

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146807.7 - phase: 0 /pseudo
(97 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
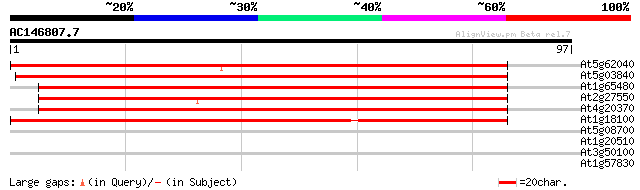
At5g62040 Fdr1 Cen - like protein 122 2e-29
At5g03840 Terminal flower1 (TFL1) 119 3e-28
At1g65480 flowering time locus T (FT) 112 2e-26
At2g27550 Arabidopsis thaliana centroradialis (ATC) 111 7e-26
At4g20370 TWIN SISTER OF FT (TSF) 101 6e-23
At1g18100 terminal Flower 1 (TFL1), putative 94 2e-20
At5g08700 putative protein 29 0.46
At1g20510 4-coumarate:CoA ligase like protein 26 3.0
At3g50100 hypothetical protein 25 5.0
At1g57830 hypothetical protein 25 8.6
>At5g62040 Fdr1 Cen - like protein
Length = 177
Score = 122 bits (307), Expect = 2e-29
Identities = 56/87 (64%), Positives = 77/87 (88%), Gaps = 1/87 (1%)
Query: 1 MSRPLEPLSVGRVIGEVVDIFNPSVRMNVTYSTKQ-VANGHELMPSIVMNKPRVDIGGED 59
MSR +EPL VGRVIG+V+++FNPSV M VT+++ V+NGHEL PS++++KPRV+IGG+D
Sbjct: 1 MSREIEPLIVGRVIGDVLEMFNPSVTMRVTFNSNTIVSNGHELAPSLLLSKPRVEIGGQD 60
Query: 60 MRSAYTLIMTDPDAPSPSDPHLREHLH 86
+RS +TLIM DPDAPSPS+P++RE+LH
Sbjct: 61 LRSFFTLIMMDPDAPSPSNPYMREYLH 87
>At5g03840 Terminal flower1 (TFL1)
Length = 177
Score = 119 bits (297), Expect = 3e-28
Identities = 52/85 (61%), Positives = 69/85 (81%)
Query: 2 SRPLEPLSVGRVIGEVVDIFNPSVRMNVTYSTKQVANGHELMPSIVMNKPRVDIGGEDMR 61
+R +EPL +GRV+G+V+D F P+ +MNV+Y+ KQV+NGHEL PS V +KPRV+I G D+R
Sbjct: 6 TRVIEPLIMGRVVGDVLDFFTPTTKMNVSYNKKQVSNGHELFPSSVSSKPRVEIHGGDLR 65
Query: 62 SAYTLIMTDPDAPSPSDPHLREHLH 86
S +TL+M DPD P PSDP L+EHLH
Sbjct: 66 SFFTLVMIDPDVPGPSDPFLKEHLH 90
>At1g65480 flowering time locus T (FT)
Length = 175
Score = 112 bits (281), Expect = 2e-26
Identities = 50/81 (61%), Positives = 65/81 (79%)
Query: 6 EPLSVGRVIGEVVDIFNPSVRMNVTYSTKQVANGHELMPSIVMNKPRVDIGGEDMRSAYT 65
+PL V RV+G+V+D FN S+ + VTY ++V NG +L PS V NKPRV+IGGED+R+ YT
Sbjct: 7 DPLIVSRVVGDVLDPFNRSITLKVTYGQREVTNGLDLRPSQVQNKPRVEIGGEDLRNFYT 66
Query: 66 LIMTDPDAPSPSDPHLREHLH 86
L+M DPD PSPS+PHLRE+LH
Sbjct: 67 LVMVDPDVPSPSNPHLREYLH 87
>At2g27550 Arabidopsis thaliana centroradialis (ATC)
Length = 175
Score = 111 bits (277), Expect = 7e-26
Identities = 54/82 (65%), Positives = 65/82 (78%), Gaps = 1/82 (1%)
Query: 6 EPLSVGRVIGEVVDIFNPSVRMNVTY-STKQVANGHELMPSIVMNKPRVDIGGEDMRSAY 64
+PL VGRVIG+VVD +V+M VTY S KQV NGHEL PS+V KP+V++ G DMRS +
Sbjct: 7 DPLMVGRVIGDVVDNCLQAVKMTVTYNSDKQVYNGHELFPSVVTYKPKVEVHGGDMRSFF 66
Query: 65 TLIMTDPDAPSPSDPHLREHLH 86
TL+MTDPD P PSDP+LREHLH
Sbjct: 67 TLVMTDPDVPGPSDPYLREHLH 88
>At4g20370 TWIN SISTER OF FT (TSF)
Length = 175
Score = 101 bits (252), Expect = 6e-23
Identities = 46/81 (56%), Positives = 61/81 (74%)
Query: 6 EPLSVGRVIGEVVDIFNPSVRMNVTYSTKQVANGHELMPSIVMNKPRVDIGGEDMRSAYT 65
+PL VG V+G+V+D F V + VTY ++V NG +L PS V+NKP V+IGG+D R+ YT
Sbjct: 7 DPLVVGSVVGDVLDPFTRLVSLKVTYGHREVTNGLDLRPSQVLNKPIVEIGGDDFRNFYT 66
Query: 66 LIMTDPDAPSPSDPHLREHLH 86
L+M DPD PSPS+PH RE+LH
Sbjct: 67 LVMVDPDVPSPSNPHQREYLH 87
>At1g18100 terminal Flower 1 (TFL1), putative
Length = 173
Score = 93.6 bits (231), Expect = 2e-20
Identities = 43/86 (50%), Positives = 62/86 (72%), Gaps = 1/86 (1%)
Query: 1 MSRPLEPLSVGRVIGEVVDIFNPSVRMNVTYSTKQVANGHELMPSIVMNKPRVDIGGEDM 60
M+ ++PL VGRVIG+V+D+F P+ M+V + K + NG E+ PS +N P+V+I G
Sbjct: 1 MAASVDPLVVGRVIGDVLDMFIPTANMSVYFGPKHITNGCEIKPSTAVNPPKVNISGHS- 59
Query: 61 RSAYTLIMTDPDAPSPSDPHLREHLH 86
YTL+MTDPDAPSPS+P++RE +H
Sbjct: 60 DELYTLVMTDPDAPSPSEPNMREWVH 85
>At5g08700 putative protein
Length = 768
Score = 28.9 bits (63), Expect = 0.46
Identities = 18/69 (26%), Positives = 28/69 (40%), Gaps = 3/69 (4%)
Query: 29 VTYSTKQVANGHELMPSIVMNKPRVDIGGEDMRSAYTLIMTDPDAPSPSDPHLREHLH*Y 88
+TY K + H++ + PR G D + + D DP L+E LH
Sbjct: 89 ITYFNKNFKHSHQISVLVADPPPRRTQGKVDAYAKGDKQIQKKDPEVDHDPQLQEFLH-- 146
Query: 89 SRYHRCLFW 97
+ H+ FW
Sbjct: 147 -QEHKLKFW 154
>At1g20510 4-coumarate:CoA ligase like protein
Length = 546
Score = 26.2 bits (56), Expect = 3.0
Identities = 14/65 (21%), Positives = 32/65 (48%), Gaps = 1/65 (1%)
Query: 7 PLSVGRVIGEVVDIFNPSVRMNVTYSTKQVANGHELMPSIVMNKPRVDIGGEDMRSAYTL 66
PL+ I + + NP + + +++ + +P ++M++ RVD G D+R +
Sbjct: 114 PLNTSNEIAKQIKDSNPVLAFTTSQLLPKISAAAKKLPIVLMDEERVDSVG-DVRRLVEM 172
Query: 67 IMTDP 71
+ +P
Sbjct: 173 MKKEP 177
>At3g50100 hypothetical protein
Length = 406
Score = 25.4 bits (54), Expect = 5.0
Identities = 13/31 (41%), Positives = 17/31 (53%)
Query: 54 DIGGEDMRSAYTLIMTDPDAPSPSDPHLREH 84
D+ GE+ L + D +A SPSDP R H
Sbjct: 24 DLRGENGNWKEFLHVYDKNADSPSDPSRRSH 54
>At1g57830 hypothetical protein
Length = 164
Score = 24.6 bits (52), Expect = 8.6
Identities = 13/31 (41%), Positives = 19/31 (60%), Gaps = 2/31 (6%)
Query: 36 VANGHELMPSIVMNKPRVDIG--GEDMRSAY 64
VA H + PS++ +PRV I GED+R +
Sbjct: 2 VAISHIIQPSLMQLQPRVFINFRGEDIRYGF 32
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,296,086
Number of Sequences: 26719
Number of extensions: 86088
Number of successful extensions: 208
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 196
Number of HSP's gapped (non-prelim): 10
length of query: 97
length of database: 11,318,596
effective HSP length: 73
effective length of query: 24
effective length of database: 9,368,109
effective search space: 224834616
effective search space used: 224834616
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146807.7


