

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146817.8 + phase: 0
(831 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
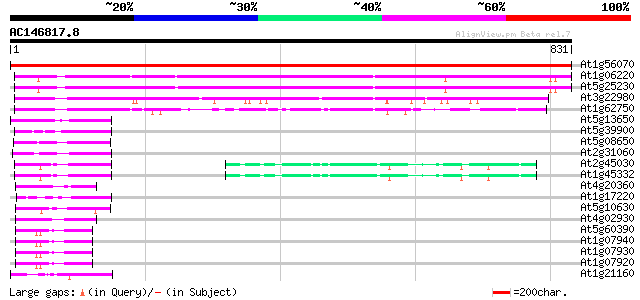
Score E
Sequences producing significant alignments: (bits) Value
At1g56070 elongation factor like protein 1466 0.0
At1g06220 elongation factor like protein 597 e-170
At5g25230 translation Elongation Factor 2 - like protein 584 e-167
At3g22980 eukaryotic translation elongation factor 2, putative 426 e-119
At1g62750 elongation factor G, putative 175 8e-44
At5g13650 GTP-binding protein typA (tyrosine phosphorylated prot... 109 6e-24
At5g39900 GTP-binding protein-like 104 2e-22
At5g08650 GTP-binding protein LepA homolog 99 1e-20
At2g31060 putative GTP-binding protein 97 3e-20
At2g45030 putative mitochondrial translation elongation factor G 84 3e-16
At1g45332 mitochondrial elongation factor, putative 84 3e-16
At4g20360 translation elongation factor EF-Tu precursor, chlorop... 60 5e-09
At1g17220 putative translation initiation factor IF2 50 4e-06
At5g10630 putative protein 50 7e-06
At4g02930 mitochondrial elongation factor Tu 49 2e-05
At5g60390 translation elongation factor eEF-1 alpha chain (gene A4) 48 3e-05
At1g07940 elongation factor 1-alpha 48 3e-05
At1g07930 elongation factor 1-alpha 48 3e-05
At1g07920 elongation factor 1-alpha 48 3e-05
At1g21160 transcription factor, putative 46 1e-04
>At1g56070 elongation factor like protein
Length = 843
Score = 1466 bits (3795), Expect = 0.0
Identities = 711/831 (85%), Positives = 771/831 (92%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD KHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTR DEAERGITIKS
Sbjct: 13 MDYKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEM+D LK+F G R+GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDC+
Sbjct: 73 TGISLYYEMTDESLKSFTGARDGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCI 132
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL +D EEAY T RVIE+ NV+MATY
Sbjct: 133 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYQTFSRVIENANVIMATY 192
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
ED LLGDVQVYPEKGTV+FSAGLHGW+FTLTNFAKMYASKFGV E KMM RLWGENFFD
Sbjct: 193 EDPLLGDVQVYPEKGTVAFSAGLHGWAFTLTNFAKMYASKFGVVESKMMERLWGENFFDP 252
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+T+KW+ K+T +PTCKRGFVQFCYEPIKQII CMNDQKDKLWPML KLGV++K++EKEL
Sbjct: 253 ATRKWSGKNTGSPTCKRGFVQFCYEPIKQIIATCMNDQKDKLWPMLAKLGVSMKNDEKEL 312
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
GK LMKRVMQ+WLPAS+ALLEMMIFHLPSP AQ+YRVENLYEGPLDD YA+AIRNCDP
Sbjct: 313 MGKPLMKRVMQTWLPASTALLEMMIFHLPSPHTAQRYRVENLYEGPLDDQYANAIRNCDP 372
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
GPLMLYVSKMIPASDKGRF+AFGRVF+GKVSTGMKVRIMGPNYIPGEKKDLY KSVQRT
Sbjct: 373 NGPLMLYVSKMIPASDKGRFFAFGRVFAGKVSTGMKVRIMGPNYIPGEKKDLYTKSVQRT 432
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGK+QETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVV VA
Sbjct: 433 VIWMGKRQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVRVA 492
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMVVCT+ E+GEHI+A AGELHLEICLKDLQDDFM G
Sbjct: 493 VQCKVASDLPKLVEGLKRLAKSDPMVVCTMEESGEHIVAGAGELHLEICLKDLQDDFMGG 552
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEI KSDP+VSFRETV ++S+ TVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRD+
Sbjct: 553 AEIIKSDPVVSFRETVCDRSTRTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDD 612
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PK KIL++EFGWDKDLAKK+W FGPETTGPNM+VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 613 PKIRSKILAEEFGWDKDLAKKIWAFGPETTGPNMVVDMCKGVQYLNEIKDSVVAGFQWAS 672
Query: 661 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 720
KEGP+A+EN+RG+CFEVCDVVLH+DAIHRGGGQ+IPTARRV YA+ +TAKPRLLEPVY+V
Sbjct: 673 KEGPLAEENMRGICFEVCDVVLHSDAIHRGGGQVIPTARRVIYASQITAKPRLLEPVYMV 732
Query: 721 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
EIQAPE ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPV+ESF F+ LRA T GQ
Sbjct: 733 EIQAPEGALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVVESFGFSSQLRAATSGQ 792
Query: 781 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
AFPQ VFDHW+M+ SDPLEPGT A+ V +IRK+KGLKE + PLSEFED+L
Sbjct: 793 AFPQCVFDHWEMMSSDPLEPGTQASVLVADIRKRKGLKEAMTPLSEFEDKL 843
>At1g06220 elongation factor like protein
Length = 987
Score = 597 bits (1538), Expect = e-170
Identities = 328/848 (38%), Positives = 503/848 (58%), Gaps = 37/848 (4%)
Query: 7 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGD---VRMTDTRQDEAERGITIKSTGI 63
+RN++++ H+ HGK+ D LV ++ A + ++ TDTR DE ER I+IK+ +
Sbjct: 138 VRNVALVGHLQHGKTVFMDMLVEQTHHMSTFNAKNEKHMKYTDTRVDEQERNISIKAVPM 197
Query: 64 SLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGV 123
SL E S YL N++D+PGHV+FS E+TA+LR+ DGA+++VD EGV
Sbjct: 198 SLVLEDS-----------RSKSYLCNIMDTPGHVNFSDEMTASLRLADGAVLIVDAAEGV 246
Query: 124 CVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYEDA 183
V TE +R A+ + + V+ +NK+DR EL L +AY ++ IE +N ++
Sbjct: 247 MVNTERAIRHAIQDHLPIVVVINKVDRLITELKLPPRDAYYKLRHTIEVINNHISA-AST 305
Query: 184 LLGDVQVY-PEKGTVSFSAGLHGWSFTLTNFAKMYASKFGV--DEEKMMNRLWGENFFDS 240
GD+ + P G V F++G GWSFTL +FAKMYA GV D +K +RLWG+ ++ S
Sbjct: 306 TAGDLPLIDPAAGNVCFASGTAGWSFTLQSFAKMYAKLHGVAMDVDKFASRLWGDVYYHS 365
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
T+ + + +R FVQF EP+ +I + + K + L +LGV L + +L
Sbjct: 366 DTRVF-KRSPPVGGGERAFVQFILEPLYKIYSQVIGEHKKSVETTLAELGVTLSNSAYKL 424
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
+ + L++ S ++S +M++ H+PSP +A +V++ Y G D P ++ CDP
Sbjct: 425 NVRPLLRLACSSVFGSASGFTDMLVKHIPSPREAAARKVDHSYTGTKDSPIYESMVECDP 484
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
GPLM+ V+K+ P SD F FGRV+SG++ TG VR++G Y P +++D+ +K V +
Sbjct: 485 SGPLMVNVTKLYPKSDTSVFDVFGRVYSGRLQTGQSVRVLGEGYSPEDEEDMTIKEVTKL 544
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEK-EVDAHPIRAMKFSVSPVVSV 479
I+ + + V P G+ V + G+D I K ATL N + D + RA++F+ PVV
Sbjct: 545 WIYQARYRIPVSSAPPGSWVLIEGVDASIMKTATLCNASYDEDVYIFRALQFNTLPVVKT 604
Query: 480 AVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMN 539
A S+LPK+VEGL++++KS P+ + + E+GEH I GEL+L+ +KDL++ +
Sbjct: 605 ATEPLNPSELPKMVEGLRKISKSYPLAITKVEESGEHTILGTGELYLDSIMKDLRELYSE 664
Query: 540 GAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRD 599
E+ +DP+VSF ETV+E SS +++PNK N++ M A P++ GLAE I++G +
Sbjct: 665 -VEVKVADPVVSFCETVVESSSMKCFAETPNKKNKITMIAEPLDRGLAEDIENGVVSIDW 723
Query: 600 EPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQY----LNEIKDSVVAG 655
K ++ WD A+ +W FGP+ GPN+L+D + + +KDS+V G
Sbjct: 724 NRKQLGDFFRTKYDWDLLAARSIWAFGPDKQGPNILLDDTLPTEVDRNLMMAVKDSIVQG 783
Query: 656 FQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLE 715
FQ ++EGP+ DE +R V F++ D + + +HRG GQ+IPTARRV Y+A L A PRL+E
Sbjct: 784 FQWGAREGPLCDEPIRNVKFKIVDARIAPEPLHRGSGQMIPTARRVAYSAFLMATPRLME 843
Query: 716 PVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRA 775
PVY VEIQ P + IY+VL+++RGHV ++ +P TP Y VKA+LPVIESF F LR
Sbjct: 844 PVYYVEIQTPIDCVTAIYTVLSRRRGHVTSDVPQPGTPAYIVKAFLPVIESFGFETDLRY 903
Query: 776 QTGGQAFPQLVFDHWDMVPSDPLE------PGTPAAAR------VVEIRKKKGLKEQLIP 823
T GQAF VFDHW +VP DPL+ P PA + +V+ R++KG+ E +
Sbjct: 904 HTQGQAFCLSVFDHWAIVPGDPLDKAIQLRPLEPAPIQHLAREFMVKTRRRKGMSEDVSG 963
Query: 824 LSEFEDRL 831
F++ +
Sbjct: 964 NKFFDEAM 971
>At5g25230 translation Elongation Factor 2 - like protein
Length = 973
Score = 584 bits (1505), Expect = e-167
Identities = 324/849 (38%), Positives = 499/849 (58%), Gaps = 39/849 (4%)
Query: 7 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGD---VRMTDTRQDEAERGITIKSTGI 63
+RN++++ H+ HGK+ D LV ++ A + +R TDTR DE ER I+IK+ +
Sbjct: 124 VRNVALVGHLQHGKTVFMDMLVEQTHRMSTFNAENDKHMRYTDTRVDEQERNISIKAVPM 183
Query: 64 SLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGV 123
SL E S YL N++D+PG+V+FS E+TA+LR+ DGA+ +VD +GV
Sbjct: 184 SLVLEDS-----------RSKSYLCNIMDTPGNVNFSDEMTASLRLADGAVFIVDAAQGV 232
Query: 124 CVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYEDA 183
V TE +R A+ + + V+ +NK+DR EL L +AY ++ IE +N ++
Sbjct: 233 MVNTERAIRHAIQDHLPIVVVINKVDRLITELKLPPRDAYYKLRYTIEVINNHISAASTN 292
Query: 184 LLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGV--DEEKMMNRLWGENFFDSS 241
+ P G V F++G GWSFTL +FA+MYA GV D +K +RLWG+ ++
Sbjct: 293 AADLPLIDPAAGNVCFASGTAGWSFTLQSFARMYAKLHGVAMDVDKFASRLWGDVYYHPD 352
Query: 242 TKKWTNKHTSTPTC--KRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKE 299
T+ + +TS P +R FVQF EP+ +I + + K + L +LGV L + +
Sbjct: 353 TRVF---NTSPPVGGGERAFVQFILEPLYKIYSQVIGEHKKSVETTLAELGVTLSNSAYK 409
Query: 300 LSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCD 359
L+ + L++ S ++S +M++ H+PSP +A +V++ Y G D P ++ CD
Sbjct: 410 LNVRPLLRLACSSVFGSASGFTDMLVKHIPSPREAAARKVDHSYTGTKDSPIYESMVECD 469
Query: 360 PEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQR 419
P GPLM+ V+K+ P SD F FGRV+SG++ TG VR++G Y P +++D+ +K V +
Sbjct: 470 PSGPLMVNVTKLYPKSDTSVFDVFGRVYSGRLQTGQSVRVLGEGYSPEDEEDMTIKEVTK 529
Query: 420 TVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEK-EVDAHPIRAMKFSVSPVVS 478
I+ + + V P G+ V + G+D I K ATL N + D + RA+KF+ PVV
Sbjct: 530 LWIYQARYRIPVSSAPPGSWVLIEGVDASIMKTATLCNASYDEDVYIFRALKFNTLPVVK 589
Query: 479 VAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFM 538
A S+LPK+VEGL++++KS P+ + + E+GEH I GEL+L+ +KDL++ +
Sbjct: 590 TATEPLNPSELPKMVEGLRKISKSYPLAITKVEESGEHTILGTGELYLDSIIKDLRELYS 649
Query: 539 NGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPR 598
++ +DP+VSF ETV+E SS +++PNK N+L M A P++ GLAE I++G +
Sbjct: 650 E-VQVKVADPVVSFCETVVESSSMKCFAETPNKKNKLTMIAEPLDRGLAEDIENGVVSID 708
Query: 599 DEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQY----LNEIKDSVVA 654
++ WD A+ +W FGP+ G N+L+D + + +KDS+V
Sbjct: 709 WNRVQLGDFFRTKYDWDLLAARSIWAFGPDKQGTNILLDDTLPTEVDRNLMMGVKDSIVQ 768
Query: 655 GFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLL 714
GFQ ++EGP+ DE +R V F++ D + + +HRG GQ+IPTARRV Y+A L A PRL+
Sbjct: 769 GFQWGAREGPLCDEPIRNVKFKIVDARIAPEPLHRGSGQMIPTARRVAYSAFLMATPRLM 828
Query: 715 EPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLR 774
EPVY VEIQ P + IY+VL+++RG+V ++ +P TP Y VKA+LPVIESF F LR
Sbjct: 829 EPVYYVEIQTPIDCVTAIYTVLSRRRGYVTSDVPQPGTPAYIVKAFLPVIESFGFETDLR 888
Query: 775 AQTGGQAFPQLVFDHWDMVPSDPLE------PGTPAAAR------VVEIRKKKGLKEQLI 822
T GQAF VFDHW +VP DPL+ P PA + +V+ R++KG+ E +
Sbjct: 889 YHTQGQAFCLSVFDHWAIVPGDPLDKAIQLRPLEPAPIQHLAREFMVKTRRRKGMSEDVS 948
Query: 823 PLSEFEDRL 831
F++ +
Sbjct: 949 GNKFFDEAM 957
>At3g22980 eukaryotic translation elongation factor 2, putative
Length = 1015
Score = 426 bits (1095), Expect = e-119
Identities = 304/972 (31%), Positives = 464/972 (47%), Gaps = 203/972 (20%)
Query: 7 IRNMSVIAHVDHGKSTLTDSLVAAAG--IIAQEVAGDVRMTDTRQDEAERGITIKSTGIS 64
+RN+ ++AHVDHGK+TL D L+A++G ++ +AG +R D +E R IT+KS+ IS
Sbjct: 9 VRNICILAHVDHGKTTLADHLIASSGGGVLHPRLAGKLRFMDYLDEEQRRAITMKSSSIS 68
Query: 65 LYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVC 124
L Y+ Y +NLIDSPGH+DF SEV+ A R++DGALV+VD VEGV
Sbjct: 69 LKYK----------------DYSLNLIDSPGHMDFCSEVSTAARLSDGALVLVDAVEGVH 112
Query: 125 VQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYE--- 181
+QT VLRQA E++ P L +NK+DR EL L EAY+ + R++ VN +++ Y+
Sbjct: 113 IQTHAVLRQAWIEKLTPCLVLNKIDRLIFELRLSPMEAYTRLIRIVHEVNGIVSAYKSEK 172
Query: 182 -----DALLG------------------DVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYA 218
D++L +V P+KG V F L GW F + FA YA
Sbjct: 173 YLSDVDSILASPSGELSAESLELLEDDEEVTFQPQKGNVVFVCALDGWGFGIAEFANFYA 232
Query: 219 SKFGVDEEKMMNRLWGENFFDSSTKKWTNKH--TSTPTCKRGFVQFCYEPIKQIIELCMN 276
SK G + LWG ++ TK K ++ K FVQF EP+ Q+ E ++
Sbjct: 233 SKLGASATALQKSLWGPRYYIPKTKMIVGKKNLSAGSKAKPMFVQFVLEPLWQVYEAALD 292
Query: 277 DQKDKLWPMLQKL--GVNLKSEEKELSG---KALMKRVMQSWLPASSALLEMMIFHLPSP 331
DK +L+K+ NL +EL K +++ VM WLP S A+L M + HLP P
Sbjct: 293 PGGDK--AVLEKVIKSFNLSIPPRELQNKDPKNVLQSVMSRWLPLSDAVLSMAVKHLPDP 350
Query: 332 TKAQKYRVENLYEGPL----DDPYAS----------AIRNCD--PEGPLMLYVS------ 369
AQ YR+ L DD +S +I CD + P +++VS
Sbjct: 351 IAAQAYRIPRLVPERKIIGGDDVDSSVLAEAELVRKSIEACDSSSDSPCVVFVSKMFAIP 410
Query: 370 -KMIPASDKGR------------------FYAFGRVFSGKVSTGMKVRIMGPNYIP--GE 408
KMIP R F AF R+FSG + G +V ++ Y P GE
Sbjct: 411 MKMIPQDGNHRERMNGLNDDDSKSESDECFLAFARIFSGVLRAGQRVFVITALYDPLKGE 470
Query: 409 KKDLYVKSVQ--RTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPI 466
Y++ + + MG+ V +V GN VA+ GL +I+K+ATL++ + + P+
Sbjct: 471 SSHKYIQEAELHSLYLMMGQGLTPVTEVKAGNVVAIRGLGPYISKSATLSSTR--NCWPL 528
Query: 467 RAMKFSVSPVVSVAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHL 526
+M+F VSP + VA+ +D+ L++GL+ L ++DP V T+S GEH++AAAGE+HL
Sbjct: 529 ASMEFQVSPTLRVAIEPSDPADMSALMKGLRLLNRADPFVEITVSARGEHVLAAAGEVHL 588
Query: 527 EICLKDLQDDFMNGAEITKSDPIVSFRETV-------LE-------KSSHTVMSKSPNKH 572
E C+KDL++ F + S P+VS+RET+ LE SS + ++PN
Sbjct: 589 ERCVKDLKERFAK-VNLEVSPPLVSYRETIEGDGSNLLESLRSLSLNSSDYIEKRTPNGR 647
Query: 573 NRLYMEARPMEEGLAEAIDDG------------------------RIGPRDEPKNHLKIL 608
+ + + L + +D+ +G +P LK
Sbjct: 648 CIIRVHVMKLPHALTKLLDENTELLGDIIGGKGSHSVKILESQKPSLGENVDPIEELKKQ 707
Query: 609 SDEFG-----------------WDKDLAKKVWCFGPETTGPNMLVD------------TC 639
E G W K L K++W GP GPN+L
Sbjct: 708 LVEAGVSSSSETEKDREKCKTEWSK-LLKRIWALGPREKGPNILFAPDGKRIAEDGSMLV 766
Query: 640 KGVQYLNE--------------------------IKDSVVAGFQIASKEGPMADENLRGV 673
+G ++++ ++ S+V+GFQ+A+ GP+ DE + G+
Sbjct: 767 RGSPHVSQRLGFTEDSTETPAEVSETALYSEALTLESSIVSGFQLATASGPLCDEPMWGL 826
Query: 674 CFEVCDVV-----LHTDAIHRGG---GQIIPTARRVFYAAMLTAKPRLLEPVYLVEIQAP 725
F + + + TD G GQ++ + AA+L PR++E +Y E+
Sbjct: 827 AFTIESHLAPAEDVETDKPENFGIFTGQVMTAVKDACRAAVLQTNPRIVEAMYFCELNTA 886
Query: 726 EHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQAFPQL 785
LG +Y+VL+++R + E + + L+ V AY+PV ESF F + LR T G A +
Sbjct: 887 PEYLGPMYAVLSRRRARILKEEMQEGSSLFTVHAYVPVSESFGFADELRKGTSGGASALM 946
Query: 786 VFDHWDMVPSDP 797
V HW+M+ DP
Sbjct: 947 VLSHWEMLEEDP 958
>At1g62750 elongation factor G, putative
Length = 783
Score = 175 bits (444), Expect = 8e-44
Identities = 211/810 (26%), Positives = 327/810 (40%), Gaps = 162/810 (20%)
Query: 8 RNMSVIAHVDHGKSTLTDSLVAAAGIIAQ--EVAGDVRMTDTRQDEAERGITIKSTGISL 65
RN+ ++AH+D GK+T T+ ++ G + EV D + E ERGITI S +
Sbjct: 97 RNIGIMAHIDAGKTTTTERILYYTGRNYKIGEVHEGTATMDWMEQEQERGITITSAATTT 156
Query: 66 YYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCV 125
+++ K+ IN+ID+PGHVDF+ EV ALR+ DGA+ + D V GV
Sbjct: 157 FWD----------------KHRINIIDTPGHVDFTLEVERALRVLDGAICLFDSVAGVEP 200
Query: 126 QTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYEDALL 185
Q+ETV RQA + + VNKMDR + + + + + + ED
Sbjct: 201 QSETVWRQADKYGVPRICFVNKMDRLGANFFRTRDMIVTNLGAKPLVLQIPIGA-EDVFK 259
Query: 186 GDVQVYPEKGTVSFSAGLHGWSFT-------LTNFAKMYASKF-----GVDEEKMMNRLW 233
G V + K V +S G F+ L + A+ Y + +D+E M N L
Sbjct: 260 GVVDLVRMKAIV-WSGEELGAKFSYEDIPEDLEDLAQEYRAAMMELIVDLDDEVMENYLE 318
Query: 234 GENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNL 293
G +++ K+ K T T + PI
Sbjct: 319 GVEPDEATVKRLVRKGTITGK---------FVPI-------------------------- 343
Query: 294 KSEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYAS 353
L G A + +Q LL+ ++ +LPSP + V + ++P +
Sbjct: 344 ------LCGSAFKNKGVQ-------PLLDAVVDYLPSPVE-----VPPMNGTDPENPEIT 385
Query: 354 AIRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLY 413
IR D + P K++ G F RV+SGK+S G V N G+K
Sbjct: 386 IIRKPDDDEPFAGLAFKIMSDPFVGSL-TFVRVYSGKISAGSYVL----NANKGKK---- 436
Query: 414 VKSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSV 473
+ + R + +E V+ G+ +A+ GL IT E V + M F
Sbjct: 437 -ERIGRLLEMHANSREDVKVALTGDIIALAGLKDTITGETLSDPENPV---VLERMDFP- 491
Query: 474 SPVVSVAVTCKVASDLPKLVEGLKRLAKSDPMV-VCTISETGEHIIAAAGELHLEICLKD 532
PV+ VA+ K +D+ K+ GL +LA+ DP E + +I GELHLEI +
Sbjct: 492 DPVIKVAIEPKTKADIDKMATGLIKLAQEDPSFHFSRDEEMNQTVIEGMGELHLEIIVDR 551
Query: 533 LQDDFMNGAEITKSDPIVSFRETVLE----KSSHTVMSKSPNKHNRLYMEARPMEEG--- 585
L+ +F E P V++RE++ + K +H S + + + P+E G
Sbjct: 552 LKREFK--VEANVGAPQVNYRESISKIAEVKYTHKKQSGGQGQFADITVRFEPLEAGSGY 609
Query: 586 -LAEAIDDGRIGPRDEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQY 644
I G + PR+ +K L + C +TG
Sbjct: 610 EFKSEIKGGAV-PREYIPGVMKGLEE-------------CM---STG------------- 639
Query: 645 LNEIKDSVVAGFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYA 704
V+AGF + + D + V V L AR F
Sbjct: 640 -------VLAGFPVVDVRACLVDGSYHDVDSSVLAFQL--------------AARGAFRE 678
Query: 705 AMLTAKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVI 764
M A PR+LEP+ VE+ PE LG + LN +RG + +P L V + +P+
Sbjct: 679 GMRKAGPRMLEPIMRVEVVTPEEHLGDVIGDLNSRRGQINSFGDKPG-GLKVVDSLVPLA 737
Query: 765 ESFQFNESLRAQTGGQAFPQLVFDHWDMVP 794
E FQ+ +LR T G+A + +D+VP
Sbjct: 738 EMFQYVSTLRGMTKGRASYTMQLAKFDVVP 767
>At5g13650 GTP-binding protein typA (tyrosine phosphorylated protein
A)
Length = 609
Score = 109 bits (273), Expect = 6e-24
Identities = 63/150 (42%), Positives = 86/150 (57%), Gaps = 16/150 (10%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
+D + N+RN++++AHVDHGK+TL DS++ A + R+ D+ E ERGITI S
Sbjct: 10 LDRRDNVRNIAIVAHVDHGKTTLVDSMLRQAKVFRDNQVMQERIMDSNDLERERGITILS 69
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
S+ Y KN K +N+ID+PGH DF EV L + DG L+VVD V
Sbjct: 70 KNTSITY-------KNTK---------VNIIDTPGHSDFGGEVERVLNMVDGVLLVVDSV 113
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDR 150
EG QT VL++AL V+ VNK+DR
Sbjct: 114 EGPMPQTRFVLKKALEFGHAVVVVVNKIDR 143
Score = 32.7 bits (73), Expect = 0.86
Identities = 12/34 (35%), Positives = 22/34 (64%)
Query: 712 RLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFD 745
+LLEP + ++ PE +G + +L ++RG +FD
Sbjct: 406 KLLEPYEIATVEVPEAHMGPVVELLGKRRGQMFD 439
>At5g39900 GTP-binding protein-like
Length = 661
Score = 104 bits (259), Expect = 2e-22
Identities = 62/144 (43%), Positives = 86/144 (59%), Gaps = 10/144 (6%)
Query: 7 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLY 66
IRN S+IAH+DHGKSTL D L+ G I ++ G + D Q ERGIT+K+ +++
Sbjct: 66 IRNFSIIAHIDHGKSTLADRLMELTGTI-KKGHGQPQYLDKLQ--RERGITVKAQTATMF 122
Query: 67 YEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQ 126
YE N ++E + YL+NLID+PGHVDFS EV+ +L GAL+VVD +GV Q
Sbjct: 123 YE-------NKVEDQEASGYLLNLIDTPGHVDFSYEVSRSLSACQGALLVVDAAQGVQAQ 175
Query: 127 TETVLRQALGERIKPVLTVNKMDR 150
T A + V +NK+D+
Sbjct: 176 TVANFYLAFEANLTIVPVINKIDQ 199
>At5g08650 GTP-binding protein LepA homolog
Length = 675
Score = 98.6 bits (244), Expect = 1e-20
Identities = 58/144 (40%), Positives = 79/144 (54%), Gaps = 13/144 (9%)
Query: 6 NIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISL 65
NIRN S+IAH+DHGKSTL D L+ G + + + D E ERGITIK +
Sbjct: 79 NIRNFSIIAHIDHGKSTLADKLLQVTGTVQNRDMKE-QFLDNMDLERERGITIKLQAARM 137
Query: 66 YYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCV 125
Y D + +NLID+PGHVDFS EV+ +L +GAL+VVD +GV
Sbjct: 138 RYVYED------------TPFCLNLIDTPGHVDFSYEVSRSLAACEGALLVVDASQGVEA 185
Query: 126 QTETVLRQALGERIKPVLTVNKMD 149
QT + AL ++ + +NK+D
Sbjct: 186 QTLANVYLALENNLEIIPVLNKID 209
>At2g31060 putative GTP-binding protein
Length = 664
Score = 97.4 bits (241), Expect = 3e-20
Identities = 57/147 (38%), Positives = 86/147 (57%), Gaps = 22/147 (14%)
Query: 5 HNIRNMSVIAHVDHGKSTLTDSLVAAAGI-IAQEVAGDVRMTDTRQDEAERGITIKSTGI 63
+ +RN++VIAHVDHGK+TL D L+ G I E R D+ E ERGITI S
Sbjct: 57 NRLRNVAVIAHVDHGKTTLMDRLLRQCGADIPHE-----RAMDSINLERERGITISSKVT 111
Query: 64 SLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGV 123
S++++ ++ +N++D+PGH DF EV + + +GA++VVD EG
Sbjct: 112 SIFWKDNE----------------LNMVDTPGHADFGGEVERVVGMVEGAILVVDAGEGP 155
Query: 124 CVQTETVLRQALGERIKPVLTVNKMDR 150
QT+ VL +AL ++P+L +NK+DR
Sbjct: 156 LAQTKFVLAKALKYGLRPILLLNKVDR 182
>At2g45030 putative mitochondrial translation elongation factor G
Length = 754
Score = 84.0 bits (206), Expect = 3e-16
Identities = 57/149 (38%), Positives = 83/149 (55%), Gaps = 21/149 (14%)
Query: 7 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQ--EVAGDVRM---TDTRQDEAERGITIKST 61
+RN+ + AH+D GK+TLT+ ++ G I + EV G + D+ E E+GITI+S
Sbjct: 65 LRNIGISAHIDSGKTTLTERVLFYTGRIHEIHEVRGRDGVGAKMDSMDLEREKGITIQSA 124
Query: 62 GISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVE 121
+ Y D Y +N+ID+PGHVDF+ EV ALR+ DGA++V+ V
Sbjct: 125 --ATYCTWKD--------------YKVNIIDTPGHVDFTIEVERALRVLDGAILVLCSVG 168
Query: 122 GVCVQTETVLRQALGERIKPVLTVNKMDR 150
GV Q+ TV RQ + V +NK+DR
Sbjct: 169 GVQSQSITVDRQMRRYEVPRVAFINKLDR 197
Score = 80.9 bits (198), Expect = 3e-15
Identities = 109/474 (22%), Positives = 191/474 (39%), Gaps = 92/474 (19%)
Query: 320 LLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDPEGPLMLYVSKMIPASDKGR 379
LL+ ++ LPSP + Y ++ ++ P+GPL+ K+ ++GR
Sbjct: 328 LLDGVVSFLPSPNEVNNYALDQ------NNNEERVTLTGSPDGPLVALAFKL----EEGR 377
Query: 380 F--YAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRTVIWMGKKQETVEDVPCG 437
F + RV+ G + G ++I +K V R V E +++ G
Sbjct: 378 FGQLTYLRVYEGVIKKG--------DFIINVNTGKRIK-VPRLVRMHSNDMEDIQEAHAG 428
Query: 438 NTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVAVTCKVASDLPKLVEGLK 497
VA+ G++ T T+ + + +M PV+S+AV + + L
Sbjct: 429 QIVAVFGIE--CASGDTFTDGSV--KYTMTSMNVP-EPVMSLAVQPVSKDSGGQFSKALN 483
Query: 498 RLAKSDPMVVCTIS-ETGEHIIAAAGELHLEICLKDLQDDFMNGAEITKSDPIVSFRETV 556
R K DP + E+G+ II+ GELHL+I ++ ++ ++ A + K P V+FRET+
Sbjct: 484 RFQKEDPTFRVGLDPESGQTIISGMGELHLDIYVERMRREYKVDATVGK--PRVNFRETI 541
Query: 557 LEKSS----HTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDEPKNHLKILSDEF 612
+++ H S ++ R+ P+ G E EF
Sbjct: 542 TQRAEFDYLHKKQSGGAGQYGRVTGYVEPLPPGSKEKF--------------------EF 581
Query: 613 GWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIASKEGPMAD---EN 669
NM+V G + ++ GF+ A+ G + EN
Sbjct: 582 -------------------ENMIV----GQAIPSGFIPAIEKGFKEAANSGSLIGHPVEN 618
Query: 670 LRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAML---TAKPRLLEPVYLVEIQAPE 726
LR +VL A H + YA L A+P +LEPV LVE++ P
Sbjct: 619 LR--------IVLTDGASHAVDSSELAFKMAAIYAFRLCYTAARPVILEPVMLVELKVPT 670
Query: 727 HALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
G + +N+++G + Q + + + A +P+ F ++ SLR+ T G+
Sbjct: 671 EFQGTVAGDINKRKGIIVGNDQEGDDSV--ITANVPLNNMFGYSTSLRSMTQGK 722
>At1g45332 mitochondrial elongation factor, putative
Length = 754
Score = 84.0 bits (206), Expect = 3e-16
Identities = 57/149 (38%), Positives = 83/149 (55%), Gaps = 21/149 (14%)
Query: 7 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQ--EVAGDVRM---TDTRQDEAERGITIKST 61
+RN+ + AH+D GK+TLT+ ++ G I + EV G + D+ E E+GITI+S
Sbjct: 65 LRNIGISAHIDSGKTTLTERVLFYTGRIHEIHEVRGRDGVGAKMDSMDLEREKGITIQSA 124
Query: 62 GISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVE 121
+ Y D Y +N+ID+PGHVDF+ EV ALR+ DGA++V+ V
Sbjct: 125 --ATYCTWKD--------------YKVNIIDTPGHVDFTIEVERALRVLDGAILVLCSVG 168
Query: 122 GVCVQTETVLRQALGERIKPVLTVNKMDR 150
GV Q+ TV RQ + V +NK+DR
Sbjct: 169 GVQSQSITVDRQMRRYEVPRVAFINKLDR 197
Score = 80.9 bits (198), Expect = 3e-15
Identities = 109/474 (22%), Positives = 191/474 (39%), Gaps = 92/474 (19%)
Query: 320 LLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDPEGPLMLYVSKMIPASDKGR 379
LL+ ++ LPSP + Y ++ ++ P+GPL+ K+ ++GR
Sbjct: 328 LLDGVVSFLPSPNEVNNYALDQ------NNNEERVTLTGSPDGPLVALAFKL----EEGR 377
Query: 380 F--YAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRTVIWMGKKQETVEDVPCG 437
F + RV+ G + G ++I +K V R V E +++ G
Sbjct: 378 FGQLTYLRVYEGVIKKG--------DFIINVNTGKRIK-VPRLVRMHSNDMEDIQEAHAG 428
Query: 438 NTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVAVTCKVASDLPKLVEGLK 497
VA+ G++ T T+ + + +M PV+S+AV + + L
Sbjct: 429 QIVAVFGIE--CASGDTFTDGSV--KYTMTSMNVP-EPVMSLAVQPVSKDSGGQFSKALN 483
Query: 498 RLAKSDPMVVCTIS-ETGEHIIAAAGELHLEICLKDLQDDFMNGAEITKSDPIVSFRETV 556
R K DP + E+G+ II+ GELHL+I ++ ++ ++ A + K P V+FRET+
Sbjct: 484 RFQKEDPTFRVGLDPESGQTIISGMGELHLDIYVERMRREYKVDATVGK--PRVNFRETI 541
Query: 557 LEKSS----HTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDEPKNHLKILSDEF 612
+++ H S ++ R+ P+ G E EF
Sbjct: 542 TQRAEFDYLHKKQSGGAGQYGRVTGYVEPLPPGSKEKF--------------------EF 581
Query: 613 GWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIASKEGPMAD---EN 669
NM+V G + ++ GF+ A+ G + EN
Sbjct: 582 -------------------ENMIV----GQAIPSGFIPAIEKGFKEAANSGSLIGHPVEN 618
Query: 670 LRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAML---TAKPRLLEPVYLVEIQAPE 726
LR +VL A H + YA L A+P +LEPV LVE++ P
Sbjct: 619 LR--------IVLTDGASHAVDSSELAFKMAAIYAFRLCYTAARPVILEPVMLVELKVPT 670
Query: 727 HALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
G + +N+++G + Q + + + A +P+ F ++ SLR+ T G+
Sbjct: 671 EFQGTVAGDINKRKGIIVGNDQEGDDSV--ITANVPLNNMFGYSTSLRSMTQGK 722
>At4g20360 translation elongation factor EF-Tu precursor,
chloroplast
Length = 476
Score = 60.1 bits (144), Expect = 5e-09
Identities = 39/120 (32%), Positives = 52/120 (42%), Gaps = 16/120 (13%)
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLYYE 68
N+ I HVDHGK+TLT +L A I VA D +E RGITI + +
Sbjct: 81 NIGTIGHVDHGKTTLTAALTMALASIGSSVAKKYDEIDAAPEERARGITINTATVEY--- 137
Query: 69 MSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQTE 128
E E Y +D PGH D+ + DGA++VV +G QT+
Sbjct: 138 -----------ETENRHYA--HVDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQTK 184
>At1g17220 putative translation initiation factor IF2
Length = 1026
Score = 50.4 bits (119), Expect = 4e-06
Identities = 36/141 (25%), Positives = 63/141 (44%), Gaps = 28/141 (19%)
Query: 10 MSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLYYEM 69
++++ HVDHGK+TL D + + + A E G +GI + +
Sbjct: 504 ITIMGHVDHGKTTLLD-YIRKSKVAASEAGG-----------ITQGIGAYKVSVPV---- 547
Query: 70 SDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQTET 129
DG L++ +D+PGH F + R+TD A++VV +G+ QT
Sbjct: 548 -DGKLQSCV-----------FLDTPGHEAFGAMRARGARVTDIAIIVVAADDGIRPQTNE 595
Query: 130 VLRQALGERIKPVLTVNKMDR 150
+ A + V+ +NK+D+
Sbjct: 596 AIAHAKAAAVPIVIAINKIDK 616
>At5g10630 putative protein
Length = 804
Score = 49.7 bits (117), Expect = 7e-06
Identities = 45/165 (27%), Positives = 70/165 (42%), Gaps = 40/165 (24%)
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMT---------------DTRQDEAE 53
N++++ HVD GKSTL+ L+ G I+Q+ D +E E
Sbjct: 378 NLAIVGHVDSGKSTLSGRLLHLLGRISQKQMHKYEKEAKLQGKGSFAYAWALDESAEERE 437
Query: 54 RGITIKSTGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGA 113
RGIT+ +++ Y S ++ + L+DSPGH DF + A D A
Sbjct: 438 RGITMT---VAVAYFNS-------------KRHHVVLLDSPGHKDFVPNMIAGATQADAA 481
Query: 114 LVVVDCVEGVCV--------QTETVLRQALGERIKPVL-TVNKMD 149
++V+D G QT R G ++ V+ +NKMD
Sbjct: 482 ILVIDASVGAFEAGFDNLKGQTREHARVLRGFGVEQVIVAINKMD 526
>At4g02930 mitochondrial elongation factor Tu
Length = 454
Score = 48.5 bits (114), Expect = 2e-05
Identities = 34/120 (28%), Positives = 50/120 (41%), Gaps = 16/120 (13%)
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLYYE 68
N+ I HVDHGK+TLT ++ + A D +E +RGITI + +
Sbjct: 69 NVGTIGHVDHGKTTLTAAITKVLAEEGKAKAIAFDEIDKAPEEKKRGITIATAHV----- 123
Query: 69 MSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQTE 128
E E K +D PGH D+ + DG ++VV +G QT+
Sbjct: 124 -----------EYETAKRHYAHVDCPGHADYVKNMITGAAQMDGGILVVSGPDGPMPQTK 172
>At5g60390 translation elongation factor eEF-1 alpha chain (gene A4)
Length = 449
Score = 47.8 bits (112), Expect = 3e-05
Identities = 40/129 (31%), Positives = 57/129 (44%), Gaps = 31/129 (24%)
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEV-------AGDVR--------MTDTRQDEAE 53
N+ VI HVD GKST T L+ G I + V A ++ + D + E E
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 68
Query: 54 RGITIKSTGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGA 113
RGITI I+L+ + E KY +ID+PGH DF + D A
Sbjct: 69 RGITID---IALW-------------KFETTKYYCTVIDAPGHRDFIKNMITGTSQADCA 112
Query: 114 LVVVDCVEG 122
++++D G
Sbjct: 113 VLIIDSTTG 121
>At1g07940 elongation factor 1-alpha
Length = 449
Score = 47.8 bits (112), Expect = 3e-05
Identities = 40/129 (31%), Positives = 57/129 (44%), Gaps = 31/129 (24%)
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEV-------AGDVR--------MTDTRQDEAE 53
N+ VI HVD GKST T L+ G I + V A ++ + D + E E
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 68
Query: 54 RGITIKSTGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGA 113
RGITI I+L+ + E KY +ID+PGH DF + D A
Sbjct: 69 RGITID---IALW-------------KFETTKYYCTVIDAPGHRDFIKNMITGTSQADCA 112
Query: 114 LVVVDCVEG 122
++++D G
Sbjct: 113 VLIIDSTTG 121
>At1g07930 elongation factor 1-alpha
Length = 449
Score = 47.8 bits (112), Expect = 3e-05
Identities = 40/129 (31%), Positives = 57/129 (44%), Gaps = 31/129 (24%)
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEV-------AGDVR--------MTDTRQDEAE 53
N+ VI HVD GKST T L+ G I + V A ++ + D + E E
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 68
Query: 54 RGITIKSTGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGA 113
RGITI I+L+ + E KY +ID+PGH DF + D A
Sbjct: 69 RGITID---IALW-------------KFETTKYYCTVIDAPGHRDFIKNMITGTSQADCA 112
Query: 114 LVVVDCVEG 122
++++D G
Sbjct: 113 VLIIDSTTG 121
>At1g07920 elongation factor 1-alpha
Length = 449
Score = 47.8 bits (112), Expect = 3e-05
Identities = 40/129 (31%), Positives = 57/129 (44%), Gaps = 31/129 (24%)
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEV-------AGDVR--------MTDTRQDEAE 53
N+ VI HVD GKST T L+ G I + V A ++ + D + E E
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 68
Query: 54 RGITIKSTGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGA 113
RGITI I+L+ + E KY +ID+PGH DF + D A
Sbjct: 69 RGITID---IALW-------------KFETTKYYCTVIDAPGHRDFIKNMITGTSQADCA 112
Query: 114 LVVVDCVEG 122
++++D G
Sbjct: 113 VLIIDSTTG 121
>At1g21160 transcription factor, putative
Length = 1088
Score = 45.8 bits (107), Expect = 1e-04
Identities = 41/157 (26%), Positives = 76/157 (48%), Gaps = 26/157 (16%)
Query: 2 DLKHNIRN--MSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIK 59
+++ N+R+ ++ HVD GK+ L D +R T+ ++ EA GIT +
Sbjct: 486 EVEENLRSPICCIMGHVDSGKTKLLDC---------------IRGTNVQEGEAG-GIT-Q 528
Query: 60 STGISLYYEMSDGDLKNFKGEREGNKYL----INLIDSPGHVDFSSEVTAALRITDGALV 115
G + + +++ E + N L I +ID+PGH F++ + + D A++
Sbjct: 529 QIGATFF---PAENIRERTKELQANAKLKVPGILVIDTPGHESFTNLRSRGSNLCDLAIL 585
Query: 116 VVDCVEGVCVQTETVLRQALGERIKPVLTVNKMDRCF 152
VVD + G+ QT L +K ++ +NK+DR +
Sbjct: 586 VVDIMRGLEPQTIESLNLLRRRNVKFIIALNKVDRLY 622
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,577,065
Number of Sequences: 26719
Number of extensions: 810732
Number of successful extensions: 2141
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 2042
Number of HSP's gapped (non-prelim): 57
length of query: 831
length of database: 11,318,596
effective HSP length: 108
effective length of query: 723
effective length of database: 8,432,944
effective search space: 6097018512
effective search space used: 6097018512
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146817.8


