

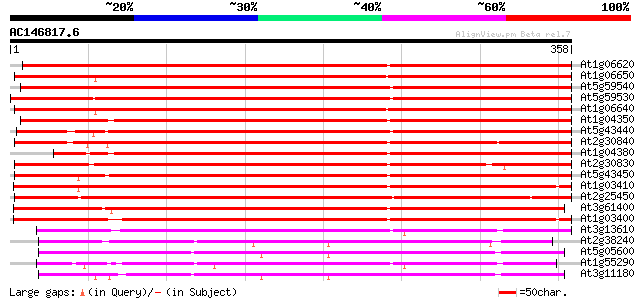
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146817.6 - phase: 0
(358 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g06620 oxidoreductase-like protein 429 e-120
At1g06650 oxidoreductase, putative 410 e-115
At5g59540 1-aminocyclopropane-1-carboxylate oxidase - like protein 400 e-112
At5g59530 1-aminocyclopropane-1-carboxylate oxidase - like protein 395 e-110
At1g06640 putative Iron/Ascorbate oxidoreductase family protein 393 e-110
At1g04350 putative 1-aminocyclopropane-1-carboxylate oxidase 388 e-108
At5g43440 1-aminocyclopropane-1-carboxylate oxidase 384 e-107
At2g30840 putative dioxygenase 382 e-106
At1g04380 putative 1-aminocyclopropane-1-carboxylate oxidase 381 e-106
At2g30830 putative dioxygenase 379 e-105
At5g43450 1-aminocyclopropane-1-carboxylate oxidase 371 e-103
At1g03410 putative 1-aminocyclopropane-1-carboxylate oxidase 368 e-102
At2g25450 putative dioxygenase 358 3e-99
At3g61400 1-aminocyclopropane-1-carboxylate oxidase-like protein 346 1e-95
At1g03400 putative 1-aminocyclopropane-1-carboxylate oxidase 335 3e-92
At3g13610 unknown protein 206 1e-53
At2g38240 putative anthocyanidin synthase 197 1e-50
At5g05600 leucoanthocyanidin dioxygenase-like protein 195 3e-50
At1g55290 leucoanthocyanidin dioxygenase 2, putative 194 8e-50
At3g11180 putative leucoanthocyanidin dioxygenase 190 1e-48
>At1g06620 oxidoreductase-like protein
Length = 365
Score = 429 bits (1103), Expect = e-120
Identities = 202/350 (57%), Positives = 254/350 (71%), Gaps = 1/350 (0%)
Query: 9 LKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDFANFGNDPKT 68
LKAFD+TKTGVKGL+D G+ +IP++F P +++ IP ID G D T
Sbjct: 17 LKAFDETKTGVKGLIDAGITEIPSIFRAPPATLTSPKPPSSSDFSIPTIDLKGGGTDSIT 76
Query: 69 RHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMRPF 128
R + KI +A E WGFFQV+NHGIP++VLE M +G+ F EQD EVKK YSRDP
Sbjct: 77 RRSLVEKIGDAAEKWGFFQVINHGIPMDVLEKMIDGIREFHEQDTEVKKGFYSRDPASKM 136
Query: 129 SYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLGMTLFELL 188
Y+SNF+L+SS A NWRDT C AP+ P+P+DLP C + ++EY + V KLG LFELL
Sbjct: 137 VYSSNFDLFSSPAANWRDTLGCYTAPDPPRPEDLPATCGEMMIEYSKEVMKLGKLLFELL 196
Query: 189 SEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGL 248
SEALGL+ NHLKDM C +L L HYYP CP+P+LT+G TKHSD+ FLT+LLQ DHIGGL
Sbjct: 197 SEALGLNTNHLKDMDCTNSLLLLGHYYPPCPQPDLTLGLTKHSDNSFLTILLQ-DHIGGL 255
Query: 249 QVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNADL 308
QV H W+D+PP+P ALVVNVGDLLQL+TNDKF SVEH+VLAN+ GPR+SVACFF++ L
Sbjct: 256 QVLHDQYWVDVPPVPGALVVNVGDLLQLITNDKFISVEHRVLANVAGPRISVACFFSSYL 315
Query: 309 NSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
+ YGPIKE LSE+NPP Y++ TI+EY +Y +KG DG S L + ++
Sbjct: 316 MANPRVYGPIKEILSEENPPNYRDTTITEYAKFYRSKGFDGTSGLLYLKI 365
>At1g06650 oxidoreductase, putative
Length = 369
Score = 410 bits (1053), Expect = e-115
Identities = 203/360 (56%), Positives = 255/360 (70%), Gaps = 6/360 (1%)
Query: 4 NRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATN-SNNTQHI--IPIIDFA 60
+R ELKAFD+TKTGVKGLVD GV ++P +FHHP K ++ H+ IP ID
Sbjct: 11 DRASELKAFDETKTGVKGLVDSGVSQVPRIFHHPTVKLSTPKPLPSDLLHLKTIPTIDLG 70
Query: 61 NFG-NDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKEL 119
D R+ +I+EA WGFFQV+NHG+ L +LE M+ GV F EQ EV+KE
Sbjct: 71 GRDFQDAIKRNNAIEEIKEAAAKWGFFQVINHGVSLELLEKMKKGVRDFHEQSQEVRKEF 130
Query: 120 YSRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTK 179
YSRD R F Y SNF+L+SS A NWRDTF C++AP+ PKPQDLP +CRD ++EY + V
Sbjct: 131 YSRDFSRRFLYLSNFDLFSSPAANWRDTFSCTMAPDTPKPQDLPEICRDIMMEYSKQVMN 190
Query: 180 LGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLL 239
LG LFELLSEALGL PNHL DM C+KG+L LSHYYP CPEP+LT+GT++HSD+ FLT+L
Sbjct: 191 LGKFLFELLSEALGLEPNHLNDMDCSKGLLMLSHYYPPCPEPDLTLGTSQHSDNSFLTVL 250
Query: 240 LQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLAN-IIGPRV 298
L D I GLQV + W D+P + AL++N+GDLLQL+TNDKF S+EH+VLAN RV
Sbjct: 251 L-PDQIEGLQVRREGHWFDVPHVSGALIINIGDLLQLITNDKFISLEHRVLANRATRARV 309
Query: 299 SVACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
SVACFF + YGPI+E +SE+NPPKY+E TI +Y Y+N KG+DG SAL HF++
Sbjct: 310 SVACFFTTGVRPNPRMYGPIRELVSEENPPKYRETTIKDYATYFNAKGLDGTSALLHFKI 369
>At5g59540 1-aminocyclopropane-1-carboxylate oxidase - like protein
Length = 366
Score = 400 bits (1028), Expect = e-112
Identities = 196/354 (55%), Positives = 255/354 (71%), Gaps = 4/354 (1%)
Query: 8 ELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHI-IPIIDFANFGNDP 66
E KAFD+TK GVKGLVD + ++P +FHH D S + + IPIIDFA+ D
Sbjct: 14 ERKAFDETKQGVKGLVDAKITEVPRIFHHRQDILTNKKPSASVSDLEIPIIDFASVHADT 73
Query: 67 KTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDP-M 125
+R I K++ A E WGFFQV+NH IPLNVLE++++GV RF E+D EVKK +SRD
Sbjct: 74 ASREAIVEKVKYAVENWGFFQVINHSIPLNVLEEIKDGVRRFHEEDPEVKKSFFSRDAGN 133
Query: 126 RPFSYNSNFNLYSSS-ALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLGMTL 184
+ F YNSNF+LYSSS ++NWRD+F C +AP+ P P+++P CRD + EY ++V G L
Sbjct: 134 KKFVYNSNFDLYSSSPSVNWRDSFSCYIAPDPPAPEEIPETCRDAMFEYSKHVLSFGGLL 193
Query: 185 FELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDDH 244
FELLSEALGL L+ M C K +L + HYYP CP+P+LT+G TKHSD+ FLTLLLQD+
Sbjct: 194 FELLSEALGLKSQTLESMDCVKTLLMICHYYPPCPQPDLTLGITKHSDNSFLTLLLQDN- 252
Query: 245 IGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFF 304
IGGLQ+ HQ+ W+D+ PI ALVVN+GD LQL+TNDKF SVEH+VLAN GPR+SVA FF
Sbjct: 253 IGGLQILHQDSWVDVSPIHGALVVNIGDFLQLITNDKFVSVEHRVLANRQGPRISVASFF 312
Query: 305 NADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
++ + S YGP+KE +SE+NPPKY++ITI EY + KG+DG S L + R+
Sbjct: 313 SSSMRPNSRVYGPMKELVSEENPPKYRDITIKEYSKIFFEKGLDGTSHLSNIRI 366
>At5g59530 1-aminocyclopropane-1-carboxylate oxidase - like protein
Length = 364
Score = 395 bits (1014), Expect = e-110
Identities = 196/361 (54%), Positives = 258/361 (71%), Gaps = 5/361 (1%)
Query: 1 MDSNRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDFA 60
++ +R E KAFD+TK GVKGL+D + +IP +FH P D S + I P IDFA
Sbjct: 6 VEFDRYIERKAFDNTKEGVKGLIDAKITEIPRIFHVPQDTLPDKKRSVSDLEI-PTIDFA 64
Query: 61 NFGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQ-DVEVKKEL 119
+ D +R I K++ A E WGFFQV+NHG+PLNVLE++++GV RF E+ D EVKK
Sbjct: 65 SVNVDTPSREAIVEKVKYAVENWGFFQVINHGVPLNVLEEIKDGVRRFHEEEDPEVKKSY 124
Query: 120 YSRD-PMRPFSYNSNFNLYSSS-ALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYV 177
YS D F+Y+SNF+LYSSS +L WRD+ C +AP+ P P++LP CRD ++EY ++V
Sbjct: 125 YSLDFTKNKFAYSSNFDLYSSSPSLTWRDSISCYMAPDPPTPEELPETCRDAMIEYSKHV 184
Query: 178 TKLGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLT 237
LG LFELLSEALGL LK M C K +L + HYYP CP+P+LT+G +KHSD+ FLT
Sbjct: 185 LSLGDLLFELLSEALGLKSEILKSMDCLKSLLMICHYYPPCPQPDLTLGISKHSDNSFLT 244
Query: 238 LLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPR 297
+LLQD+ IGGLQ+ HQ+ W+D+ P+P ALVVNVGD LQL+TNDKF SVEH+VLAN GPR
Sbjct: 245 VLLQDN-IGGLQILHQDSWVDVSPLPGALVVNVGDFLQLITNDKFISVEHRVLANTRGPR 303
Query: 298 VSVACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFR 357
+SVA FF++ + S YGP+KE +SE+NPPKY++ T+ EY Y KG+DG S L +FR
Sbjct: 304 ISVASFFSSSIRENSTVYGPMKELVSEENPPKYRDTTLREYSEGYFKKGLDGTSHLSNFR 363
Query: 358 V 358
+
Sbjct: 364 I 364
>At1g06640 putative Iron/Ascorbate oxidoreductase family protein
Length = 369
Score = 393 bits (1009), Expect = e-110
Identities = 198/360 (55%), Positives = 255/360 (70%), Gaps = 6/360 (1%)
Query: 4 NRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHP-LDKFGKATNSNNTQHI--IPIIDFA 60
+R ELKAFD+TKTGVKGLVD G+ KIP +FHH ++ ++ H+ IP ID
Sbjct: 11 DRASELKAFDETKTGVKGLVDSGISKIPRIFHHSSVELANPKPLPSDLLHLKTIPTIDLG 70
Query: 61 NFGNDPKTRHEITSK-IREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKEL 119
+H+ + I+EA WGFFQV+NHG+ L +LE M++GV F EQ EV+K+L
Sbjct: 71 GRDFQDAIKHKNAIEGIKEAAAKWGFFQVINHGVSLELLEKMKDGVRDFHEQPPEVRKDL 130
Query: 120 YSRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTK 179
YSRD R F Y SNF+LY+++A NWRDTF C +AP+ P+PQDLP +CRD ++EY + V
Sbjct: 131 YSRDFGRKFIYLSNFDLYTAAAANWRDTFYCYMAPDPPEPQDLPEICRDVMMEYSKQVMI 190
Query: 180 LGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLL 239
LG LFELLSEALGL+PNHLKDM C KG+ L HY+P CPEP+LT GT+KHSD FLT+L
Sbjct: 191 LGEFLFELLSEALGLNPNHLKDMECLKGLRMLCHYFPPCPEPDLTFGTSKHSDGSFLTVL 250
Query: 240 LQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLAN-IIGPRV 298
L D+I GLQV + W D+P +P AL++N+GDLLQL+TNDKF S++H+VLAN RV
Sbjct: 251 L-PDNIEGLQVCREGYWFDVPHVPGALIINIGDLLQLITNDKFISLKHRVLANRATRARV 309
Query: 299 SVACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
SVACFF+ + YGPIKE +SE+NPPKY+E TI +Y Y+N KG+ G SAL F+V
Sbjct: 310 SVACFFHTHVKPNPRVYGPIKELVSEENPPKYRETTIRDYATYFNGKGLGGTSALLDFKV 369
>At1g04350 putative 1-aminocyclopropane-1-carboxylate oxidase
Length = 360
Score = 388 bits (997), Expect = e-108
Identities = 192/354 (54%), Positives = 251/354 (70%), Gaps = 7/354 (1%)
Query: 8 ELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFG-KATNSNNTQHIIPIIDFANFGNDP 66
E KAFD+TKTGVKGL+D + +IP +F P K + T IPIIDF
Sbjct: 11 ERKAFDETKTGVKGLIDAHITEIPRIFCLPQGSLSDKKPFVSTTDFAIPIIDFEGLH--- 67
Query: 67 KTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMR 126
+R +I KI++A WGFFQV+NHG+PLNVL+++++GV RF E+ EVKK ++RD +
Sbjct: 68 VSREDIVGKIKDAASNWGFFQVINHGVPLNVLQEIQDGVRRFHEEAPEVKKTYFTRDATK 127
Query: 127 PFSYNSNFNLYSSSA-LNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLGMTLF 185
F YNSNF+LYSSS+ +NWRD+F C +AP+ P P+DLP+ CR + EY +++ +LG LF
Sbjct: 128 RFVYNSNFDLYSSSSCVNWRDSFACYMAPDPPNPEDLPVACRVAMFEYSKHMMRLGDLLF 187
Query: 186 ELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDDHI 245
ELLSEALGL + LK M C KG+L L HYYP CP+P+LT+GT HSD+ FLT+LLQ D I
Sbjct: 188 ELLSEALGLRSDKLKSMDCMKGLLLLCHYYPPCPQPDLTIGTNNHSDNSFLTILLQ-DQI 246
Query: 246 GGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLAN-IIGPRVSVACFF 304
GGLQ+FHQ+ W+D+ PIP ALV+N+GD LQL+TNDK SVEH+VLAN PR+SVA FF
Sbjct: 247 GGLQIFHQDCWVDVSPIPGALVINMGDFLQLITNDKVISVEHRVLANRAATPRISVASFF 306
Query: 305 NADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
+ + S YGPIKE LSE+NP KY+ I + EY Y KG+DG S L H+++
Sbjct: 307 STSMRPNSTVYGPIKELLSEENPSKYRVIDLKEYTEGYFKKGLDGTSYLSHYKI 360
>At5g43440 1-aminocyclopropane-1-carboxylate oxidase
Length = 365
Score = 384 bits (987), Expect = e-107
Identities = 189/362 (52%), Positives = 255/362 (70%), Gaps = 14/362 (3%)
Query: 5 RLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQH-------IIPII 57
RL+ELKAF TK GVKGLVD + ++P +FH P +T SNN +PII
Sbjct: 10 RLNELKAFVSTKAGVKGLVDTKITEVPRIFHIP----SSSTLSNNKPSDIFGLNLTVPII 65
Query: 58 DFANFGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKK 117
D + GN R+ + SKI+EA E WGFFQV+NHGIPL VL+D++ GV RF E+D EVKK
Sbjct: 66 DLGD-GNTSAARNVLVSKIKEAAENWGFFQVINHGIPLTVLKDIKQGVRRFHEEDPEVKK 124
Query: 118 ELYSRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAP-KPQDLPLVCRDELLEYGEY 176
+ ++ D F+YN+NF+++ SS +NW+D+F C P P KP+++PL CRD ++EY ++
Sbjct: 125 QYFATDFNTRFAYNTNFDIHYSSPMNWKDSFTCYTCPQDPLKPEEIPLACRDVVIEYSKH 184
Query: 177 VTKLGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFL 236
V +LG LF+LLSEALGL LK+M C KG+L L HYYP CP+P+LT+G +KH+D+ F+
Sbjct: 185 VMELGGLLFQLLSEALGLDSEILKNMDCLKGLLMLCHYYPPCPQPDLTLGISKHTDNSFI 244
Query: 237 TLLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGP 296
T+LLQD IGGLQV HQ+ W+D+ P+P ALV+++GD +QL+TNDKF S+EH+V AN GP
Sbjct: 245 TILLQDQ-IGGLQVLHQDSWVDVTPVPGALVISIGDFMQLITNDKFLSMEHRVRANRDGP 303
Query: 297 RVSVACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHF 356
R+SVACF ++ + S YGPIKE LS++NP KY++ITI EY Y DG S L F
Sbjct: 304 RISVACFVSSGVFPNSTVYGPIKELLSDENPAKYRDITIPEYTVGYLASIFDGKSHLSKF 363
Query: 357 RV 358
R+
Sbjct: 364 RI 365
>At2g30840 putative dioxygenase
Length = 362
Score = 382 bits (981), Expect = e-106
Identities = 196/362 (54%), Positives = 255/362 (70%), Gaps = 12/362 (3%)
Query: 4 NRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSN----NTQHIIPIIDF 59
+R E+KAFD+ K GVKGL+D GV +IP +FHHP T+SN +T +IP ID
Sbjct: 6 DRASEVKAFDELKIGVKGLLDAGVTQIPRIFHHP---HLNLTDSNLLLSSTTMVIPTIDL 62
Query: 60 AN--FGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKK 117
F TR + + IR+A E +GFFQV+NHGI +V+E M++G+ F EQD +V+K
Sbjct: 63 KGGVFDEYTVTRESVIAMIRDAVERFGFFQVINHGISNDVMEKMKDGIRGFHEQDSDVRK 122
Query: 118 ELYSRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYV 177
+ Y+RD + YNSNF+LYSS + NWRDT C +AP+ P+ +DLP +C + +LEY + V
Sbjct: 123 KFYTRDVTKTVKYNSNFDLYSSPSANWRDTLSCFMAPDVPETEDLPDICGEIMLEYAKRV 182
Query: 178 TKLGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLT 237
KLG +FELLSEALGL+PNHLK+M C KG+L LSHYYP CPEP LT GT+ HSD FLT
Sbjct: 183 MKLGELIFELLSEALGLNPNHLKEMDCTKGLLMLSHYYPPCPEPGLTFGTSPHSDRSFLT 242
Query: 238 LLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANI-IGP 296
+LLQ DHIGGLQV W+D+PP+P AL+VN+GDLLQL+TND+F SVEH+VLAN P
Sbjct: 243 ILLQ-DHIGGLQVRQNGYWVDVPPVPGALLVNLGDLLQLMTNDQFVSVEHRVLANKGEKP 301
Query: 297 RVSVACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHF 356
R+SVA FF L S YGPIKE LSE N PKY++ T++EY ++Y +G+ G S L F
Sbjct: 302 RISVASFFVHPLPSL-RVYGPIKELLSEQNLPKYRDTTVTEYTSHYMARGLYGNSVLLDF 360
Query: 357 RV 358
++
Sbjct: 361 KI 362
>At1g04380 putative 1-aminocyclopropane-1-carboxylate oxidase
Length = 345
Score = 381 bits (978), Expect = e-106
Identities = 177/330 (53%), Positives = 242/330 (72%), Gaps = 6/330 (1%)
Query: 29 KIPTLFHHPLDKFGKATNSNNTQHIIPIIDFANFGNDPKTRHEITSKIREACETWGFFQV 88
K+P +F P D +++ +PIIDFA K+R + KI+ A E WG FQV
Sbjct: 22 KVPPIFGLPPDALDDKKPTSD--FAVPIIDFAGVH---KSREAVVEKIKAAAENWGIFQV 76
Query: 89 VNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMRPFSYNSNFNLYSSSALNWRDTF 148
+NHG+PL+VLE+++NGV+RF E+D EVKK +S D + F Y++NF LYSSSA NWRD+F
Sbjct: 77 INHGVPLSVLEEIQNGVVRFHEEDPEVKKSYFSLDLTKTFIYHNNFELYSSSAGNWRDSF 136
Query: 149 VCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLGMTLFELLSEALGLHPNHLKDMGCAKGI 208
VC + P+ P+DLP+ CRD ++ Y ++V LG LFELLSEALGL+ + LK MGC KG+
Sbjct: 137 VCYMDPDPSNPEDLPVACRDAMIGYSKHVMSLGGLLFELLSEALGLNSDTLKSMGCMKGL 196
Query: 209 LSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQVFHQNKWIDIPPIPDALVV 268
+ HYYP CP+P+ T+GT+KHSD+ F+T+LLQ D+IGGLQ+ HQ+ W+D+ P+P AL++
Sbjct: 197 HMICHYYPPCPQPDQTLGTSKHSDNTFITILLQ-DNIGGLQILHQDCWVDVSPLPGALII 255
Query: 269 NVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNADLNSFSNQYGPIKEFLSEDNPP 328
N+GD LQL+TNDKF SV+H+VL N +GPR+S+ACFF++ +N S YGPIKE LSE+NPP
Sbjct: 256 NIGDFLQLMTNDKFISVDHRVLTNRVGPRISIACFFSSSMNPNSTVYGPIKELLSEENPP 315
Query: 329 KYKEITISEYVAYYNTKGIDGISALQHFRV 358
KY++ TI EY Y KG+DG S L H+R+
Sbjct: 316 KYRDFTIPEYSKGYIEKGLDGTSHLSHYRI 345
>At2g30830 putative dioxygenase
Length = 358
Score = 379 bits (972), Expect = e-105
Identities = 191/359 (53%), Positives = 247/359 (68%), Gaps = 10/359 (2%)
Query: 4 NRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDFAN-F 62
+R E+KAFD K GVKGLVD G+ K+P +FHH S+ + IP ID
Sbjct: 6 DRAGEVKAFDQMKIGVKGLVDAGITKVPRIFHHQDVAVTNPKPSSTLE--IPTIDVGGGV 63
Query: 63 GNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSR 122
TR + +K+R A E +GFFQV+NHGIPL V+E M++G+ F EQD EVKK YSR
Sbjct: 64 FESTVTRKSVIAKVRAAVEKFGFFQVINHGIPLEVMESMKDGIRGFHEQDSEVKKTFYSR 123
Query: 123 DPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLGM 182
D + YN+NF+LYSS A NWRDT +AP+ P+ DLP++CR+ +LEY + + KLG
Sbjct: 124 DITKKVKYNTNFDLYSSQAANWRDTLTMVMAPDVPQAGDLPVICREIMLEYSKRMMKLGE 183
Query: 183 TLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQD 242
+FELLSEALGL PNHLK++ CAK + LSHYYP CPEP+ T G + H+D F+T+LLQ
Sbjct: 184 LIFELLSEALGLKPNHLKELNCAKSLSLLSHYYPPCPEPDRTFGISSHTDISFITILLQ- 242
Query: 243 DHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANI-IGPRVSVA 301
DHIGGLQV H WID+PP P+AL+VN+GDLLQL+TNDKF SVEH+VLAN PR+S A
Sbjct: 243 DHIGGLQVLHDGYWIDVPPNPEALIVNLGDLLQLITNDKFVSVEHRVLANRGEEPRISSA 302
Query: 302 CFFNADLNSFSNQ--YGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
FF +++ N+ YGP+KE LS+ NPPKY+ T +E +Y +G+DG S L HFR+
Sbjct: 303 SFF---MHTIPNEQVYGPMKELLSKQNPPKYRNTTTTEMARHYLARGLDGTSPLLHFRI 358
>At5g43450 1-aminocyclopropane-1-carboxylate oxidase
Length = 362
Score = 371 bits (952), Expect = e-103
Identities = 178/357 (49%), Positives = 249/357 (68%), Gaps = 5/357 (1%)
Query: 4 NRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFG--KATNSNNTQHIIPIIDFAN 61
+RL++L F TKTGVKGLVD + ++P++FH P + ++ + +PIID +
Sbjct: 9 DRLNDLTTFISTKTGVKGLVDAEITEVPSMFHVPSSILSNNRPSDISGLNLTVPIIDLGD 68
Query: 62 FGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYS 121
+ +R+ + SKI++A E WGFFQV+NH +PL VLE+++ V RF EQD VK +
Sbjct: 69 --RNTSSRNVVISKIKDAAENWGFFQVINHDVPLTVLEEIKESVRRFHEQDPVVKNQYLP 126
Query: 122 RDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLG 181
D + F YN++F+LY SS LNWRD+F C +AP+ P P+++PL CR ++EY ++V +LG
Sbjct: 127 TDNNKRFVYNNDFDLYHSSPLNWRDSFTCYIAPDPPNPEEIPLACRSAVIEYTKHVMELG 186
Query: 182 MTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQ 241
LF+LLSEALGL LK + C KG+ L HYYP CP+P+LT+G +KH+D+ FLTLLLQ
Sbjct: 187 AVLFQLLSEALGLDSETLKRIDCLKGLFMLCHYYPPCPQPDLTLGISKHTDNSFLTLLLQ 246
Query: 242 DDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVA 301
D IGGLQV H++ W+D+PP+P ALVVN+GD +QL+TNDKF SVEH+V N PR+SVA
Sbjct: 247 -DQIGGLQVLHEDYWVDVPPVPGALVVNIGDFMQLITNDKFLSVEHRVRPNKDRPRISVA 305
Query: 302 CFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
CFF++ L+ S YGPIK+ LS++NP KYK+ITI EY A + D S L ++ +
Sbjct: 306 CFFSSSLSPNSTVYGPIKDLLSDENPAKYKDITIPEYTAGFLASIFDEKSYLTNYMI 362
>At1g03410 putative 1-aminocyclopropane-1-carboxylate oxidase
Length = 398
Score = 368 bits (944), Expect = e-102
Identities = 186/360 (51%), Positives = 244/360 (67%), Gaps = 6/360 (1%)
Query: 3 SNRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFG--KATNSNNTQHIIPIIDFA 60
S+R + KAFD+TKTGVKGLV G+ +IP +FH P D K T + Q IP +D
Sbjct: 41 SDRSSQAKAFDETKTGVKGLVASGIKEIPAMFHTPPDTLTSLKQTAPPSQQLTIPTVDLK 100
Query: 61 NFGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELY 120
D +R + KI +A E WGFFQVVNHGI + V+E M+ G+ RF EQD EVKK Y
Sbjct: 101 GGSMDLISRRSVVEKIGDAAERWGFFQVVNHGISVEVMERMKEGIRRFHEQDPEVKKRFY 160
Query: 121 SRDPMRPFSYNSNFNLYS-SSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTK 179
SRD R Y SN +L++ + A NWRDT C +AP+ PK QDLP VC + ++EY + +
Sbjct: 161 SRDHTRDVLYYSNIDLHTCNKAANWRDTLACYMAPDPPKLQDLPAVCGEIMMEYSKQLMT 220
Query: 180 LGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLL 239
LG LFELLSEALGL+PNHLKDMGCAK + YYP CP+P+LT+G +KH+D F+T+L
Sbjct: 221 LGEFLFELLSEALGLNPNHLKDMGCAKSHIMFGQYYPPCPQPDLTLGISKHTDFSFITIL 280
Query: 240 LQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLAN-IIGPRV 298
LQ D+IGGLQV H W+D+ P+P ALV+N+GDLLQL++NDKF S EH+V+AN PR+
Sbjct: 281 LQ-DNIGGLQVIHDQCWVDVSPVPGALVINIGDLLQLISNDKFISAEHRVIANGSSEPRI 339
Query: 299 SVACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
S+ CF + + YGPIKE LSE NP KY+++TI+E+ + ++ I AL HFR+
Sbjct: 340 SMPCFVSTFMKPNPRIYGPIKELLSEQNPAKYRDLTITEFSNTFRSQTISH-PALHHFRI 398
>At2g25450 putative dioxygenase
Length = 359
Score = 358 bits (918), Expect = 3e-99
Identities = 182/357 (50%), Positives = 241/357 (66%), Gaps = 5/357 (1%)
Query: 4 NRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDFANFG 63
+R ELKAFD+ K GVKGLVD GV K+P +FH+P +T +IP ID
Sbjct: 6 DRASELKAFDEMKIGVKGLVDAGVTKVPRIFHNPHVNVANP-KPTSTVVMIPTIDLGGVF 64
Query: 64 NDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRD 123
R + +K+++A E +GFFQ +NHG+PL+V+E M NG+ RF +QD EV+K Y+RD
Sbjct: 65 ESTVVRESVVAKVKDAMEKFGFFQAINHGVPLDVMEKMINGIRRFHDQDPEVRKMFYTRD 124
Query: 124 PMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLGMT 183
+ Y+SN +LY S A +WRDT C +AP+ PK QDLP VC + +LEY + V KL
Sbjct: 125 KTKKLKYHSNADLYESPAASWRDTLSCVMAPDVPKAQDLPEVCGEIMLEYSKEVMKLAEL 184
Query: 184 LFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDD 243
+FE+LSEALGL PNHLK+M CAKG+ L H +P CPEP T G +H+D FLT+LL D+
Sbjct: 185 MFEILSEALGLSPNHLKEMDCAKGLWMLCHCFPPCPEPNRTFGGAQHTDRSFLTILLNDN 244
Query: 244 HIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLAN-IIGPRVSVAC 302
+ GGLQV + WID+PP P+AL+ NVGD LQL++NDKF S+EH++LAN PR+SVAC
Sbjct: 245 N-GGLQVLYDGYWIDVPPNPEALIFNVGDFLQLISNDKFVSMEHRILANGGEEPRISVAC 303
Query: 303 FFNADLNSFSNQ-YGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
FF S S++ YGPIKE LSE NPPKY++ T SE +Y + +G S+L H R+
Sbjct: 304 FFVHTFTSPSSRVYGPIKELLSELNPPKYRD-TTSESSNHYVARKPNGNSSLDHLRI 359
>At3g61400 1-aminocyclopropane-1-carboxylate oxidase-like protein
Length = 370
Score = 346 bits (887), Expect = 1e-95
Identities = 178/361 (49%), Positives = 239/361 (65%), Gaps = 11/361 (3%)
Query: 3 SNRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDFANF 62
S+RL +L +F++T TGVKGLVD G+ ++P +F P Q IP ID N
Sbjct: 9 SDRLSQLNSFEETMTGVKGLVDSGIKEVPAMFREPPAILASRKPPLALQFTIPTIDL-NG 67
Query: 63 G------NDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVK 116
G D TR + KI +A E WGFFQVVNHGIPL+VLE ++ G+ F EQD E+K
Sbjct: 68 GVVYYKNQDSVTRRSMVEKIGDAAEKWGFFQVVNHGIPLDVLEKVKEGIRAFHEQDAELK 127
Query: 117 KELYSRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEY 176
K YSRD R Y SN +L+++ +WRDT +AP+ P +DLP VC + ++EY +
Sbjct: 128 KRFYSRDHTRKMVYYSNLDLFTAMKASWRDTMCAYMAPDPPTSEDLPEVCGEIMMEYAKE 187
Query: 177 VTKLGMTLFELLSEALGL-HPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCF 235
+ LG +FELLSEALGL + NHLKDM C+K ++ YYP CP+P+ T+G +KH+D F
Sbjct: 188 IMNLGELIFELLSEALGLNNSNHLKDMDCSKSLVLFGQYYPPCPQPDHTLGLSKHTDFSF 247
Query: 236 LTLLLQDDHIGGLQVFHQNK-WIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLAN-I 293
LT++LQ ++GGLQV H + WIDIPP+P ALVVN+GDLLQL++N KF SVEH+V+AN
Sbjct: 248 LTIVLQ-GNLGGLQVLHDKQYWIDIPPVPGALVVNLGDLLQLISNGKFISVEHRVIANRA 306
Query: 294 IGPRVSVACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISAL 353
PR+SV CFF+ + YGPIKE LSE NPPKY++ TISE+ + Y +K I+ + L
Sbjct: 307 AEPRISVPCFFSTVMRESHRVYGPIKELLSEQNPPKYRDTTISEFASMYASKEINTSALL 366
Query: 354 Q 354
+
Sbjct: 367 R 367
>At1g03400 putative 1-aminocyclopropane-1-carboxylate oxidase
Length = 351
Score = 335 bits (858), Expect = 3e-92
Identities = 170/358 (47%), Positives = 230/358 (63%), Gaps = 12/358 (3%)
Query: 3 SNRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHI-IPIIDFAN 61
++R + KAFD+ K GVKGLVD G+ +IP LF + +H+ IP +D
Sbjct: 4 TDRSSQAKAFDEAKIGVKGLVDSGITEIPALFRATPATLASLKSPPPPKHLTIPTVDLKG 63
Query: 62 FGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYS 121
+ KI EA E WG F +VNHGIP+ VLE M G+ F EQ+ E KK YS
Sbjct: 64 AS--------VVEKIGEAAEKWGLFHLVNHGIPVEVLERMIQGIRGFHEQEPEAKKRFYS 115
Query: 122 RDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLG 181
RD R Y SN +L +S A +WRDT C AP P+ +DLP VC + +LEY + + LG
Sbjct: 116 RDHTRDVLYFSNHDLQNSEAASWRDTLGCYTAPEPPRLEDLPAVCGEIMLEYSKEIMSLG 175
Query: 182 MTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQ 241
LFELLSEALGL+ +HLKDM CAK + +YP CP+P+LT+G KH+D FLT+LLQ
Sbjct: 176 ERLFELLSEALGLNSHHLKDMDCAKSQYMVGQHYPPCPQPDLTIGINKHTDISFLTVLLQ 235
Query: 242 DDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLAN-IIGPRVSV 300
D++GGLQVFH+ WID+ P+P ALV+N+GD LQL+TNDKF S EH+V+AN PR SV
Sbjct: 236 -DNVGGLQVFHEQYWIDVTPVPGALVINIGDFLQLITNDKFISAEHRVIANGSSEPRTSV 294
Query: 301 ACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
A F+ + ++S YGPIK+ LS +NP KY++ T++E+ +++K +D L HF++
Sbjct: 295 AIVFSTFMRAYSRVYGPIKDLLSAENPAKYRDCTLTEFSTIFSSKTLDA-PKLHHFKI 351
>At3g13610 unknown protein
Length = 361
Score = 206 bits (525), Expect = 1e-53
Identities = 117/345 (33%), Positives = 182/345 (51%), Gaps = 13/345 (3%)
Query: 18 GVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDFANFGNDPKTRHEITSKIR 77
GVKGL + G+ +P + PL++ N T IP+ID +N D + +
Sbjct: 26 GVKGLSETGIKALPEQYIQPLEERLINKFVNETDEAIPVIDMSNPDED-----RVAEAVC 80
Query: 78 EACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMRP-FSYNSNFNL 136
+A E WGFFQV+NHG+PL VL+D++ +FF VE K++ + + + ++F+
Sbjct: 81 DAAEKWGFFQVINHGVPLEVLDDVKAATHKFFNLPVEEKRKFTKENSLSTTVRFGTSFSP 140
Query: 137 YSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLGMTLFELLSEALGLHP 196
+ AL W+D A Q P +CR+E LEY K+ L E L + L +
Sbjct: 141 LAEQALEWKDYLSLFFVSEAEAEQFWPDICRNETLEYINKSKKMVRRLLEYLGKNLNVKE 200
Query: 197 NHLKDMGCAKGILSLS-HYYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQV--FHQ 253
G + ++ +YYP CP P+LT+G +HSD LT+LLQD IGGL V
Sbjct: 201 LDETKESLFMGSIRVNLNYYPICPNPDLTVGVGRHSDVSSLTILLQDQ-IGGLHVRSLAS 259
Query: 254 NKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNADLNSFSN 313
W+ +PP+ + V+N+GD +Q+++N +KSVEH+VLAN R+SV F N S
Sbjct: 260 GNWVHVPPVAGSFVINIGDAMQIMSNGLYKSVEHRVLANGYNNRISVPIFVNPKPESV-- 317
Query: 314 QYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
GP+ E ++ P Y+++ S+YV Y+ K DG + + ++
Sbjct: 318 -IGPLPEVIANGEEPIYRDVLYSDYVKYFFRKAHDGKKTVDYAKI 361
>At2g38240 putative anthocyanidin synthase
Length = 353
Score = 197 bits (500), Expect = 1e-50
Identities = 122/338 (36%), Positives = 182/338 (53%), Gaps = 20/338 (5%)
Query: 19 VKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHI-IPIIDFANFGNDPKTRHEITSKIR 77
V+ L GV +P + P + + + I IP++D ND + E +R
Sbjct: 12 VQSLSQTGVPTVPNRYVKPAHQRPVFNTTQSDAGIEIPVLDM----NDVWGKPEGLRLVR 67
Query: 78 EACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMRPFSYNSNFNLY 137
ACE WGFFQ+VNHG+ +++E +R FFE +E K++ Y+ P Y S +
Sbjct: 68 SACEEWGFFQMVNHGVTHSLMERVRGAWREFFELPLEEKRK-YANSPDTYEGYGSRLGVV 126
Query: 138 SSSALNWRDTFVCSLAP----NAPKPQDLPLVCRDELLEYGEYVTKLGMTLFELLSEALG 193
+ L+W D F + P N K P R+ + +YGE V KL L E LSE+LG
Sbjct: 127 KDAKLDWSDYFFLNYLPSSIRNPSKWPSQPPKIRELIEKYGEEVRKLCERLTETLSESLG 186
Query: 194 LHPNHLKDM---GCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQV 250
L PN L G G +++YP CP+P+LT+G + HSD +T+LL D+ + GLQV
Sbjct: 187 LKPNKLMQALGGGDKVGASLRTNFYPKCPQPQLTLGLSSHSDPGGITILLPDEKVAGLQV 246
Query: 251 FHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFN--ADL 308
+ W+ I +P+AL+VN+GD LQ+++N +KSVEH+V+ N RVS+A F+N +D+
Sbjct: 247 RRGDGWVTIKSVPNALIVNIGDQLQILSNGIYKSVEHQVIVNSGMERVSLAFFYNPRSDI 306
Query: 309 NSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKG 346
GPI+E ++ + P YK I EY + KG
Sbjct: 307 -----PVGPIEELVTANRPALYKPIRFDEYRSLIRQKG 339
>At5g05600 leucoanthocyanidin dioxygenase-like protein
Length = 371
Score = 195 bits (496), Expect = 3e-50
Identities = 117/343 (34%), Positives = 173/343 (50%), Gaps = 11/343 (3%)
Query: 19 VKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDFAN-FGNDPKTRHEITSKIR 77
V+ L + + +P + P T T IPIID F + + I ++I
Sbjct: 27 VQSLAESNLSSLPDRYIKPASLRPTTTEDAPTATNIPIIDLEGLFSEEGLSDDVIMARIS 86
Query: 78 EACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMRPFSYNSNFNLY 137
EAC WGFFQVVNHG+ +++ R FF V K E YS P Y S +
Sbjct: 87 EACRGWGFFQVVNHGVKPELMDAARENWREFFHMPVNAK-ETYSNSPRTYEGYGSRLGVE 145
Query: 138 SSSALNWRDTFVCSLAPNAPKP----QDLPLVCRDELLEYGEYVTKLGMTLFELLSEALG 193
++L+W D + L P+ K P R+ + EYGE + KL + +LS LG
Sbjct: 146 KGASLDWSDYYFLHLLPHHLKDFNKWPSFPPTIREVIDEYGEELVKLSGRIMRVLSTNLG 205
Query: 194 LHPNHLKDM--GCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQVF 251
L + ++ G G +YYP CP PEL +G + HSD +T+LL DD + GLQV
Sbjct: 206 LKEDKFQEAFGGENIGACLRVNYYPKCPRPELALGLSPHSDPGGMTILLPDDQVFGLQVR 265
Query: 252 HQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNADLNSF 311
+ WI + P P A +VN+GD +Q+++N +KSVEH+V+ N RVS+A F+N +
Sbjct: 266 KDDTWITVKPHPHAFIVNIGDQIQILSNSTYKSVEHRVIVNSDKERVSLAFFYNPKSDI- 324
Query: 312 SNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQ 354
P++E +S NPP Y +T +Y + T+G G S ++
Sbjct: 325 --PIQPLQELVSTHNPPLYPPMTFDQYRLFIRTQGPQGKSHVE 365
>At1g55290 leucoanthocyanidin dioxygenase 2, putative
Length = 361
Score = 194 bits (492), Expect = 8e-50
Identities = 120/340 (35%), Positives = 183/340 (53%), Gaps = 20/340 (5%)
Query: 18 GVKGLVDKGVVKIPTLFHHPLDKFGKATN---SNNTQHIIPIIDFANFGNDPKTRHEITS 74
GVKGL + G+ +P + P ++ + N ++ IP+ID +N D K+ ++
Sbjct: 25 GVKGLSETGIKVLPDQYIQPFEE--RLINFHVKEDSDESIPVIDISNL--DEKS---VSK 77
Query: 75 KIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMRPFS--YNS 132
+ +A E WGFFQV+NHG+ + VLE+M+ RFF VE K++ +SR+ + + +
Sbjct: 78 AVCDAAEEWGFFQVINHGVSMEVLENMKTATHRFFGLPVEEKRK-FSREKSLSTNVRFGT 136
Query: 133 NFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLGMTLFELLSEAL 192
+F+ ++ AL W+D A Q P CR E LEY L L L E L
Sbjct: 137 SFSPHAEKALEWKDYLSLFFVSEAEASQLWPDSCRSETLEYMNETKPLVKKLLRFLGENL 196
Query: 193 GLHPNHLKDMGCAKGILSLS-HYYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQV- 250
+ G ++ +YYP CP PELT+G +HSD LT+LLQD+ IGGL V
Sbjct: 197 NVKELDKTKESFFMGSTRINLNYYPICPNPELTVGVGRHSDVSSLTILLQDE-IGGLHVR 255
Query: 251 -FHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNADLN 309
+W+ +PPI +LV+N+GD +Q+++N ++KSVEH+VLAN R+SV F +
Sbjct: 256 SLTTGRWVHVPPISGSLVINIGDAMQIMSNGRYKSVEHRVLANGSYNRISVPIFVSPKPE 315
Query: 310 SFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDG 349
S GP+ E + P YK+I ++YV ++ K DG
Sbjct: 316 SV---IGPLLEVIENGEKPVYKDILYTDYVKHFFRKAHDG 352
>At3g11180 putative leucoanthocyanidin dioxygenase
Length = 400
Score = 190 bits (482), Expect = 1e-48
Identities = 117/348 (33%), Positives = 180/348 (51%), Gaps = 21/348 (6%)
Query: 19 VKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHI----IPIIDFANF--GNDPKTRHEI 72
V+ L + + +P + P + + T ++ + IPIID + GN+ +
Sbjct: 56 VQSLAESNLTSLPDRYIKPPSQRPQTTIIDHQPEVADINIPIIDLDSLFSGNEDDKK--- 112
Query: 73 TSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMRPFSYNS 132
+I EAC WGFFQV+NHG+ +++ R FF VE K E+YS P Y S
Sbjct: 113 --RISEACREWGFFQVINHGVKPELMDAARETWKSFFNLPVEAK-EVYSNSPRTYEGYGS 169
Query: 133 NFNLYSSSALNWRDTFVCSLAPNAPKP----QDLPLVCRDELLEYGEYVTKLGMTLFELL 188
+ + L+W D + P A K LP R+ EYG+ + KLG L +L
Sbjct: 170 RLGVEKGAILDWNDYYYLHFLPLALKDFNKWPSLPSNIREMNDEYGKELVKLGGRLMTIL 229
Query: 189 SEALGLHPNHLKDM--GCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDDHIG 246
S LGL L++ G G +YYP CP+PEL +G + HSD +T+LL DD +
Sbjct: 230 SSNLGLRAEQLQEAFGGEDVGACLRVNYYPKCPQPELALGLSPHSDPGGMTILLPDDQVV 289
Query: 247 GLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNA 306
GLQV H + WI + P+ A +VN+GD +Q+++N K+KSVEH+V+ N RVS+A F+N
Sbjct: 290 GLQVRHGDTWITVNPLRHAFIVNIGDQIQILSNSKYKSVEHRVIVNSEKERVSLAFFYNP 349
Query: 307 DLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQ 354
+ P+++ ++ PP Y +T +Y + T+G G S ++
Sbjct: 350 KSDI---PIQPMQQLVTSTMPPLYPPMTFDQYRLFIRTQGPRGKSHVE 394
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,825,883
Number of Sequences: 26719
Number of extensions: 396215
Number of successful extensions: 1096
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 98
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 770
Number of HSP's gapped (non-prelim): 116
length of query: 358
length of database: 11,318,596
effective HSP length: 100
effective length of query: 258
effective length of database: 8,646,696
effective search space: 2230847568
effective search space used: 2230847568
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146817.6


