHAPWKNYRVLQGHLGWVRSVAFDPSNEWFCTGSADRTIKIWDVATGV 203
Query: 178 SRLQSFRQIEPSEGHQIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRD 237
+L I Q+R L+ S Q K +D + +
Sbjct: 204 LKLTLTGHI-----GQVRGLAVSNRHTYMFSAGDDKQVKCWDLE------------QNKV 246
Query: 238 LKNTKGHITGLTCGEWHPKTKETILTSSEDGSLRIWDV 275
+++ GH+ G+ C HP T + +LT D R+WD+
Sbjct: 247 IRSYHGHLHGVYCLALHP-TLDVVLTGGRDSVCRVWDI 283
>At5g16750 WD40-repeat protein
Length = 876
Score = 46.6 bits (109), Expect = 2e-05
Identities = 40/172 (23%), Positives = 78/172 (45%), Gaps = 27/172 (15%)
Query: 140 LKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSW 199
++G + ++ALA+ + S + +R++D L++ + I +GH+ +
Sbjct: 56 IEGESDTLTALALSPDDKLLFSAGHSRQIRVWD-------LETLKCIRSWKGHEGPVMGM 108
Query: 200 SPTADRFLCVTGSAQAKI--YDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKT 257
+ A L T A K+ +D DG + +G KG ++ + +HP +
Sbjct: 109 ACHASGGLLATAGADRKVLVWDVDGGFCTHYFRGH---------KGVVSSIL---FHPDS 156
Query: 258 KETILTS-SEDGSLRIWDVNDFTTQKQVWKIFTLLFFHGHFHFIFHLWLGEE 308
+ IL S S+D ++R+WD+N T+K+ L HF + + L E+
Sbjct: 157 NKNILISGSDDATVRVWDLNAKNTEKK-----CLAIMEKHFSAVTSIALSED 203
Score = 30.0 bits (66), Expect = 1.7
Identities = 33/148 (22%), Positives = 60/148 (40%), Gaps = 24/148 (16%)
Query: 132 IPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEG 191
I L V+ H K ++++AV S V +GS D ++ RL + +G
Sbjct: 487 INLKTRSVVAAHDKDINSVAVARNDSLVCTGSEDRTASIW-------RLPDLVHVVTLKG 539
Query: 192 HQIR--NLSWSPTADRFLCVTGSAQAKIYD-RDGLTLGEFVKGDMYIRDLKNTKGHITGL 248
H+ R ++ +S + +G KI+ DG LK +GH + +
Sbjct: 540 HKRRIFSVEFSTVDQCVMTASGDKTVKIWAISDGSC-------------LKTFEGHTSSV 586
Query: 249 TCGEWHPKTKETILTSSEDGSLRIWDVN 276
+ + ++ DG L++W+VN
Sbjct: 587 LRASFITDGTQ-FVSCGADGLLKLWNVN 613
>At2g46630 putative extensin
Length = 394
Score = 46.2 bits (108), Expect = 2e-05
Identities = 26/80 (32%), Positives = 36/80 (44%), Gaps = 13/80 (16%)
Query: 37 RRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPP-----PPPPHHE 91
R+ P S P+ P ++ + PP+ PP+ P Q+ SP PP PP P
Sbjct: 50 RQRQPRSPPRQQDPPSPPRQQQQPLTPPRQKAPPTSPPQERSPYHSPPSRHMSPPTPPKA 109
Query: 92 ADDDDGEMIGPPPPPPGSNF 111
A PPPPPP S++
Sbjct: 110 AT--------PPPPPPRSSY 121
Score = 31.2 bits (69), Expect = 0.77
Identities = 27/94 (28%), Positives = 36/94 (37%), Gaps = 4/94 (4%)
Query: 37 RRSVPTSNPKTTSD-DFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDD 95
+++ PTS P+ S P + PPK PP PP + S PP P EA
Sbjct: 79 QKAPPTSPPQERSPYHSPPSRHMSPPTPPKAATPPPPPPR--SSYTSPPSPKEVQEA-LP 135
Query: 96 DGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNR 129
+ PP P S SE S + +R
Sbjct: 136 PRKPNSPPSPAHSSRSTTSESVKTRSPSESENHR 169
>At5g25150 transcription initiation factor IID-associated
factor-like protein
Length = 700
Score = 45.8 bits (107), Expect = 3e-05
Identities = 39/158 (24%), Positives = 69/158 (42%), Gaps = 24/158 (15%)
Query: 118 DNDSDQDELGNRFRIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMN 177
+NDS +G R + +L GH+ V + G VLS S D +R++ + +N
Sbjct: 395 ENDSSDQSIGPNGR---RSYTLLLGHSGPVYSATFSPPGDFVLSSSADTTIRLWSTK-LN 450
Query: 178 SRLQSFRQIEPSEGHQIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRD 237
+ L ++ + + + +SP F + A+I+ D I+
Sbjct: 451 ANLVCYK----GHNYPVWDAQFSPFGHYFASCSHDRTARIWSMD------------RIQP 494
Query: 238 LKNTKGHITGLTCGEWHPKTKETILTSSEDGSLRIWDV 275
L+ GH++ + +WHP I T S D ++R+WDV
Sbjct: 495 LRIMAGHLSDV---DWHPNC-NYIATGSSDKTVRLWDV 528
>At4g15900 PRL1 protein
Length = 486
Score = 45.8 bits (107), Expect = 3e-05
Identities = 37/155 (23%), Positives = 67/155 (42%), Gaps = 21/155 (13%)
Query: 133 PLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGH 192
P N V++GH V ++A D + +GS D ++++D +L IE
Sbjct: 165 PWKNYRVIQGHLGWVRSVAFDPSNEWFCTGSADRTIKIWDVATGVLKLTLTGHIE----- 219
Query: 193 QIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGE 252
Q+R L+ S Q K +D + + +++ GH++G+ C
Sbjct: 220 QVRGLAVSNRHTYMFSAGDDKQVKCWDLE------------QNKVIRSYHGHLSGVYCLA 267
Query: 253 WHPKTKETILTSSEDGSLRIWDVNDFTTQKQVWKI 287
HP T + +LT D R+WD+ T+ Q++ +
Sbjct: 268 LHP-TLDVLLTGGRDSVCRVWDIR---TKMQIFAL 298
Score = 28.5 bits (62), Expect = 5.0
Identities = 30/135 (22%), Positives = 54/135 (39%), Gaps = 19/135 (14%)
Query: 140 LKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSW 199
L GH V ++ T +V++GS+D ++ +D + + + + S +R ++
Sbjct: 298 LSGHDNTVCSVFTRPTDPQVVTGSHDTTIKFWDLR-YGKTMSTLTHHKKS----VRAMTL 352
Query: 200 SPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKE 259
P + F SA A + L GEF L K I + E
Sbjct: 353 HPKENAF----ASASADNTKKFSLPKGEFCH-----NMLSQQKTIINAMAVNE-----DG 398
Query: 260 TILTSSEDGSLRIWD 274
++T ++GS+ WD
Sbjct: 399 VMVTGGDNGSIWFWD 413
>At4g13340 extensin-like protein
Length = 760
Score = 45.8 bits (107), Expect = 3e-05
Identities = 22/55 (40%), Positives = 26/55 (47%), Gaps = 10/55 (18%)
Query: 53 PSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P ++S PP P PP PP+ P PPPPPP + PPPPPP
Sbjct: 458 PPPPPVYSPPPPPPPPPPPPPVYSPPPPSPPPPPPP----------VYSPPPPPP 502
Score = 43.1 bits (100), Expect = 2e-04
Identities = 20/45 (44%), Positives = 22/45 (48%), Gaps = 4/45 (8%)
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP+ P PPPPPP + PPPPPP
Sbjct: 452 PPPPPPPPPPPVYSPPPPPPPPPPPPPVYSPPPP----SPPPPPP 492
Score = 42.7 bits (99), Expect = 3e-04
Identities = 20/45 (44%), Positives = 23/45 (50%), Gaps = 2/45 (4%)
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP +P PP PP+ SP PPPPPP + PPPPP
Sbjct: 483 PPPSPPPPPPPVY--SPPPPPPPPPPPPVYSPPPPPVYSSPPPPP 525
Score = 42.4 bits (98), Expect = 3e-04
Identities = 23/67 (34%), Positives = 28/67 (41%), Gaps = 1/67 (1%)
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMI 100
P +P P ++S PP P PP PP+ P PPPPPP +
Sbjct: 462 PVYSPPPPPPPPPPPPPVYSPPPPSPP-PPPPPVYSPPPPPPPPPPPPVYSPPPPPVYSS 520
Query: 101 GPPPPPP 107
PPPP P
Sbjct: 521 PPPPPSP 527
Score = 42.4 bits (98), Expect = 3e-04
Identities = 20/49 (40%), Positives = 24/49 (48%), Gaps = 1/49 (2%)
Query: 59 HSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
HS PP +P PP P P + PPPPP + + PPPPPP
Sbjct: 567 HSSPPPHSPPPPHSPPPPIYPYLSPPPPPT-PVSSPPPTPVYSPPPPPP 614
Score = 42.4 bits (98), Expect = 3e-04
Identities = 24/63 (38%), Positives = 27/63 (42%), Gaps = 12/63 (19%)
Query: 45 PKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPP 104
P TS PS + P P PP PP+ P PPPPPP + PPP
Sbjct: 421 PTLTSPPPPSPP--PPVYSPPPPPPPPPPVYSPPPPPPPPPPPP----------VYSPPP 468
Query: 105 PPP 107
PPP
Sbjct: 469 PPP 471
Score = 40.8 bits (94), Expect = 0.001
Identities = 20/45 (44%), Positives = 21/45 (46%), Gaps = 18/45 (40%)
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P+ PPPPPP PPPPPP
Sbjct: 451 PPPPPPPPPPP-----PVYSPPPPPP-------------PPPPPP 477
Score = 38.9 bits (89), Expect = 0.004
Identities = 19/47 (40%), Positives = 22/47 (46%), Gaps = 6/47 (12%)
Query: 63 PPKNPNPPSPPLQDDSPL--VGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP +PP PP +P+ PPPPPPH PPPP P
Sbjct: 515 PPVYSSPPPPPSPAPTPVYCTRPPPPPPH----SPPPPQFSPPPPEP 557
Score = 38.5 bits (88), Expect = 0.005
Identities = 22/73 (30%), Positives = 29/73 (39%), Gaps = 11/73 (15%)
Query: 53 PSKECLHSIRPPKNP----NPPSPPLQDDSP-------LVGPPPPPPHHEADDDDGEMIG 101
P +H PP P +PP PP+ SP PPPP H+ +
Sbjct: 631 PPPPVVHYSSPPPPPVYYSSPPPPPVYYSSPPPPPPVHYSSPPPPEVHYHSPPPSPVHYS 690
Query: 102 PPPPPPGSNFNDS 114
PPPPP + +S
Sbjct: 691 SPPPPPSAPCEES 703
Score = 37.4 bits (85), Expect = 0.011
Identities = 21/57 (36%), Positives = 26/57 (44%), Gaps = 9/57 (15%)
Query: 53 PSKECLHSIRPPKNPNPPSP-PLQDDSP-LVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P + + PP P+PP P P+ P L PPPP P + PPPPPP
Sbjct: 394 PRPPVVTPLPPPSLPSPPPPAPIFSTPPTLTSPPPPSP-------PPPVYSPPPPPP 443
Score = 37.0 bits (84), Expect = 0.014
Identities = 23/78 (29%), Positives = 33/78 (41%), Gaps = 10/78 (12%)
Query: 41 PTSNPKTTS--DDFPSKECLHSIRPPK----NPNPPSPPLQDDSPLVGPPPPPPHHEADD 94
P S+P T P C+ PP +P PP P + SP PPPP ++ +
Sbjct: 597 PVSSPPPTPVYSPPPPPPCIEPPPPPPCIEYSPPPPPPVVHYSSP----PPPPVYYSSPP 652
Query: 95 DDGEMIGPPPPPPGSNFN 112
PPPPP +++
Sbjct: 653 PPPVYYSSPPPPPPVHYS 670
Score = 37.0 bits (84), Expect = 0.014
Identities = 22/59 (37%), Positives = 28/59 (47%), Gaps = 7/59 (11%)
Query: 53 PSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPP--PPPPGS 109
P+ ++ RPP P P SPP P PPPP P++ + PP PPPP S
Sbjct: 527 PAPTPVYCTRPPPPP-PHSPP----PPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHS 580
Score = 33.5 bits (75), Expect = 0.16
Identities = 19/61 (31%), Positives = 25/61 (40%), Gaps = 5/61 (8%)
Query: 53 PSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPP--HHEADDDDGEMIGPPPPPPGSN 110
P +H PP P+ P +P+V PPPP HH +I PPPP
Sbjct: 682 PPPSPVHYSSPPPPPSAPCEESPPPAPVVHHSPPPPMVHHSPPP---PVIHQSPPPPSPE 738
Query: 111 F 111
+
Sbjct: 739 Y 739
Score = 32.0 bits (71), Expect = 0.45
Identities = 22/71 (30%), Positives = 26/71 (35%), Gaps = 5/71 (7%)
Query: 39 SVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPP--HHEADDDD 96
S P P P E + PP + SPP +P PPP P HH
Sbjct: 659 SSPPPPPPVHYSSPPPPEVHYHSPPPSPVHYSSPPPPPSAPCEESPPPAPVVHHSPPP-- 716
Query: 97 GEMIGPPPPPP 107
M+ PPPP
Sbjct: 717 -PMVHHSPPPP 726
Score = 31.6 bits (70), Expect = 0.59
Identities = 22/62 (35%), Positives = 26/62 (41%), Gaps = 9/62 (14%)
Query: 53 PSKECLHSIRPPKNP---NPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGS 109
PS C S PP P + P PP+ SP PPP H E GP PP G
Sbjct: 696 PSAPCEES--PPPAPVVHHSPPPPMVHHSP----PPPVIHQSPPPPSPEYEGPLPPVIGV 749
Query: 110 NF 111
++
Sbjct: 750 SY 751
Score = 27.7 bits (60), Expect = 8.5
Identities = 18/54 (33%), Positives = 20/54 (36%), Gaps = 7/54 (12%)
Query: 54 SKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
S C S+ P P PP L PPPP P + PPPP P
Sbjct: 385 SFSCGRSVSPRPPVVTPLPP----PSLPSPPPPAPIFSTPP---TLTSPPPPSP 431
>At3g18810 protein kinase, putative
Length = 700
Score = 45.4 bits (106), Expect = 4e-05
Identities = 29/98 (29%), Positives = 42/98 (42%), Gaps = 14/98 (14%)
Query: 35 ATRRSVPTSNPKTTSDDFPSKECLHSIRPP----KNPNPPSPPLQDDSPLVGPPPPPPHH 90
A +S S P T S + PP +P+PP+PP DDS P PP
Sbjct: 2 AEGQSPENSPPSPTPPSPSSSDNQQQSSPPPSDSSSPSPPAPPPPDDSSNGSPQPPSSDS 61
Query: 91 EADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGN 128
++ PP P G+N ND + +N++D + N
Sbjct: 62 QS----------PPSPQGNNNNDGNNGNNNNDNNNNNN 89
Score = 31.6 bits (70), Expect = 0.59
Identities = 18/42 (42%), Positives = 20/42 (46%), Gaps = 9/42 (21%)
Query: 83 PPPPPPHHEADDDDGEMIGPPPPPP---GSNFNDSEDEDNDS 121
PPPPPP + PPPPPP G N N S+ N S
Sbjct: 267 PPPPPP------GSWQPSPPPPPPPVSGGMNGNSSDFSSNYS 302
Score = 29.6 bits (65), Expect = 2.2
Identities = 19/52 (36%), Positives = 22/52 (41%), Gaps = 13/52 (25%)
Query: 66 NPNPPSPPLQDDSPLVGPPPPPP-----HHEADDDDGEMIGP-----PPPPP 107
+P PP P SP PPPPPP + + D GP PPP P
Sbjct: 266 SPPPPPPGSWQPSP---PPPPPPVSGGMNGNSSDFSSNYSGPHGPSVPPPHP 314
>At3g18060 WD40-repeat protein
Length = 609
Score = 45.4 bits (106), Expect = 4e-05
Identities = 34/144 (23%), Positives = 64/144 (43%), Gaps = 16/144 (11%)
Query: 135 SNEIVLKGHTKVVSALAVDHTGS-RVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQ 193
SN GH++ V + A+ T R+++ D++V Y+ +L S +
Sbjct: 135 SNVGEFDGHSRRVLSCAIKPTRPFRIVTCGEDFLVNFYEGPPFKFKLSS-----REHSNF 189
Query: 194 IRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEW 253
+ + ++P +F+ V+ + IYD + + +L + GH + W
Sbjct: 190 VNCVRFAPDGSKFITVSSDKKGIIYDGKTCEI---------LGELSSDDGHKGSIYAVSW 240
Query: 254 HPKTKETILTSSEDGSLRIWDVND 277
P K+ +LT S D S +IWD++D
Sbjct: 241 SPDGKQ-VLTVSADKSAKIWDISD 263
Score = 33.5 bits (75), Expect = 0.16
Identities = 22/89 (24%), Positives = 41/89 (45%), Gaps = 6/89 (6%)
Query: 133 PLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGH 192
P ++ + H+ V+ + GS+ ++ S D +YD + ++ +GH
Sbjct: 176 PFKFKLSSREHSNFVNCVRFAPDGSKFITVSSDKKGIIYD----GKTCEILGELSSDDGH 231
Query: 193 Q--IRNLSWSPTADRFLCVTGSAQAKIYD 219
+ I +SWSP + L V+ AKI+D
Sbjct: 232 KGSIYAVSWSPDGKQVLTVSADKSAKIWD 260
>At5g52820 Notchless protein homolog
Length = 473
Score = 45.1 bits (105), Expect = 5e-05
Identities = 39/142 (27%), Positives = 63/142 (43%), Gaps = 24/142 (16%)
Query: 140 LKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQ--IRNL 197
+ GH + V ++ G ++ SGS D VR++D L + + +GH+ + +
Sbjct: 105 IAGHAEAVLCVSFSPDGKQLASGSGDTTVRLWD-------LYTETPLFTCKGHKNWVLTV 157
Query: 198 SWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHP-- 255
+WSP V+GS +I KG++ L K ITG++ W P
Sbjct: 158 AWSPDGKHL--VSGSKSGEI------CCWNPKKGELEGSPLTGHKKWITGIS---WEPVH 206
Query: 256 --KTKETILTSSEDGSLRIWDV 275
+TSS+DG RIWD+
Sbjct: 207 LSSPCRRFVTSSKDGDARIWDI 228
Score = 31.2 bits (69), Expect = 0.77
Identities = 32/136 (23%), Positives = 59/136 (42%), Gaps = 22/136 (16%)
Query: 140 LKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSW 199
L GH ++V+ + G + S S+D VR++ N F + + +SW
Sbjct: 356 LTGHQQLVNHVYFSPDGKWIASASFDKSVRLW-----NGITGQFVTVFRGHVGPVYQVSW 410
Query: 200 SPTADRFLCVTGSAQA--KIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKT 257
S AD L ++GS + KI++ ++ +DL GH + +W P
Sbjct: 411 S--ADSRLLLSGSKDSTLKIWE---------IRTKKLKQDL---PGHADEVFAVDWSP-D 455
Query: 258 KETILTSSEDGSLRIW 273
E +++ +D L++W
Sbjct: 456 GEKVVSGGKDRVLKLW 471
>At3g13965 unknown protein
Length = 209
Score = 45.1 bits (105), Expect = 5e-05
Identities = 28/105 (26%), Positives = 44/105 (41%), Gaps = 13/105 (12%)
Query: 21 KQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPL 80
+Q Q+P + P + P+ + PSK ++ + PP P P SPP P
Sbjct: 50 QQQFMQSPPFPPESGNTQPPPLTPPQFNTPPPPSKSTINVMSPPPPPLP-SPPPPPPPPT 108
Query: 81 VGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDE 125
PPPPP I PPPPP + + + +S +++
Sbjct: 109 PSPPPPP------------ISKPPPPPRAQASPPHSNNRESHRNQ 141
Score = 30.0 bits (66), Expect = 1.7
Identities = 20/70 (28%), Positives = 29/70 (40%), Gaps = 11/70 (15%)
Query: 45 PKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPP--------PPPHHEADDDD 96
P+ S P + PP+ SPP +S PPP PPP ++ +
Sbjct: 30 PQLQSPPPPPSPLQNLSPPPQQQFMQSPPFPPESGNTQPPPLTPPQFNTPPPPSKSTIN- 88
Query: 97 GEMIGPPPPP 106
++ PPPPP
Sbjct: 89 --VMSPPPPP 96
Score = 28.9 bits (63), Expect = 3.8
Identities = 26/89 (29%), Positives = 34/89 (37%), Gaps = 15/89 (16%)
Query: 22 QSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNP---NPPSPPLQDDS 78
Q + Q+P S P S FP E ++ PP P N P PP +
Sbjct: 29 QPQLQSPPPPPSPLQNLSPPPQQQFMQSPPFPP-ESGNTQPPPLTPPQFNTPPPPSKSTI 87
Query: 79 PLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
++ PPPP + PPPPPP
Sbjct: 88 NVM--SPPPP---------PLPSPPPPPP 105
>At1g59910 hypothetical protein
Length = 929
Score = 45.1 bits (105), Expect = 5e-05
Identities = 29/85 (34%), Positives = 35/85 (41%), Gaps = 13/85 (15%)
Query: 25 SQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPP 84
+ TPL T + T + P + P + P PP PP PP P PP
Sbjct: 357 ASTPL-TPGQFTTANAPPAPPGPANQTSPPPP-----PPPSAAAPPPPPPPKKGPAAPPP 410
Query: 85 PPPPHHEADDDDGEMIGPPPPPPGS 109
PPPP + GPPPPPP S
Sbjct: 411 PPPPGKKG-------AGPPPPPPMS 428
>At1g24150 unknown protein
Length = 725
Score = 45.1 bits (105), Expect = 5e-05
Identities = 38/120 (31%), Positives = 51/120 (41%), Gaps = 18/120 (15%)
Query: 10 GVRAEFPLSFGKQSKSQTPLETIHKATRRSVPTSNP---KTTSDDFPSKECLHSIRPPKN 66
G E PL G+ S S + IH R+ T++P KT S +F + PP
Sbjct: 194 GPVTETPLLRGRSSTSHS---VIHNDNYRNATTTHPPHVKTDSFEFVKPDPTPPPPPPPP 250
Query: 67 ---------PNPPSPPLQDDSPLVGPPPPPP-HHEADDDDGEMIGPPPPPPGSNFNDSED 116
P PP P L+++ P PPPPPP A PPP P GS+ +S +
Sbjct: 251 IPVKQSATPPPPPPPKLKNNGP--SPPPPPPLKKTAALSSSASKKPPPAPRGSSSGESSN 308
>At2g01330 putative WD-40-repeat protein
Length = 611
Score = 44.3 bits (103), Expect = 9e-05
Identities = 37/152 (24%), Positives = 63/152 (41%), Gaps = 18/152 (11%)
Query: 142 GHTKVVSALAVDHTGS-RVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWS 200
GH++ V + A T R+ + D++V YD F + + + +S
Sbjct: 143 GHSRRVLSCAFKPTRPFRIATCGEDFLVNFYDGPPFK-----FHSSHREHSNFVNCIRYS 197
Query: 201 PTADRFLCVTGSAQAKIYD-RDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKE 259
P +F+ V+ + IYD + G +GE D GH + W P +K
Sbjct: 198 PDGTKFITVSSDKKGMIYDGKTGDKVGELASED----------GHKGSIYAVSWSPDSKR 247
Query: 260 TILTSSEDGSLRIWDVNDFTTQKQVWKIFTLL 291
+LT S D S ++W+V + T V K + +
Sbjct: 248 -VLTVSADKSAKVWEVAEDGTIGSVIKTLSFM 278
>At2g15880 unknown protein
Length = 727
Score = 43.9 bits (102), Expect = 1e-04
Identities = 29/71 (40%), Positives = 33/71 (45%), Gaps = 19/71 (26%)
Query: 37 RRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDD 96
RRS P P S PS +HS PP +PP PP P+ PPPPPP
Sbjct: 490 RRSPPP--PPVHSPPPPSP--IHSPPPPPVYSPPPPP-----PVYSPPPPPP-------- 532
Query: 97 GEMIGPPPPPP 107
+ PPPPPP
Sbjct: 533 --VYSPPPPPP 541
Score = 43.9 bits (102), Expect = 1e-04
Identities = 26/82 (31%), Positives = 36/82 (43%), Gaps = 6/82 (7%)
Query: 28 PLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLV---GPP 84
P + H P S+P T+ + +H +PPK P+ P D SP+ PP
Sbjct: 436 PQQPHHHVVHSPPPASSPPTSPPVHSTPSPVHKPQPPKESPQPNDPY-DQSPVKFRRSPP 494
Query: 85 PPPPHHEADDDDGEMIGPPPPP 106
PPP H + + PPPPP
Sbjct: 495 PPPVH--SPPPPSPIHSPPPPP 514
Score = 41.2 bits (95), Expect = 7e-04
Identities = 19/45 (42%), Positives = 22/45 (48%), Gaps = 4/45 (8%)
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP +PP PP+ P V PPPP H + PPPPPP
Sbjct: 581 PPPVYSPPPPPVHSPPPPVHSPPPPVH----SPPPPVYSPPPPPP 621
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/52 (38%), Positives = 23/52 (43%), Gaps = 3/52 (5%)
Query: 58 LHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEAD---DDDGEMIGPPPPP 106
+HS PP PP PP+ P V PPPP H + PPPPP
Sbjct: 606 VHSPPPPVYSPPPPPPVHSPPPPVFSPPPPVHSPPPPVYSPPPPVYSPPPPP 657
Score = 38.5 bits (88), Expect = 0.005
Identities = 20/53 (37%), Positives = 25/53 (46%), Gaps = 4/53 (7%)
Query: 53 PSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPP 105
P +HS PP + +PP P P+ PPPPPP H + PPPP
Sbjct: 587 PPPPPVHSPPPPVH-SPPPPVHSPPPPVYSPPPPPPVHSPPP---PVFSPPPP 635
Score = 36.6 bits (83), Expect = 0.018
Identities = 21/59 (35%), Positives = 25/59 (41%), Gaps = 9/59 (15%)
Query: 53 PSKECLHSIRPPKNPNPPS-----PPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPP 106
P +HS PP + PP PP+ P V PPPP H + PPPPP
Sbjct: 537 PPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVH----SPPPPVYSPPPPP 591
Score = 35.8 bits (81), Expect = 0.031
Identities = 24/82 (29%), Positives = 30/82 (36%), Gaps = 5/82 (6%)
Query: 25 SQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPP 84
S P IH V + P P ++S PP + P PP+ P V P
Sbjct: 500 SPPPPSPIHSPPPPPVYSPPPPPPVYSPPPPPPVYSPPPPPPVHSPPPPVHSPPPPVHSP 559
Query: 85 PPPPHHEADDDDGEMIGPPPPP 106
PPP H + PPPP
Sbjct: 560 PPPVHSPPPP-----VHSPPPP 576
Score = 34.7 bits (78), Expect = 0.070
Identities = 21/68 (30%), Positives = 28/68 (40%), Gaps = 4/68 (5%)
Query: 43 SNPKTTSDDFPSKECLHSIRPPK-NPNPPSPPLQDDSPLVGPPPPP--PHHEADDDDGEM 99
+ P + P++ +H +PPK +P P P Q PPPP PHH
Sbjct: 392 ATPSKSPSPVPTRP-VHKPQPPKESPQPNDPYNQSPVKFRRSPPPPQQPHHHVVHSPPPA 450
Query: 100 IGPPPPPP 107
PP PP
Sbjct: 451 SSPPTSPP 458
Score = 33.1 bits (74), Expect = 0.20
Identities = 20/60 (33%), Positives = 25/60 (41%), Gaps = 6/60 (10%)
Query: 53 PSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPP-----PHHEADDDDGEMIGPPPPPP 107
P +HS P +P P SPP+ V P PP P+ D + PPPPP
Sbjct: 439 PHHHVVHSPPPASSP-PTSPPVHSTPSPVHKPQPPKESPQPNDPYDQSPVKFRRSPPPPP 497
Score = 31.6 bits (70), Expect = 0.59
Identities = 24/81 (29%), Positives = 29/81 (35%), Gaps = 7/81 (8%)
Query: 28 PLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPN--PPSPPLQDDSPLVGPPP 85
P +H S S P P ++S PP PP PP+ P V PP
Sbjct: 494 PPPPVHSPPPPSPIHSPPPPPVYSPPPPPPVYSPPPPPPVYSPPPPPPVHSPPPPVHSPP 553
Query: 86 PPPHHEADDDDGEMIGPPPPP 106
PP H + PPPP
Sbjct: 554 PPVHSPPPP-----VHSPPPP 569
Score = 31.6 bits (70), Expect = 0.59
Identities = 16/45 (35%), Positives = 19/45 (41%)
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP +PP P P V PPPPP + +M PP P
Sbjct: 640 PPPVYSPPPPVYSPPPPPVKSPPPPPVYSPPLLPPKMSSPPTQTP 684
Score = 31.2 bits (69), Expect = 0.77
Identities = 18/50 (36%), Positives = 20/50 (40%), Gaps = 6/50 (12%)
Query: 63 PPKNPNPP------SPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPP 106
PPK +PP SPP + S V PPP G PPPP
Sbjct: 673 PPKMSSPPTQTPVNSPPPRTPSQTVEAPPPSEEFIIPPFIGHQYASPPPP 722
Score = 29.6 bits (65), Expect = 2.2
Identities = 22/95 (23%), Positives = 31/95 (32%), Gaps = 5/95 (5%)
Query: 23 SKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVG 82
SKS +P+ T + + P +D + PP P + P
Sbjct: 395 SKSPSPVPT--RPVHKPQPPKESPQPNDPYNQSPVKFRRSPPPPQQPHHHVVHSPPPASS 452
Query: 83 PPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDE 117
PP PP H + P PP ND D+
Sbjct: 453 PPTSPPVHST---PSPVHKPQPPKESPQPNDPYDQ 484
Score = 29.3 bits (64), Expect = 2.9
Identities = 20/71 (28%), Positives = 24/71 (33%), Gaps = 17/71 (23%)
Query: 58 LHSIRPPKNPNPPSPPLQDDSPLVGPPPP--------PPHHEADDDDGEMIGPPP----- 104
+HS PP PP P+ PPPP PP + + PPP
Sbjct: 636 VHSPPPPVYSPPPPVYSPPPPPVKSPPPPPVYSPPLLPPKMSSPPTQTPVNSPPPRTPSQ 695
Query: 105 ----PPPGSNF 111
PPP F
Sbjct: 696 TVEAPPPSEEF 706
>At2g19520 WD-40 repeat protein MSI4 (MSI4)
Length = 507
Score = 43.5 bits (101), Expect = 1e-04
Identities = 28/92 (30%), Positives = 44/92 (47%), Gaps = 9/92 (9%)
Query: 185 QIEPSEGHQIRNLSWSPTADRFLCVTGSAQ--AKIYDRDGLTLGEFVKGDMYIRDLKNTK 242
++E + + + W+P D L +TGSA +++DR LT V +Y +
Sbjct: 331 KVEKAHDADLHCVDWNPHDDN-LILTGSADNTVRLFDRRKLTANG-VGSPIY-----KFE 383
Query: 243 GHITGLTCGEWHPKTKETILTSSEDGSLRIWD 274
GH + C +W P +S+EDG L IWD
Sbjct: 384 GHKAAVLCVQWSPDKSSVFGSSAEDGLLNIWD 415
Score = 38.5 bits (88), Expect = 0.005
Identities = 47/186 (25%), Positives = 77/186 (41%), Gaps = 38/186 (20%)
Query: 137 EIVLKGHTKVVS-ALAVDHTGSRVLSGSYDYMVRMYDFQ------GMNSRL------QSF 183
+++L GH ALA+ T VLSG D V ++ Q G +S+ Q+
Sbjct: 212 DLILTGHQDNAEFALAMCPTEPFVLSGGKDKSVVLWSIQDHITTIGTDSKSSGSIIKQTG 271
Query: 184 RQIEPSE-----------GHQ--IRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVK 230
+ +E GH+ + ++++SPT+ + C G D L L +
Sbjct: 272 EGTDKNESPTVGPRGVYHGHEDTVEDVAFSPTSAQEFCSVG-------DDSCLILWDART 324
Query: 231 GDMYIRDLKNTKGHITGLTCGEWHPKTKETILTSSEDGSLRIWDVNDFTTQKQVWKIFTL 290
G + ++ K H L C +W+P ILT S D ++R++D T I+
Sbjct: 325 GTNPVTKVE--KAHDADLHCVDWNPHDDNLILTGSADNTVRLFDRRKLTANGVGSPIYK- 381
Query: 291 LFFHGH 296
F GH
Sbjct: 382 --FEGH 385
Score = 31.6 bits (70), Expect = 0.59
Identities = 38/160 (23%), Positives = 65/160 (39%), Gaps = 25/160 (15%)
Query: 154 HTGSRVLSGSYDYMVRMYD-----FQGMNSRLQSFRQIEPSEGHQ--IRNLSWSPTADRF 206
H + +L+GS D VR++D G+ S + F EGH+ + + WSP D+
Sbjct: 348 HDDNLILTGSADNTVRLFDRRKLTANGVGSPIYKF------EGHKAAVLCVQWSP--DKS 399
Query: 207 LCVTGSAQ---AKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILT 263
SA+ I+D D ++ GH + W+ TI++
Sbjct: 400 SVFGSSAEDGLLNIWDYDRVSKKSDRAAKSPAGLFFQHAGHRDKVVDFHWNASDPWTIVS 459
Query: 264 SSED-------GSLRIWDVNDFTTQKQVWKIFTLLFFHGH 296
S+D G+L+IW ++D + + + L F H
Sbjct: 460 VSDDCETTGGGGTLQIWRMSDLIYRPEEEVVAELEKFKSH 499
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,315,701
Number of Sequences: 26719
Number of extensions: 526620
Number of successful extensions: 9448
Number of sequences better than 10.0: 466
Number of HSP's better than 10.0 without gapping: 223
Number of HSP's successfully gapped in prelim test: 249
Number of HSP's that attempted gapping in prelim test: 3255
Number of HSP's gapped (non-prelim): 2579
length of query: 316
length of database: 11,318,596
effective HSP length: 99
effective length of query: 217
effective length of database: 8,673,415
effective search space: 1882131055
effective search space used: 1882131055
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146819.11


