

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146819.10 - phase: 0
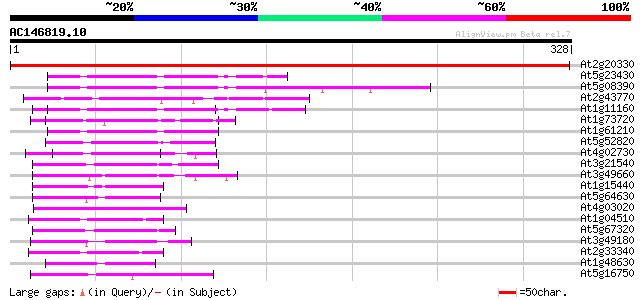
(328 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g20330 putative WD-40 repeat protein 561 e-160
At5g23430 unknown protein 55 4e-08
At5g08390 katanin p80 subunit - like protein 55 4e-08
At2g43770 putative splicing factor 53 3e-07
At1g11160 hypothetical protein 52 3e-07
At1g73720 unknown protein 51 8e-07
At1g61210 50 1e-06
At5g52820 Notchless protein homolog 49 4e-06
At4g02730 putative WD-repeat protein 45 7e-05
At3g21540 WD repeat like protein 44 9e-05
At3g49660 putative WD-40 repeat - protein 44 1e-04
At1g15440 unknown protein 44 1e-04
At5g64630 FAS2 like protein 44 2e-04
At4g03020 putative WD-repeat protein 44 2e-04
At1g04510 putative pre-mRNA splicing factor PRP19 43 2e-04
At5g67320 unknown protein 42 6e-04
At3g49180 unknown protein 42 6e-04
At2g33340 putative PRP19-like spliceosomal protein 42 6e-04
At1g48630 unknown protein 42 6e-04
At5g16750 WD40-repeat protein 41 8e-04
>At2g20330 putative WD-40 repeat protein
Length = 648
Score = 561 bits (1447), Expect = e-160
Identities = 265/327 (81%), Positives = 295/327 (90%)
Query: 1 VIKPKLARPGRVPVTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDI 60
VIKPKLARPGRVPVTTCAWD DGK IAGG+GDGSIQ+WS+KPGWGSRPDI+ K H DDI
Sbjct: 317 VIKPKLARPGRVPVTTCAWDRDGKRIAGGVGDGSIQIWSLKPGWGSRPDIYVGKAHTDDI 376
Query: 61 TSLKFSADGRILLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTNVGFSPDEQLFFTG 120
TS+KFS+DGRILLSRSFD SLKVWDLR+ KEALKVFE LPN+Y QTNV FSPDEQ+ TG
Sbjct: 377 TSVKFSSDGRILLSRSFDGSLKVWDLRQMKEALKVFEGLPNYYPQTNVAFSPDEQIILTG 436
Query: 121 TSVEKDSTTGGLLCFFDRVNLELVSKVGISPTCSVVQCSWHPKLNQIFASVGDKSQGGTH 180
TSVEKDSTTGGLLCF+DR LE+V KVGISPT SVVQC+WHP+LNQIFA+ GDKSQGGTH
Sbjct: 437 TSVEKDSTTGGLLCFYDRTKLEIVQKVGISPTSSVVQCAWHPRLNQIFATSGDKSQGGTH 496
Query: 181 ILYDPTISERGALVCVARAPRKKSVDDFVANPVIHNPHALPLFRDQPNRKRQREKVLKDP 240
ILYDP+ SERGA VCVARAPRKKSVDD+ PVIHNPHALPLFRD P+RKR+REK LKDP
Sbjct: 497 ILYDPSQSERGACVCVARAPRKKSVDDYQPEPVIHNPHALPLFRDAPSRKREREKALKDP 556
Query: 241 LKSHKPEMPMTGPGFGGRVGTSQGSLLTQYLLKKGGMIKETWMDEDPREAILKYADVAAK 300
+KSHKPE+PMTGPG GGRVGT+ LLTQYLLK+GGMIKETWM+EDPREAILKYA+VA K
Sbjct: 557 MKSHKPEIPMTGPGHGGRVGTTGSGLLTQYLLKQGGMIKETWMEEDPREAILKYAEVAVK 616
Query: 301 EPKFIAPAYAETQPEPLFAKSDSEDEE 327
+PKFIAPAY++TQP+ +FAKSD E+EE
Sbjct: 617 DPKFIAPAYSQTQPKTIFAKSDDEEEE 643
>At5g23430 unknown protein
Length = 837
Score = 55.5 bits (132), Expect = 4e-08
Identities = 46/142 (32%), Positives = 73/142 (51%), Gaps = 19/142 (13%)
Query: 23 GKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDDSLK 82
G+ A G D ++++W I+ + IH K H + L+F+ DGR ++S D+ +K
Sbjct: 113 GEFFASGSLDTNLKIWDIR----KKGCIHTYKGHTRGVNVLRFTPDGRWVVSGGEDNIVK 168
Query: 83 VWDLRKTKEALKVFEELPNHYGQ-TNVGFSPDEQLFFTGTSVEKDSTTGGLLCFFDRVNL 141
VWDL A K+ E +H GQ ++ F P E L TG++ D T + F+D
Sbjct: 169 VWDL----TAGKLLTEFKSHEGQIQSLDFHPHEFLLATGSA---DRT----VKFWDLETF 217
Query: 142 ELVSKVGISPTCSVVQC-SWHP 162
EL+ G P + V+C S++P
Sbjct: 218 ELIGSGG--PETAGVRCLSFNP 237
Score = 39.3 bits (90), Expect = 0.003
Identities = 34/128 (26%), Positives = 57/128 (43%), Gaps = 13/128 (10%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILL 73
+ + +D +A G G+I++W ++ + H + S+ F G
Sbjct: 62 IDSVTFDASEVLVAAGAASGTIKLWDLEEA----KIVRTLTGHRSNCISVDFHPFGEFFA 117
Query: 74 SRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTNV-GFSPDEQLFFTG--TSVEK--DST 128
S S D +LK+WD+RK K + ++ H NV F+PD + +G ++ K D T
Sbjct: 118 SGSLDTNLKIWDIRK-KGCIHTYK---GHTRGVNVLRFTPDGRWVVSGGEDNIVKVWDLT 173
Query: 129 TGGLLCFF 136
G LL F
Sbjct: 174 AGKLLTEF 181
>At5g08390 katanin p80 subunit - like protein
Length = 823
Score = 55.5 bits (132), Expect = 4e-08
Identities = 61/248 (24%), Positives = 103/248 (40%), Gaps = 39/248 (15%)
Query: 23 GKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDDSLK 82
G+ A G D ++++W I+ + IH K H + L+F+ DGR ++S D+ +K
Sbjct: 206 GEFFASGSLDTNLKIWDIR----KKGCIHTYKGHTRGVNVLRFTPDGRWIVSGGEDNVVK 261
Query: 83 VWDLRKTKEALKVFEELPNHYGQ-TNVGFSPDEQLFFTGTSVEKDSTTGGLLCFFDRVNL 141
VWDL A K+ E +H G+ ++ F P E L TG++ D T + F+D
Sbjct: 262 VWDL----TAGKLLHEFKSHEGKIQSLDFHPHEFLLATGSA---DKT----VKFWDLETF 310
Query: 142 ELVSKVG----------ISPTCSVVQCSWHPKLNQIFASVGDKSQGGTHI-------LYD 184
EL+ G +P V C L + G +Q +H
Sbjct: 311 ELIGSGGTETTGVRCLTFNPDGKSVLCGLQESLKRTEPMSGGATQSNSHPEKTSGSGRDQ 370
Query: 185 PTISERGALVCVARAPRKKSVDDFV-----ANPVIHNPHALPLFRDQPNRKRQREKVLKD 239
+++ + V + + P + VD + + + ++ PL +D + R D
Sbjct: 371 AGLNDNSSKVILGKLPGSQKVDPLLKETKSLGKLSVSQNSDPLPKDTKSTGRSSVSQSSD 430
Query: 240 PL-KSHKP 246
PL K KP
Sbjct: 431 PLVKEPKP 438
Score = 40.0 bits (92), Expect = 0.002
Identities = 34/125 (27%), Positives = 56/125 (44%), Gaps = 13/125 (10%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILL 73
+ + +D +A G G+I++W ++ + H + S+ F G
Sbjct: 155 IDSVTFDASEGLVAAGAASGTIKLWDLEEA----KVVRTLTGHRSNCVSVNFHPFGEFFA 210
Query: 74 SRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTNV-GFSPDEQLFFTG--TSVEK--DST 128
S S D +LK+WD+RK K + ++ H NV F+PD + +G +V K D T
Sbjct: 211 SGSLDTNLKIWDIRK-KGCIHTYK---GHTRGVNVLRFTPDGRWIVSGGEDNVVKVWDLT 266
Query: 129 TGGLL 133
G LL
Sbjct: 267 AGKLL 271
>At2g43770 putative splicing factor
Length = 343
Score = 52.8 bits (125), Expect = 3e-07
Identities = 46/173 (26%), Positives = 80/173 (45%), Gaps = 19/173 (10%)
Query: 9 PGRVPVTTCAW-DYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSA 67
P + +T ++ D K GG+ D ++VW ++ G + + H+D IT + S
Sbjct: 178 PDKYQITAVSFSDAADKIFTGGV-DNDVKVWDLRKGEATMT----LEGHQDTITGMSLSP 232
Query: 68 DGRILLSRSFDDSLKVWDLR---KTKEALKVFEELPNHYGQT--NVGFSPDEQLFFTGTS 122
DG LL+ D+ L VWD+R +K+FE +++ + +SPD GT
Sbjct: 233 DGSYLLTNGMDNKLCVWDMRPYAPQNRCVKIFEGHQHNFEKNLLKCSWSPD------GTK 286
Query: 123 VEKDSTTGGLLCFFDRVNLELVSKVGISPTCSVVQCSWHPKLNQIFASVGDKS 175
V S + ++ +D + + K+ T SV +C +HP I + DK+
Sbjct: 287 VTAGS-SDRMVHIWDTTSRRTIYKLP-GHTGSVNECVFHPTEPIIGSCSSDKN 337
Score = 38.1 bits (87), Expect = 0.007
Identities = 24/84 (28%), Positives = 42/84 (49%), Gaps = 3/84 (3%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILL 73
V T ++ G IA G D I +W + G + K H++ I L +++DG ++
Sbjct: 56 VYTMKFNPAGTLIASGSHDREIFLWRVH---GDCKNFMVLKGHKNAILDLHWTSDGSQIV 112
Query: 74 SRSFDDSLKVWDLRKTKEALKVFE 97
S S D +++ WD+ K+ K+ E
Sbjct: 113 SASPDKTVRAWDVETGKQIKKMAE 136
Score = 37.7 bits (86), Expect = 0.009
Identities = 29/103 (28%), Positives = 47/103 (45%), Gaps = 9/103 (8%)
Query: 19 WDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGR-ILLSRSF 77
W DG I D +++ W ++ G I H + S + G +++S S
Sbjct: 104 WTSDGSQIVSASPDKTVRAWDVETG----KQIKKMAEHSSFVNSCCPTRRGPPLIISGSD 159
Query: 78 DDSLKVWDLRKTKEALKVFEELPNHYGQTNVGFSPDEQLFFTG 120
D + K+WD+R+ + A++ F P+ Y T V FS FTG
Sbjct: 160 DGTAKLWDMRQ-RGAIQTF---PDKYQITAVSFSDAADKIFTG 198
>At1g11160 hypothetical protein
Length = 974
Score = 52.4 bits (124), Expect = 3e-07
Identities = 45/152 (29%), Positives = 72/152 (46%), Gaps = 17/152 (11%)
Query: 23 GKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDDSLK 82
G+ +A G D +++VW + + I K H I++++FS DGR ++S D+ +K
Sbjct: 61 GEFLASGSSDTNLRVWDTR----KKGCIQTYKGHTRGISTIEFSPDGRWVVSGGLDNVVK 116
Query: 83 VWDLRKTKEALKVFEELPNHYGQ-TNVGFSPDEQLFFTGTSVEKDSTTGGLLCFFDRVNL 141
VWDL A K+ E H G ++ F P E L TG++ D T + F+D
Sbjct: 117 VWDL----TAGKLLHEFKCHEGPIRSLDFHPLEFLLATGSA---DRT----VKFWDLETF 165
Query: 142 ELVSKVGISPTCSVVQCSWHPKLNQIFASVGD 173
EL+ T V ++HP +F + D
Sbjct: 166 ELIGTTRPEAT-GVRAIAFHPDGQTLFCGLDD 196
Score = 49.3 bits (116), Expect = 3e-06
Identities = 34/107 (31%), Positives = 50/107 (45%), Gaps = 7/107 (6%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILL 73
++T + DG+ + G D ++VW + G +H KCHE I SL F +L
Sbjct: 94 ISTIEFSPDGRWVVSGGLDNVVKVWDLTAG----KLLHEFKCHEGPIRSLDFHPLEFLLA 149
Query: 74 SRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTNVGFSPDEQLFFTG 120
+ S D ++K WDL +T E + P G + F PD Q F G
Sbjct: 150 TGSADRTVKFWDL-ETFELIGTTR--PEATGVRAIAFHPDGQTLFCG 193
>At1g73720 unknown protein
Length = 511
Score = 51.2 bits (121), Expect = 8e-07
Identities = 36/111 (32%), Positives = 52/111 (46%), Gaps = 9/111 (8%)
Query: 13 PVTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRIL 72
PV + D + +A G DG I++W I+ G R F+ H +TSL FS DG L
Sbjct: 265 PVLCIDFSRDSEMLASGSQDGKIKIWRIRTGVCIR---RFD-AHSQGVTSLSFSRDGSQL 320
Query: 73 LSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTNVG-FSPDEQLFFTGTS 122
LS SFD + ++ L+ K+ +E H N F+ D T +S
Sbjct: 321 LSTSFDQTARIHGLKSG----KLLKEFRGHTSYVNHAIFTSDGSRIITASS 367
Score = 44.7 bits (104), Expect = 7e-05
Identities = 33/116 (28%), Positives = 57/116 (48%), Gaps = 10/116 (8%)
Query: 22 DGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEK-----CHEDDITSLKFSADGRILLSRS 76
DG+ +A DG I+VW G + D+ ++ H+D + + FS D +L S S
Sbjct: 224 DGQFLASSSVDGFIEVWDYISG-KLKKDLQYQADESFMMHDDPVLCIDFSRDSEMLASGS 282
Query: 77 FDDSLKVWDLRKTKEALKVFEELPNHYGQTNVGFSPDEQLFFTGTSVEKDSTTGGL 132
D +K+W +R T ++ F+ + G T++ FS D + TS ++ + GL
Sbjct: 283 QDGKIKIWRIR-TGVCIRRFD--AHSQGVTSLSFSRDGSQLLS-TSFDQTARIHGL 334
Score = 27.7 bits (60), Expect = 8.9
Identities = 11/22 (50%), Positives = 15/22 (68%)
Query: 64 KFSADGRILLSRSFDDSLKVWD 85
+FS DG+ L S S D ++VWD
Sbjct: 220 RFSPDGQFLASSSVDGFIEVWD 241
>At1g61210
Length = 282
Score = 50.4 bits (119), Expect = 1e-06
Identities = 32/101 (31%), Positives = 54/101 (52%), Gaps = 9/101 (8%)
Query: 23 GKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDDSLK 82
G+ +A G D ++++W I+ + I K H I++++F+ DGR ++S D+ +K
Sbjct: 142 GEFLASGSSDANLKIWDIR----KKGCIQTYKGHSRGISTIRFTPDGRWVVSGGLDNVVK 197
Query: 83 VWDLRKTKEALKVFEELPNHYGQ-TNVGFSPDEQLFFTGTS 122
VWDL A K+ E H G ++ F P E L TG++
Sbjct: 198 VWDL----TAGKLLHEFKFHEGPIRSLDFHPLEFLLATGSA 234
Score = 40.4 bits (93), Expect = 0.001
Identities = 34/124 (27%), Positives = 60/124 (47%), Gaps = 11/124 (8%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILL 73
V + A+D + G G I++W ++ R H + ++++F G L
Sbjct: 91 VDSVAFDSAEVLVLAGASSGVIKLWDVEEAKMVRAFTG----HRSNCSAVEFHPFGEFLA 146
Query: 74 SRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTNVGFSPDEQLFFTG--TSVEK--DSTT 129
S S D +LK+WD+RK K ++ ++ + G + + F+PD + +G +V K D T
Sbjct: 147 SGSSDANLKIWDIRK-KGCIQTYK--GHSRGISTIRFTPDGRWVVSGGLDNVVKVWDLTA 203
Query: 130 GGLL 133
G LL
Sbjct: 204 GKLL 207
Score = 37.7 bits (86), Expect = 0.009
Identities = 23/73 (31%), Positives = 36/73 (48%), Gaps = 4/73 (5%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILL 73
++T + DG+ + G D ++VW + G +H K HE I SL F +L
Sbjct: 175 ISTIRFTPDGRWVVSGGLDNVVKVWDLTAG----KLLHEFKFHEGPIRSLDFHPLEFLLA 230
Query: 74 SRSFDDSLKVWDL 86
+ S D ++K WDL
Sbjct: 231 TGSADRTVKFWDL 243
>At5g52820 Notchless protein homolog
Length = 473
Score = 48.9 bits (115), Expect = 4e-06
Identities = 34/100 (34%), Positives = 55/100 (55%), Gaps = 9/100 (9%)
Query: 22 DGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDDSL 81
DGK IA D S+++W+ G + + H + + +SAD R+LLS S D +L
Sbjct: 371 DGKWIASASFDKSVRLWNGITG----QFVTVFRGHVGPVYQVSWSADSRLLLSGSKDSTL 426
Query: 82 KVWDLRKTKEALKVFEELPNHYGQT-NVGFSPDEQLFFTG 120
K+W++R TK K+ ++LP H + V +SPD + +G
Sbjct: 427 KIWEIR-TK---KLKQDLPGHADEVFAVDWSPDGEKVVSG 462
Score = 36.6 bits (83), Expect = 0.019
Identities = 19/72 (26%), Positives = 37/72 (51%), Gaps = 4/72 (5%)
Query: 13 PVTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRIL 72
PV +W D + + G D ++++W I+ + D+ H D++ ++ +S DG +
Sbjct: 404 PVYQVSWSADSRLLLSGSKDSTLKIWEIRTK-KLKQDL---PGHADEVFAVDWSPDGEKV 459
Query: 73 LSRSFDDSLKVW 84
+S D LK+W
Sbjct: 460 VSGGKDRVLKLW 471
Score = 35.8 bits (81), Expect = 0.033
Identities = 25/74 (33%), Positives = 40/74 (53%), Gaps = 8/74 (10%)
Query: 56 HEDDITSLKFSADGRILLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQT-NVGFSPDE 114
H+ + + FS DG+ + S SFD S+++W+ T + + VF H G V +S D
Sbjct: 359 HQQLVNHVYFSPDGKWIASASFDKSVRLWN-GITGQFVTVFR---GHVGPVYQVSWSADS 414
Query: 115 QLFFTGTSVEKDST 128
+L +G+ KDST
Sbjct: 415 RLLLSGS---KDST 425
Score = 34.3 bits (77), Expect = 0.095
Identities = 18/67 (26%), Positives = 37/67 (54%), Gaps = 4/67 (5%)
Query: 22 DGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDDSL 81
DGK +A G GD ++++W + + + K H++ + ++ +S DG+ L+S S +
Sbjct: 120 DGKQLASGSGDTTVRLWDLY----TETPLFTCKGHKNWVLTVAWSPDGKHLVSGSKSGEI 175
Query: 82 KVWDLRK 88
W+ +K
Sbjct: 176 CCWNPKK 182
Score = 32.0 bits (71), Expect = 0.47
Identities = 23/82 (28%), Positives = 34/82 (41%), Gaps = 2/82 (2%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPG--WGSRPDIHFEKCHEDDITSLKFSADGRI 71
V T AW DGK + G G I W+ K G GS H + + S+ R
Sbjct: 154 VLTVAWSPDGKHLVSGSKSGEICCWNPKKGELEGSPLTGHKKWITGISWEPVHLSSPCRR 213
Query: 72 LLSRSFDDSLKVWDLRKTKEAL 93
++ S D ++WD+ K +
Sbjct: 214 FVTSSKDGDARIWDITLKKSII 235
Score = 30.8 bits (68), Expect = 1.1
Identities = 33/143 (23%), Positives = 62/143 (43%), Gaps = 13/143 (9%)
Query: 56 HEDDITSLKFSADGRILLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTNVGFSPDEQ 115
H + + + FS DG+ L S S D ++++WDL T+ L + N V +SPD +
Sbjct: 108 HAEAVLCVSFSPDGKQLASGSGDTTVRLWDL-YTETPLFTCKGHKNWV--LTVAWSPDGK 164
Query: 116 LFFTGTSVEKDSTTGGLLCFFDRVNLELVSKVGISPTCSVVQCSWHP-KLNQIFASVGDK 174
+G+ G +C ++ EL + SW P L+
Sbjct: 165 HLVSGSK-------SGEICCWNPKKGELEGSPLTGHKKWITGISWEPVHLSSPCRRFVTS 217
Query: 175 SQGGTHILYDPTISERGALVCVA 197
S+ G ++D T+ + +++C++
Sbjct: 218 SKDGDARIWDITLKK--SIICLS 238
>At4g02730 putative WD-repeat protein
Length = 333
Score = 44.7 bits (104), Expect = 7e-05
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 4/63 (6%)
Query: 26 IAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDDSLKVWD 85
I G D +I++W +K G + K H I+S+ F+ DG +++S S D S K+WD
Sbjct: 143 IVSGSFDETIRIWEVKTG----KCVRMIKAHSMPISSVHFNRDGSLIVSASHDGSCKIWD 198
Query: 86 LRK 88
++
Sbjct: 199 AKE 201
Score = 44.7 bits (104), Expect = 7e-05
Identities = 34/117 (29%), Positives = 51/117 (43%), Gaps = 15/117 (12%)
Query: 10 GRVPVTTCA-WDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSAD 68
G +C + DG +A D ++ +WS + IH + H I+ L +S+D
Sbjct: 41 GHTAAISCVKFSNDGNLLASASVDKTMILWSAT----NYSLIHRYEGHSSGISDLAWSSD 96
Query: 69 GRILLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTN----VGFSPDEQLFFTGT 121
S S D +L++WD R E LKV G TN V F+P L +G+
Sbjct: 97 SHYTCSASDDCTLRIWDARSPYECLKVLR------GHTNFVFCVNFNPPSNLIVSGS 147
Score = 40.4 bits (93), Expect = 0.001
Identities = 37/166 (22%), Positives = 72/166 (43%), Gaps = 11/166 (6%)
Query: 5 KLARPGRVPVTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLK 64
++ + +P+++ ++ DG I DGS ++W K G + I + ++ K
Sbjct: 164 RMIKAHSMPISSVHFNRDGSLIVSASHDGSCKIWDAKEGTCLKTLIDDK---SPAVSFAK 220
Query: 65 FSADGRILLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTNVGFSPDEQLFFTGTSVE 124
FS +G+ +L + D +LK+ + T + LKV+ G TN F T
Sbjct: 221 FSPNGKFILVATLDSTLKLSNY-ATGKFLKVYT------GHTNKVFCITSAFSVTNGKYI 273
Query: 125 KDSTTGGLLCFFDRVNLELVSKVGISPTCSVVQCSWHPKLNQIFAS 170
+ + +D ++ ++ T +V+ S HP N+I +S
Sbjct: 274 VSGSEDNCVYLWDLQARNILQRLE-GHTDAVISVSCHPVQNEISSS 318
Score = 33.9 bits (76), Expect = 0.12
Identities = 13/77 (16%), Positives = 38/77 (48%), Gaps = 3/77 (3%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILL 73
++ AW D D ++++W + + + + H + + + F+ +++
Sbjct: 88 ISDLAWSSDSHYTCSASDDCTLRIWDARSPYEC---LKVLRGHTNFVFCVNFNPPSNLIV 144
Query: 74 SRSFDDSLKVWDLRKTK 90
S SFD+++++W+++ K
Sbjct: 145 SGSFDETIRIWEVKTGK 161
>At3g21540 WD repeat like protein
Length = 949
Score = 44.3 bits (103), Expect = 9e-05
Identities = 31/110 (28%), Positives = 55/110 (49%), Gaps = 9/110 (8%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILL 73
VT ++ G +A G D I +W + G + + H D +T L F G+ L+
Sbjct: 109 VTALRYNKVGSMLASGSKDNDIILWDVVGESG----LFRLRGHRDQVTDLVFLDGGKKLV 164
Query: 74 SRSFDDSLKVWDLRKTKEALKVFEELPNHYGQT-NVGFSPDEQLFFTGTS 122
S S D L+VWDL +T+ +++ + H+ + +V P+E+ TG++
Sbjct: 165 SSSKDKFLRVWDL-ETQHCMQI---VSGHHSEVWSVDTDPEERYVVTGSA 210
Score = 40.4 bits (93), Expect = 0.001
Identities = 23/85 (27%), Positives = 43/85 (50%), Gaps = 4/85 (4%)
Query: 2 IKPKLARPGRVPVTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDIT 61
+ P +R G T +A G DGSI++W + G +++F H+ +T
Sbjct: 55 LTPSSSRGGPSLAVTSIASSASSLVAVGYADGSIRIWDTEKG---TCEVNFNS-HKGAVT 110
Query: 62 SLKFSADGRILLSRSFDDSLKVWDL 86
+L+++ G +L S S D+ + +WD+
Sbjct: 111 ALRYNKVGSMLASGSKDNDIILWDV 135
Score = 33.5 bits (75), Expect = 0.16
Identities = 23/84 (27%), Positives = 36/84 (42%), Gaps = 6/84 (7%)
Query: 11 RVPVTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEK-CHEDDITSLKFSADG 69
++PV DG+ I G D ++++W + G D H H D + +KF +
Sbjct: 575 KLPVMCIDISSDGELIVTGSQDKNLKIWGLDFG-----DCHKSIFAHGDSVMGVKFVRNT 629
Query: 70 RILLSRSFDDSLKVWDLRKTKEAL 93
L S D +K WD K + L
Sbjct: 630 HYLFSIGKDRLVKYWDADKFEHLL 653
Score = 27.7 bits (60), Expect = 8.9
Identities = 10/31 (32%), Positives = 20/31 (64%)
Query: 56 HEDDITSLKFSADGRILLSRSFDDSLKVWDL 86
H+ + + S+DG ++++ S D +LK+W L
Sbjct: 574 HKLPVMCIDISSDGELIVTGSQDKNLKIWGL 604
>At3g49660 putative WD-40 repeat - protein
Length = 317
Score = 43.9 bits (102), Expect = 1e-04
Identities = 38/130 (29%), Positives = 63/130 (48%), Gaps = 18/130 (13%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWG--SRPDIHFEKCHEDDITSLKFSADGRI 71
V++ + DG+ +A D +I+ ++I + P F HE+ I+ + FS+D R
Sbjct: 27 VSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPVQEFTG-HENGISDVAFSSDARF 85
Query: 72 LLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTN----VGFSPDEQLFFTGTSVEK-- 125
++S S D +LK+WD+ +T +K G TN V F+P + +G+ E
Sbjct: 86 IVSASDDKTLKLWDV-ETGSLIKTL------IGHTNYAFCVNFNPQSNMIVSGSFDETVR 138
Query: 126 --DSTTGGLL 133
D TTG L
Sbjct: 139 IWDVTTGKCL 148
Score = 40.8 bits (94), Expect = 0.001
Identities = 25/96 (26%), Positives = 46/96 (47%), Gaps = 6/96 (6%)
Query: 26 IAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDDSLKVWD 85
I G D ++++W + G + H D +T++ F+ DG +++S S+D ++WD
Sbjct: 128 IVSGSFDETVRIWDVTTG----KCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRIWD 183
Query: 86 LRKTKEALKVFEELPNHYGQTNVGFSPDEQLFFTGT 121
T +K + N + V FSP+ + GT
Sbjct: 184 -SGTGHCVKTLIDDENP-PVSFVRFSPNGKFILVGT 217
Score = 38.9 bits (89), Expect = 0.004
Identities = 38/168 (22%), Positives = 69/168 (40%), Gaps = 21/168 (12%)
Query: 13 PVTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDD----ITSLKFSAD 68
PVT ++ DG I DG ++W G H K DD ++ ++FS +
Sbjct: 157 PVTAVDFNRDGSLIVSSSYDGLCRIWDSGTG-------HCVKTLIDDENPPVSFVRFSPN 209
Query: 69 GRILLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTNVGFSPDEQLFFTGTSVEKDST 128
G+ +L + D++L++W++ K L + G N + T +
Sbjct: 210 GKFILVGTLDNTLRLWNISSAK-------FLKTYTGHVNAQYCISSAFSVTNGKRIVSGS 262
Query: 129 TGGLLCFFDRVNLELVSKV-GISPTCSVVQCSWHPKLNQIFASVGDKS 175
+ ++ + +L+ K+ G + T V C HP N I + DK+
Sbjct: 263 EDNCVHMWELNSKKLLQKLEGHTETVMNVAC--HPTENLIASGSLDKT 308
Score = 37.7 bits (86), Expect = 0.009
Identities = 26/85 (30%), Positives = 45/85 (52%), Gaps = 8/85 (9%)
Query: 56 HEDDITSLKFSADGRILLSRSFDDSLKVWDLRKTKEAL-KVFEELPNH-YGQTNVGFSPD 113
H ++S+KFS+DGR+L S S D +++ + + + + + +E H G ++V FS D
Sbjct: 23 HNRAVSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPVQEFTGHENGISDVAFSSD 82
Query: 114 EQLFFTGTSVEK-----DSTTGGLL 133
+ F S +K D TG L+
Sbjct: 83 AR-FIVSASDDKTLKLWDVETGSLI 106
>At1g15440 unknown protein
Length = 900
Score = 43.9 bits (102), Expect = 1e-04
Identities = 24/77 (31%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILL 73
V + D + +A G D ++VW++ G I F + H + +T+L F AD LL
Sbjct: 392 VNCVTYSPDSQLLATGADDNKVKVWNVMSGTCF---ITFTE-HTNAVTALHFMADNHSLL 447
Query: 74 SRSFDDSLKVWDLRKTK 90
S S D +++ WD ++ K
Sbjct: 448 SASLDGTVRAWDFKRYK 464
Score = 36.6 bits (83), Expect = 0.019
Identities = 34/114 (29%), Positives = 54/114 (46%), Gaps = 16/114 (14%)
Query: 27 AGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDDSLKVWDL 86
AG + I VWS K G DI HE + L FS ++L S S+D ++++WD+
Sbjct: 491 AGTLDSFEIFVWSKKTG--QIKDIL--SGHEAPVHGLMFSPLTQLLASSSWDYTVRLWDV 546
Query: 87 RKTKEALKVFEELPNHYGQTNVGFSPD-EQLFFTGTSVEKDSTTGGLLCFFDRV 139
+K ++ F +++ V F PD +QL ST G + F+D +
Sbjct: 547 FASKGTVETFR---HNHDVLTVAFRPDGKQL--------ASSTLDGQINFWDTI 589
Score = 28.5 bits (62), Expect = 5.2
Identities = 32/117 (27%), Positives = 42/117 (35%), Gaps = 16/117 (13%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPD---IHFEKCHEDDITSLKFSADGR 70
VT C + + G +G ++ + PD IH +T+ F+ G
Sbjct: 307 VTACDYHQGLDMVVVGFSNGVFGLYQM-------PDFICIHLLSISRQKLTTAVFNERGN 359
Query: 71 ILLSRSFD-DSLKVWDLRKTKEALKVFEELPNHYGQTN-VGFSPDEQLFFTGTSVEK 125
L L VWD R LK HY N V +SPD QL TG K
Sbjct: 360 WLTFGCAKLGQLLVWDWRTETYILKQ----QGHYFDVNCVTYSPDSQLLATGADDNK 412
>At5g64630 FAS2 like protein
Length = 397
Score = 43.5 bits (101), Expect = 2e-04
Identities = 23/80 (28%), Positives = 37/80 (45%), Gaps = 8/80 (10%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPG-----WGSRPDIHFEKCHEDDITSLKFSAD 68
V T + G+ +A G G + +W + P W + F H D+ L++S D
Sbjct: 67 VNTIRFSPSGELLASGADGGELFIWKLHPSETNQSWKVHKSLSF---HRKDVLDLQWSPD 123
Query: 69 GRILLSRSFDDSLKVWDLRK 88
L+S S D+S +WD+ K
Sbjct: 124 DAYLISGSVDNSCIIWDVNK 143
Score = 37.7 bits (86), Expect = 0.009
Identities = 40/172 (23%), Positives = 73/172 (42%), Gaps = 18/172 (10%)
Query: 13 PVTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSR--PDIHFEKC---HEDDITSLKFSA 67
PV T + +A D I++W I G + P + ++ H + +++FS
Sbjct: 15 PVLTVDFHPISGLLATAGADYDIKLWLINSGQAEKKVPSVSYQSSLTYHGCAVNTIRFSP 74
Query: 68 DGRILLSRSFDDSLKVWDLR--KTKEALKVFEELPNHYGQT-NVGFSPDEQLFFTGTSVE 124
G +L S + L +W L +T ++ KV + L H ++ +SPD+ +G SV+
Sbjct: 75 SGELLASGADGGELFIWKLHPSETNQSWKVHKSLSFHRKDVLDLQWSPDDAYLISG-SVD 133
Query: 125 KDSTTGGLLCFFDRVNLELVSKVGISPTCSVVQ-CSWHPKLNQIFASVGDKS 175
C VN V ++ + C VQ +W P + + D++
Sbjct: 134 NS-------CIIWDVNKGSVHQI-LDAHCHYVQGVAWDPLAKYVASLSSDRT 177
>At4g03020 putative WD-repeat protein
Length = 493
Score = 43.5 bits (101), Expect = 2e-04
Identities = 24/89 (26%), Positives = 37/89 (40%)
Query: 15 TTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLS 74
T C D G I G D +VW + G H + +T + DGR +S
Sbjct: 276 TVCFADESGNLILSGSDDNLCKVWDRRCFIGRDKPAGVLVGHLEGVTFIDSRGDGRYFIS 335
Query: 75 RSFDDSLKVWDLRKTKEALKVFEELPNHY 103
D ++K+WD+RK + E+ +Y
Sbjct: 336 NGKDQTIKLWDIRKMSSSAPARHEVLRNY 364
Score = 34.3 bits (77), Expect = 0.095
Identities = 16/35 (45%), Positives = 25/35 (70%)
Query: 60 ITSLKFSADGRILLSRSFDDSLKVWDLRKTKEALK 94
I S+KFS DGR +++ S DDS+ V+DL + +L+
Sbjct: 232 IFSVKFSTDGREVVAGSSDDSIYVYDLEANRVSLR 266
>At1g04510 putative pre-mRNA splicing factor PRP19
Length = 523
Score = 43.1 bits (100), Expect = 2e-04
Identities = 24/79 (30%), Positives = 41/79 (51%), Gaps = 5/79 (6%)
Query: 12 VPVTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRI 71
V T A+ DG + G +++W +K S+ ++ H +ITS+ FS +G
Sbjct: 355 VNYTAAAFHPDGLILGTGTAQSIVKIWDVK----SQANVAKFGGHNGEITSISFSENGYF 410
Query: 72 LLSRSFDDSLKVWDLRKTK 90
L + + D +++WDLRK K
Sbjct: 411 LATAALD-GVRLWDLRKLK 428
Score = 34.3 bits (77), Expect = 0.095
Identities = 24/109 (22%), Positives = 44/109 (40%), Gaps = 1/109 (0%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILL 73
VT+ + D + D ++++W G+ H K H ++ ++ A + +
Sbjct: 267 VTSIKFVGDTDLVLTASSDKTVRIWGCSED-GNYTSRHTLKDHSAEVRAVTVHATNKYFV 325
Query: 74 SRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTNVGFSPDEQLFFTGTS 122
S S D + +DL +V + N T F PD + TGT+
Sbjct: 326 SASLDSTWCFYDLSSGLCLAQVTDASENDVNYTAAAFHPDGLILGTGTA 374
Score = 28.9 bits (63), Expect = 4.0
Identities = 10/29 (34%), Positives = 19/29 (65%)
Query: 56 HEDDITSLKFSADGRILLSRSFDDSLKVW 84
H +TS+KF D ++L+ S D ++++W
Sbjct: 263 HSKKVTSIKFVGDTDLVLTASSDKTVRIW 291
>At5g67320 unknown protein
Length = 613
Score = 41.6 bits (96), Expect = 6e-04
Identities = 28/84 (33%), Positives = 43/84 (50%), Gaps = 6/84 (7%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILL 73
VTT W+ +G +A G DG ++W++ I H+ I SLK++ G LL
Sbjct: 327 VTTLDWNGEGTLLATGSCDGQARIWTLNGEL-----ISTLSKHKGPIFSLKWNKKGDYLL 381
Query: 74 SRSFDDSLKVWDLRKTKEALKVFE 97
+ S D + VWD+ K +E + FE
Sbjct: 382 TGSVDRTAVVWDV-KAEEWKQQFE 404
Score = 40.0 bits (92), Expect = 0.002
Identities = 25/88 (28%), Positives = 40/88 (45%), Gaps = 17/88 (19%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHF-------------EKCHE--D 58
V CAW +A G GD + ++WSI GS +H K +E
Sbjct: 268 VCACAWSPSASLLASGSGDATARIWSIPE--GSFKAVHTGRNINALILKHAKGKSNEKSK 325
Query: 59 DITSLKFSADGRILLSRSFDDSLKVWDL 86
D+T+L ++ +G +L + S D ++W L
Sbjct: 326 DVTTLDWNGEGTLLATGSCDGQARIWTL 353
Score = 35.8 bits (81), Expect = 0.033
Identities = 22/81 (27%), Positives = 37/81 (45%), Gaps = 13/81 (16%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADG---- 69
V WD G +A D + ++W+IK +H + H +I ++++S G
Sbjct: 451 VNCVKWDPTGSLLASCSDDSTAKIWNIK----QSTFVHDLREHTKEIYTIRWSPTGPGTN 506
Query: 70 -----RILLSRSFDDSLKVWD 85
L S SFD ++K+WD
Sbjct: 507 NPNKQLTLASASFDSTVKLWD 527
Score = 32.3 bits (72), Expect = 0.36
Identities = 16/33 (48%), Positives = 21/33 (63%)
Query: 11 RVPVTTCAWDYDGKCIAGGIGDGSIQVWSIKPG 43
R PV + A+ +G+ IA G D SI +WSIK G
Sbjct: 541 REPVYSLAFSPNGEYIASGSLDKSIHIWSIKEG 573
>At3g49180 unknown protein
Length = 438
Score = 41.6 bits (96), Expect = 6e-04
Identities = 27/99 (27%), Positives = 43/99 (43%), Gaps = 20/99 (20%)
Query: 13 PVTTCAWDYDGKCIAGGIGDGSIQVWSIKPG-----WGSRPDIHFEKCHEDDITSLKFSA 67
P+ A + +G + GG G I +W + G W H +T L FS
Sbjct: 80 PIKALAANNEGTYLVGGGISGDIYLWEVATGKLLKKWHG---------HYRSVTCLVFSG 130
Query: 68 DGRILLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQT 106
D +L+S S D S++VW L +++F++ G T
Sbjct: 131 DDSLLVSGSQDGSIRVWSL------IRLFDDFQRQQGNT 163
Score = 32.0 bits (71), Expect = 0.47
Identities = 24/86 (27%), Positives = 37/86 (42%), Gaps = 9/86 (10%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSI-------KPGWGSRPDIHFEKCHEDDITSLKFS 66
VT + D + G DGSI+VWS+ + G+ H H +T +
Sbjct: 123 VTCLVFSGDDSLLVSGSQDGSIRVWSLIRLFDDFQRQQGNTLYEHNFNEHTMSVTDIVID 182
Query: 67 ADG--RILLSRSFDDSLKVWDLRKTK 90
G +++S S D + KVW L + K
Sbjct: 183 YGGCNAVIISSSEDRTCKVWSLSRGK 208
Score = 29.6 bits (65), Expect = 2.3
Identities = 15/48 (31%), Positives = 23/48 (47%)
Query: 60 ITSLKFSADGRILLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTN 107
IT L + ADG +L+S S D + VWD + + + + G N
Sbjct: 266 ITCLAYCADGNLLISGSEDGVVCVWDPKSLRHVRTLIHAKGSRKGPVN 313
>At2g33340 putative PRP19-like spliceosomal protein
Length = 565
Score = 41.6 bits (96), Expect = 6e-04
Identities = 21/79 (26%), Positives = 42/79 (52%), Gaps = 5/79 (6%)
Query: 12 VPVTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRI 71
V T A+ DG + G +++W +K S+ ++ H ++T++ FS +G
Sbjct: 354 VDYTAAAFHPDGLILGTGTSQSVVKIWDVK----SQANVAKFDGHTGEVTAISFSENGYF 409
Query: 72 LLSRSFDDSLKVWDLRKTK 90
L + + +D +++WDLRK +
Sbjct: 410 LATAA-EDGVRLWDLRKLR 427
Score = 29.3 bits (64), Expect = 3.1
Identities = 10/29 (34%), Positives = 19/29 (65%)
Query: 56 HEDDITSLKFSADGRILLSRSFDDSLKVW 84
H +TS+KF D ++L+ S D ++++W
Sbjct: 263 HSKKVTSVKFVGDSDLVLTASADKTVRIW 291
Score = 28.9 bits (63), Expect = 4.0
Identities = 20/67 (29%), Positives = 31/67 (45%), Gaps = 7/67 (10%)
Query: 59 DITSLKFSADGRILLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQ-TNVGFSPDEQLF 117
D T+ F DG IL + + +K+WD++ K H G+ T + FS E +
Sbjct: 355 DYTAAAFHPDGLILGTGTSQSVVKIWDVKSQANVAK----FDGHTGEVTAISFS--ENGY 408
Query: 118 FTGTSVE 124
F T+ E
Sbjct: 409 FLATAAE 415
>At1g48630 unknown protein
Length = 326
Score = 41.6 bits (96), Expect = 6e-04
Identities = 20/64 (31%), Positives = 35/64 (54%), Gaps = 4/64 (6%)
Query: 22 DGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDDSL 81
DG+ G DG +++W + G +R + H D+ S+ FS D R ++S S D ++
Sbjct: 74 DGQFALSGSWDGELRLWDLATGESTRRFVG----HTKDVLSVAFSTDNRQIVSASRDRTI 129
Query: 82 KVWD 85
K+W+
Sbjct: 130 KLWN 133
Score = 34.3 bits (77), Expect = 0.095
Identities = 29/110 (26%), Positives = 54/110 (48%), Gaps = 9/110 (8%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRI-- 71
V + A+ D + I D +I++W+ + I H++ ++ ++FS + +
Sbjct: 108 VLSVAFSTDNRQIVSASRDRTIKLWNTLGE--CKYTISEADGHKEWVSCVRFSPNTLVPT 165
Query: 72 LLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTN-VGFSPDEQLFFTG 120
++S S+D ++KVW+L+ K+ L H G N V SPD L +G
Sbjct: 166 IVSASWDKTVKVWNLQN----CKLRNTLAGHSGYLNTVAVSPDGSLCASG 211
Score = 30.4 bits (67), Expect = 1.4
Identities = 30/113 (26%), Positives = 52/113 (45%), Gaps = 11/113 (9%)
Query: 56 HEDDITSLKFSADGRILLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQTNVGFSPDEQ 115
H + + S+DG+ LS S+D L++WDL T E+ + F + + +V FS D +
Sbjct: 62 HSHFVQDVVLSSDGQFALSGSWDGELRLWDL-ATGESTRRF--VGHTKDVLSVAFSTDNR 118
Query: 116 LFFTGT---SVEKDSTTGGLLCFFDRV--NLELVSKVGISPTC---SVVQCSW 160
+ + +++ +T G + E VS V SP ++V SW
Sbjct: 119 QIVSASRDRTIKLWNTLGECKYTISEADGHKEWVSCVRFSPNTLVPTIVSASW 171
Score = 29.6 bits (65), Expect = 2.3
Identities = 34/124 (27%), Positives = 54/124 (43%), Gaps = 19/124 (15%)
Query: 14 VTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDD----ITSLKFSADG 69
+ T A DG A G DG I +W + G +K + + I SL FS +
Sbjct: 196 LNTVAVSPDGSLCASGGKDGVILLWDLAEG---------KKLYSLEAGSIIHSLCFSPN- 245
Query: 70 RILLSRSFDDSLKVWDL--RKTKEALKV--FEELPNHYGQTNVGFSPDEQLFFTGTSVEK 125
R L + ++S+++WDL + E LKV E G T +G + + ++ T +
Sbjct: 246 RYWLCAATENSIRIWDLESKSVVEDLKVDLKAEAEKTDGSTGIG-NKTKVIYCTSLNWSA 304
Query: 126 DSTT 129
D T
Sbjct: 305 DGNT 308
Score = 27.7 bits (60), Expect = 8.9
Identities = 11/26 (42%), Positives = 14/26 (53%)
Query: 15 TTCAWDYDGKCIAGGIGDGSIQVWSI 40
T+ W DG + G DG I+VW I
Sbjct: 298 TSLNWSADGNTLFSGYTDGVIRVWGI 323
>At5g16750 WD40-repeat protein
Length = 876
Score = 41.2 bits (95), Expect = 8e-04
Identities = 29/110 (26%), Positives = 49/110 (44%), Gaps = 7/110 (6%)
Query: 13 PVTTCAWDYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGR-- 70
PV A G +A D + VW + G+ + H+ + H+ ++S+ F D
Sbjct: 104 PVMGMACHASGGLLATAGADRKVLVWDVDGGFCT----HYFRGHKGVVSSILFHPDSNKN 159
Query: 71 ILLSRSFDDSLKVWDLRKTKEALKVFEELPNHYGQ-TNVGFSPDEQLFFT 119
IL+S S D +++VWDL K + H+ T++ S D F+
Sbjct: 160 ILISGSDDATVRVWDLNAKNTEKKCLAIMEKHFSAVTSIALSEDGLTLFS 209
Score = 34.3 bits (77), Expect = 0.095
Identities = 17/67 (25%), Positives = 30/67 (44%)
Query: 20 DYDGKCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDD 79
D + + G D +++VW + + + + H +TS+ S DG L S D
Sbjct: 155 DSNKNILISGSDDATVRVWDLNAKNTEKKCLAIMEKHFSAVTSIALSEDGLTLFSAGRDK 214
Query: 80 SLKVWDL 86
+ +WDL
Sbjct: 215 VVNLWDL 221
Score = 33.1 bits (74), Expect = 0.21
Identities = 19/75 (25%), Positives = 36/75 (47%), Gaps = 5/75 (6%)
Query: 24 KCIAGGIGDGSIQVWSIKPGWGSRPDIHFEKCHEDDITSLKFSADGRILLSRSFDDSLKV 83
+C+ GD ++++W+I G + FE H + F DG +S D LK+
Sbjct: 554 QCVMTASGDKTVKIWAISDGSCLKT---FEG-HTSSVLRASFITDGTQFVSCGADGLLKL 609
Query: 84 WDLRKTKEALKVFEE 98
W++ T E + +++
Sbjct: 610 WNV-NTSECIATYDQ 623
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,434,120
Number of Sequences: 26719
Number of extensions: 388170
Number of successful extensions: 1377
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 98
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 987
Number of HSP's gapped (non-prelim): 394
length of query: 328
length of database: 11,318,596
effective HSP length: 100
effective length of query: 228
effective length of database: 8,646,696
effective search space: 1971446688
effective search space used: 1971446688
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146819.10


