

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146806.4 + phase: 0 /pseudo
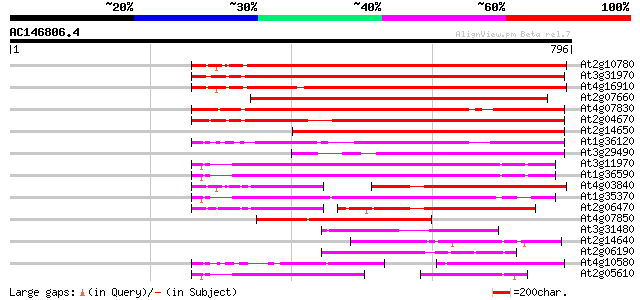
(796 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 499 e-141
At3g31970 hypothetical protein 479 e-135
At4g16910 retrotransposon like protein 475 e-134
At2g07660 putative retroelement pol polyprotein 466 e-131
At4g07830 putative reverse transcriptase 425 e-119
At2g04670 putative retroelement pol polyprotein 424 e-118
At2g14650 putative retroelement pol polyprotein 389 e-108
At1g36120 putative reverse transcriptase gb|AAD22339.1 362 e-100
At3g29490 hypothetical protein 291 1e-78
At3g11970 hypothetical protein 281 1e-75
At1g36590 hypothetical protein 276 3e-74
At4g03840 putative transposon protein 251 9e-67
At1g35370 hypothetical protein 242 5e-64
At2g06470 putative retroelement pol polyprotein 227 2e-59
At4g07850 putative polyprotein 208 1e-53
At3g31480 hypothetical protein 187 2e-47
At2g14640 putative retroelement pol polyprotein 167 2e-41
At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein 162 5e-40
At4g10580 putative reverse-transcriptase -like protein 135 7e-32
At2g05610 putative retroelement pol polyprotein 120 3e-27
>At2g10780 pseudogene
Length = 1611
Score = 499 bits (1286), Expect = e-141
Identities = 263/537 (48%), Positives = 352/537 (64%), Gaps = 12/537 (2%)
Query: 258 PGKANVVADVLSRKTLHMYALMVKELELIEQFGEL---SLVSELTPDGVRLGMLKLTSNI 314
PGKAN VAD LSR+ + A +++L+ G L +L E+ P G LG +++
Sbjct: 1057 PGKANQVADALSRRRSEVEAER-SQVDLVNMMGTLHVNALSKEVEPLG--LGAAD-QADL 1112
Query: 315 LEEIKNGQKEDLELVDRVTLVNQGKGGDFRLDENDVLMFRDRVCVPDVLDLKRQILDEGH 374
L I+ Q+ D E+ Q +++ N ++ RVCVP+ LK +IL E H
Sbjct: 1113 LSRIRLAQERDEEIKGWA----QNNKTEYQTSNNGTIVVNGRVCVPNDRALKEEILREAH 1168
Query: 375 RSSLSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVP 434
+S SIHPG+ KMY+DLKR + W GMKK++A +V C CQ EHQ PSGL+Q L +P
Sbjct: 1169 QSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPIP 1228
Query: 435 EWTWDSISMDFVGALPKTSKS-FDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIV 493
EW WD I+MDFV LP KS + +WV+VDRLTKS HF+ I +AE YI++IV
Sbjct: 1229 EWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIV 1288
Query: 494 RLHGIPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRA 553
RLHGIP SIVSDRD RFTS FW++ Q ALGT+++LS+AYHPQTD Q+ERTIQ+LED+LRA
Sbjct: 1289 RLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQSERTIQTLEDMLRA 1348
Query: 554 CVLEQGVSWDECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPEV 613
CVL+ G +W++ L L+EF YNNSF +SI M+P+EALYGR CRTPLCW GE + P +
Sbjct: 1349 CVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGERRLFGPTI 1408
Query: 614 VQETTEKVKMIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKL 673
V ETTE++K ++ K+K +Q KSY +KRRK++EFQVGD V+L G G +KL
Sbjct: 1409 VDETTERMKFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKL 1468
Query: 674 TPRFVGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHVIRVDDLEVRDN 733
+PR+VGP+ VIE+VG VAY++ LPP L+ HNVFHV +LRK + D + +++N
Sbjct: 1469 SPRYVGPYKVIERVGAVAYKLDLPPKLNAFHNVFHVSQLRKCLSDQEESVEDIPPGLKEN 1528
Query: 734 LTVETWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWEPESGMMVSYRSCFLQV 790
+TVE WPVRI DR K RGK L+KV+W E TWE E+ M ++ F ++
Sbjct: 1529 MTVEAWPVRIMDRMTKGTRGKARDLLKVLWNCRGREEYTWETENKMKANFPEWFKEM 1585
>At3g31970 hypothetical protein
Length = 1329
Score = 479 bits (1234), Expect = e-135
Identities = 249/530 (46%), Positives = 347/530 (64%), Gaps = 14/530 (2%)
Query: 259 GKANVVADVLSRKTLHMYALMVKELELIEQFGELSL-VSELTPDGVRLGMLKLTSNILEE 317
GKANVVAD LSRK + L+ + G L L V P G+ +++L
Sbjct: 807 GKANVVADALSRKRVGGSVEA-----LVSEIGALRLCVMAQEPLGLEAVD---RADLLTR 858
Query: 318 IKNGQKEDLELVDRVTLVNQGKGGDFRLDENDVLMFRDRVCVPDVLDLKRQILDEGHRSS 377
++ Q++D L+ ++ G +++ N ++ RVCVP +L+R+IL E H S
Sbjct: 859 VRLAQEKDEGLI----AASKADGSEYQFAANGTILVHGRVCVPKDEELRREILSEAHASM 914
Query: 378 LSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVPEWT 437
SIHP ATKMY+DLKR + W GMK+++A +V C VCQ EHQ P GL+Q L + EW
Sbjct: 915 FSIHPRATKMYRDLKRYYQWVGMKRDVANWVTECDVCQLVKAEHQVPGGLLQSLPILEWK 974
Query: 438 WDSISMDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLHG 497
WD I+MDFV LP S++ D IWVIVDRLTKS HF+ I+ + LA+ Y+ +IV LHG
Sbjct: 975 WDFITMDFVVGLP-VSRTKDAIWVIVDRLTKSAHFLAIRKTDGAVLLAKKYVSEIVELHG 1033
Query: 498 IPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLE 557
+P SIVSDRD +FTS FW + Q +GTK+ +S+AYHPQTDGQ+ERTIQ+LED+LR CVL+
Sbjct: 1034 VPVSIVSDRDSKFTSAFWRAFQGEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLD 1093
Query: 558 QGVSWDECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPEVVQET 617
+G W + L L+EF YNNS+ +SIRMAPFEALYGR CRTPLCW + GE ++ + V ET
Sbjct: 1094 RGGHWADHLSLVEFAYNNSYQASIRMAPFEALYGRPCRTPLCWTQVGERSIYGADYVLET 1153
Query: 618 TEKVKMIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKLTPRF 677
TE++++++ MK +Q +SY DKRR+++EF+VGD V+L + + G ++ KLTPR+
Sbjct: 1154 TERIRVLKLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLTPRY 1213
Query: 678 VGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHVIRVDDLEVRDNLTVE 737
+GPF ++E+VG VAYR+ LP + H VFHV LRK +H V+ +++ N+T+E
Sbjct: 1214 MGPFRIVERVGPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEVLAKILEDLQPNMTLE 1273
Query: 738 TWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWEPESGMMVSYRSCF 787
PVRI +R +K LR K+I L+KV+W TWEPE+ M S++ F
Sbjct: 1274 ARPVRILERRIKELRRKKIPLIKVLWNCDGVTEETWEPEARMKASFKKWF 1323
>At4g16910 retrotransposon like protein
Length = 687
Score = 475 bits (1222), Expect = e-134
Identities = 253/537 (47%), Positives = 344/537 (63%), Gaps = 20/537 (3%)
Query: 258 PGKANVVADVLSRKTLHMYALMVKELELIEQFGEL---SLVSELTPDGVRLGMLKLTSNI 314
PGKAN V D LSR + A ++ L+ G L +L E+ P G+R +++
Sbjct: 149 PGKANQVVDALSRLRTVVEAER-NQVNLVNMMGTLHLNALSKEVEPLGLRAAN---QADL 204
Query: 315 LEEIKNGQKEDLELVDRVTLVNQGKGGDFRLDENDVLMFRDRVCVPDVLDLKRQILDEGH 374
L I++ Q+ D E+ Q +++ N ++ RVC P+ LK +IL E H
Sbjct: 205 LSRIRSAQERDEEIKGWA----QNNKTEYQSSNNGTIVVNGRVCGPNDKALKEEILKEAH 260
Query: 375 RSSLSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVP 434
+S SIHPG+ KMY+DLKR + W GMKK++A +V + EHQ PSG++Q L +P
Sbjct: 261 QSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWV--------AKEEHQVPSGMLQNLPIP 312
Query: 435 EWTWDSISMDFVGALPKTSKS-FDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIV 493
EW WD I MDFV LP KS + +WV+VDRLTKS HF+ I + +AE YI++IV
Sbjct: 313 EWKWDHIMMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDAAEIIAEKYIDEIV 372
Query: 494 RLHGIPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRA 553
RLHGIP SIVSDRD RFTS FW+ Q LGT+++LS+AYHPQTDGQ+ERTIQ+LED+LRA
Sbjct: 373 RLHGIPVSIVSDRDTRFTSKFWKPFQKVLGTRVNLSTAYHPQTDGQSERTIQTLEDMLRA 432
Query: 554 CVLEQGVSWDECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPEV 613
CVL+ G +W++ L L+EF YNNSF +SI M+P+EALYGR RTPLCW GE + P V
Sbjct: 433 CVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRAGRTPLCWTPVGERRLFGPAV 492
Query: 614 VQETTEKVKMIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKL 673
V ETT+K+K ++ K+K +Q KSY +KRRK++EFQVGD V+L G G +KL
Sbjct: 493 VDETTKKMKFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKL 552
Query: 674 TPRFVGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHVIRVDDLEVRDN 733
PR+VGP+ VIE+VG VAY++ LPP L HNVFHV +LRK + + + +++N
Sbjct: 553 RPRYVGPYKVIERVGAVAYKLDLPPKLDAFHNVFHVSQLRKCLSEQEESMEDVPPGLKEN 612
Query: 734 LTVETWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWEPESGMMVSYRSCFLQV 790
+TVE WPVRI D+ K RGK + L+K++W E TWE E+ M ++ F ++
Sbjct: 613 MTVEAWPVRIMDQMKKGTRGKSMDLLKILWNCGGREEYTWETETKMKANFPEWFKEM 669
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 466 bits (1199), Expect = e-131
Identities = 228/422 (54%), Positives = 298/422 (70%), Gaps = 1/422 (0%)
Query: 342 DFRLDENDVLMFRDRVCVPDVLDLKRQILDEGHRSSLSIHPGATKMYQDLKRLFWWPGMK 401
+++ N ++ RVCVP+ LK +IL E H+S SIHPG+ KMY+DLKR + W GM+
Sbjct: 528 EYQTSNNGTIVVNGRVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMR 587
Query: 402 KEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVPEWTWDSISMDFVGALPKTSKS-FDTIW 460
K++A +V C CQ EHQ PSGL+Q L + EW WD I+MDFV LP KS + +W
Sbjct: 588 KDVARWVAKCPTCQLVKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAVW 647
Query: 461 VIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLHGIPSSIVSDRDPRFTSNFWESLQA 520
V+VDRLTKS HF+ I +AE YI++I+RLHGIP SIVSDRD RFTS FW + Q
Sbjct: 648 VVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIMRLHGIPVSIVSDRDTRFTSKFWNAFQK 707
Query: 521 ALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLEQGVSWDECLPLIEFTYNNSFHSS 580
ALGT+++LS+AYHPQTDGQ+ERTIQ+LED+LRACVL+ G +W++ L LIEF YNNSF +S
Sbjct: 708 ALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLIEFAYNNSFQAS 767
Query: 581 IRMAPFEALYGRRCRTPLCWFESGESAMLAPEVVQETTEKVKMIQEKMKASQST*KSYHD 640
I M+P+EALYGR CRTPLCW GE + P +V ETTE++K ++ K+K +Q KSY +
Sbjct: 768 IGMSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYAN 827
Query: 641 KRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKLTPRFVGPFDVIEKVGVVAYRIALPPSL 700
KRRK++EFQVGD V+L G G +KL+PR+VGP+ VIE+VG VAY++ LPP L
Sbjct: 828 KRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKL 887
Query: 701 SNLHNVFHVPKLRKYVHDASHVIRVDDLEVRDNLTVETWPVRIEDRELKRLRGKEIVLVK 760
+ HNVFHV +LRKY+ D + +++N+TVE WPVRI DR K RGK L+K
Sbjct: 888 NVFHNVFHVSQLRKYLSDQEESVEDIPPGLKENMTVEAWPVRIMDRMSKGTRGKSRDLLK 947
Query: 761 VI 762
V+
Sbjct: 948 VL 949
>At4g07830 putative reverse transcriptase
Length = 611
Score = 425 bits (1092), Expect = e-119
Identities = 233/530 (43%), Positives = 332/530 (61%), Gaps = 29/530 (5%)
Query: 258 PGKANVVADVLSRKTLHMYALMVKELELIEQFGELSLVSELTPDGVRLGMLKLTSNILEE 317
PGKANVV D LSRK + E +IE G L L + + + + L + T ++L
Sbjct: 69 PGKANVVTDALSRKRVGAAPGQSVETLVIE-IGALRLCA-VAREPLGLEAVDQT-DLLSR 125
Query: 318 IKNGQKEDLELVDRVTLVNQGKGGDFRLDENDVLMFRDRVCVPDVLDLKRQILDEGHRSS 377
++ Q++D L+ ++ +G +++ N ++ RVCVP +L+++IL E H S
Sbjct: 126 VQLAQEKDEGLI----AASKAEGFEYQFAANGTILVHGRVCVPKDKELRQEILSEAHASM 181
Query: 378 LSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVPEWT 437
SIHPGATKMY+DLKR + W GMK+++ +V C VCQ IEHQ L+Q L +PEW
Sbjct: 182 FSIHPGATKMYRDLKRYYQWVGMKRDVGNWVEECDVCQLVKIEHQVSGSLLQSLPIPEWK 241
Query: 438 WDSISMDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLHG 497
WD I+MDFV LP S++ D IWVIVDRLTKS HF+ I+ + A LA+ Y+ +IV+LHG
Sbjct: 242 WDFITMDFVVGLP-VSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHG 300
Query: 498 IPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLE 557
+P SIVSDRD +FTS FW + QA +GTK+ +S+AYHPQTDGQ+ERTIQ+LED+LR CVL+
Sbjct: 301 VPVSIVSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLD 360
Query: 558 QGVSWDECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPEVVQET 617
W + L L+EF YNNS+ +SI MAPFE LYGR CRT LCW + GE ++ + VQE
Sbjct: 361 WRGHWADHLSLVEFAYNNSYQASIGMAPFEVLYGRPCRT-LCWTQVGERSIYGADYVQEI 419
Query: 618 TEKVKMIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKLTPRF 677
TE++++++ MK +Q+ +SY DKRRK++EF+VGD V+ GH +
Sbjct: 420 TERIRVLKLNMKEAQNRQRSYADKRRKELEFEVGD-------SVSQDGHVARSE------ 466
Query: 678 VGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHVIRVDDLEVRDNLTVE 737
++VG VA+R+ L + H VFHV LRK +H V+ +++ N+T+E
Sbjct: 467 -------QRVGPVAFRLELSDVMRAFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLE 519
Query: 738 TWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWEPESGMMVSYRSCF 787
PVR+ +R +K LR K+I L+KV+ TWEPE+ + ++ F
Sbjct: 520 ARPVRVLERRIKELRRKKIPLIKVLRNCDGVTEETWEPEARLKARFKKWF 569
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 424 bits (1089), Expect = e-118
Identities = 230/531 (43%), Positives = 326/531 (61%), Gaps = 41/531 (7%)
Query: 258 PGKANVVADVLSRKTLHMYALMVKELELIEQFGELSL-VSELTPDGVRLGMLKLTSNILE 316
PGKANVVAD LSRK + E L+ + G L L V P G+ +++L
Sbjct: 915 PGKANVVADALSRKRVGAAPGQSVEA-LVSEIGALRLCVVAREPLGLEAVD---RADLLT 970
Query: 317 EIKNGQKEDLELVDRVTLVNQGKGGDFRLDENDVLMFRDRVCVPDVLDLKRQILDEGHRS 376
+ Q++D L+ ++ +G +++ N + RVCVP +L+R+IL E H S
Sbjct: 971 RARLAQEKDEGLI----AASKAEGSEYQFAANGTIFVYGRVCVPKDEELRREILSEAHAS 1026
Query: 377 SLSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVPEW 436
SIHPGATKMY+DLKR + W GMK+++A +V C VCQ EHQ P
Sbjct: 1027 MFSIHPGATKMYRDLKRYYQWVGMKRDVANWVAECDVCQLVKAEHQVP------------ 1074
Query: 437 TWDSISMDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLH 496
D IWVI+DRLTKS HF+ I+ + A LA+ Y+ +IV+LH
Sbjct: 1075 --------------------DAIWVIMDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLH 1114
Query: 497 GIPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVL 556
G+P SIVSDRD +FT FW + QA +GTK+ +S+AYHPQTDGQ+ERTIQ+LED+LR CVL
Sbjct: 1115 GVPVSIVSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVL 1174
Query: 557 EQGVSWDECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPEVVQE 616
+ G W + L L+EF YNNS+ +SI MAPFEALYGR C TPL W + E ++ + VQE
Sbjct: 1175 DWGGHWADHLSLVEFAYNNSYQASIGMAPFEALYGRPCWTPLRWTQVEERSIYGADYVQE 1234
Query: 617 TTEKVKMIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKLTPR 676
TTE++++++ MK +Q+ +SY DKRR+++EF+VGD V+L + + G ++ KL+PR
Sbjct: 1235 TTERIRVLKLNMKEAQARQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSILETKLSPR 1294
Query: 677 FVGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHVIRVDDLEVRDNLTV 736
++GPF ++E+VG VAYR+ LP + H VFHV LRK +H V+ +++ N+T+
Sbjct: 1295 YMGPFRIVERVGPVAYRLELPDVMRAFHKVFHVLMLRKCLHKDDEVLVKIPEDLQPNMTL 1354
Query: 737 ETWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWEPESGMMVSYRSCF 787
E PVR+ +R +K LR K+I L+KV+W TWEPE+ M ++ F
Sbjct: 1355 EARPVRVLERRIKELRRKKIPLIKVLWDCDGVTEETWEPEARMKARFKKWF 1405
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 389 bits (1000), Expect = e-108
Identities = 190/386 (49%), Positives = 268/386 (69%), Gaps = 1/386 (0%)
Query: 402 KEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVPEWTWDSISMDFVGALPKTSKSFDTIWV 461
+++A +V C VCQ EHQ P G++Q L +PEW WD I++DFV LP S++ D IWV
Sbjct: 941 EDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLP-VSRTKDAIWV 999
Query: 462 IVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLHGIPSSIVSDRDPRFTSNFWESLQAA 521
IVDRLTKS HF+ I+ + A LA+ Y+ +IV+LHG+P SIVSDRD +FTS FW + QA
Sbjct: 1000 IVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAE 1059
Query: 522 LGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLEQGVSWDECLPLIEFTYNNSFHSSI 581
+GTK+ +S+AYHPQT GQ+ERTIQ+LED+LR CVL+ G W + L L+EF YNNS+ +SI
Sbjct: 1060 MGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPASI 1119
Query: 582 RMAPFEALYGRRCRTPLCWFESGESAMLAPEVVQETTEKVKMIQEKMKASQST*KSYHDK 641
MAPFEALY R CRTPLC + GE ++ + VQETTE++++++ MK +Q +SY DK
Sbjct: 1120 GMAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQRSYADK 1179
Query: 642 RRKDIEFQVGDHVFLWVNPVTGVGHALKCRKLTPRFVGPFDVIEKVGVVAYRIALPPSLS 701
RR+++EF+VGD V+L + + G ++ KL+PR++GPF ++E+VG VAYR+ LP +
Sbjct: 1180 RRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVERVGPVAYRLELPDVMR 1239
Query: 702 NLHNVFHVPKLRKYVHDASHVIRVDDLEVRDNLTVETWPVRIEDRELKRLRGKEIVLVKV 761
H VFHV LRK +H V+ +++ N+T+E PVR+ +R +K LR K+I L+KV
Sbjct: 1240 AFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKV 1299
Query: 762 IWVGPTGESATWEPESGMMVSYRSCF 787
+W TWEPE+ M ++ F
Sbjct: 1300 LWDCDGVTKETWEPEARMKARFKKWF 1325
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 362 bits (929), Expect = e-100
Identities = 209/530 (39%), Positives = 293/530 (54%), Gaps = 105/530 (19%)
Query: 258 PGKANVVADVLSRKTLHMYALMVKELELIEQFGELSLVSELTPDGVRLGMLKLTSNILEE 317
PGK NVVAD LSRK + A + +E +LVSE+ G L+L E
Sbjct: 805 PGKTNVVADALSRKRVG--AAPGQSVE--------ALVSEI-------GALRLCVVAREP 847
Query: 318 IKNGQKEDLELVDRVTLVNQGKGGDFRLDENDVLMFRDRVCVPDVLDLKRQILDEGHRSS 377
+K LE VDR N ++ +RVC+P +L+R+IL E H S
Sbjct: 848 LK------LEAVDRAA--------------NGTILVHERVCLPKDEELRREILSEAHASM 887
Query: 378 LSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVPEWT 437
SIHPGATKMY+DLKR + W GMK+++A +V C VCQ EHQ P GL+Q L + EW
Sbjct: 888 FSIHPGATKMYRDLKRHYQWVGMKRDVANWVTECDVCQLVKAEHQVPGGLLQSLPISEWK 947
Query: 438 WDSISMDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLHG 497
D LPK Y+ +IV+LHG
Sbjct: 948 ----KTDGAAVLPKK-----------------------------------YVSEIVKLHG 968
Query: 498 IPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLE 557
+P SI+S RD +FTS FW + Q +GTK+ +S+AYHPQTDGQ+ERTIQ+LED+L+ CVL+
Sbjct: 969 VPVSILSHRDSKFTSAFWRAFQVEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLQMCVLD 1028
Query: 558 QGVSWDECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPEVVQET 617
G W + L L++F YNNS+ +SI MAPFEALYGR CRT LCW + GE ++ + VQET
Sbjct: 1029 WGGHWADHLSLVKFAYNNSYQASIGMAPFEALYGRPCRTLLCWTQVGEKSIYGADYVQET 1088
Query: 618 TEKVKMIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKLTPRF 677
TE++++++ MK +Q +SY DKRR+++EF+VG
Sbjct: 1089 TERIRVLKLNMKEAQDRQRSYADKRRRELEFEVGT------------------------- 1123
Query: 678 VGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHVIRVDDLEVRDNLTVE 737
+++E+VG VAYR+ LP + HNVFHV LRK +H V+ +++ N+T+E
Sbjct: 1124 ----EIVERVGPVAYRLELPDVMRAFHNVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLE 1179
Query: 738 TWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWEPESGMMVSYRSCF 787
PVR+ +R +K +R K+I ++KV+W TWEPE+ + ++ F
Sbjct: 1180 ARPVRVLERRIKEVRRKKIPMIKVLWDCDGVTEETWEPEARIKARFKKWF 1229
>At3g29490 hypothetical protein
Length = 438
Score = 291 bits (744), Expect = 1e-78
Identities = 153/388 (39%), Positives = 225/388 (57%), Gaps = 53/388 (13%)
Query: 400 MKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVPEWTWDSISMDFVGALPKTSKSFDTI 459
MK+++A +V C VCQ EHQ P G++Q L +PEW
Sbjct: 1 MKRDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWNT--------------------- 39
Query: 460 WVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLHGIPSSIVSDRDPRFTSNFWESLQ 519
F+ I+ + A LA+ Y+ +IV+LHG+P
Sbjct: 40 ------------FLAIRKTDGAAVLAKKYVSEIVKLHGVP-------------------- 67
Query: 520 AALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLEQGVSWDECLPLIEFTYNNSFHS 579
A +GTK+ +S+ YHPQTDGQ ERTIQ+LED+LR CVL+ G W + L L+EF YNNS+ +
Sbjct: 68 AEMGTKVQMSTPYHPQTDGQFERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQA 127
Query: 580 SIRMAPFEALYGRRCRTPLCWFESGESAMLAPEVVQETTEKVKMIQEKMKASQST*KSYH 639
I MAPFEALYGR CRTPLCW + GE ++ + VQETTE++++++ MK +Q SY
Sbjct: 128 GIGMAPFEALYGRPCRTPLCWTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQWSYA 187
Query: 640 DKRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKLTPRFVGPFDVIEKVGVVAYRIALPPS 699
DKRR+++EF+VGD V+L + + G ++ KL+ R++GPF ++E+VG VAY + LP
Sbjct: 188 DKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSLRYMGPFRIVERVGPVAYMLELPDV 247
Query: 700 LSNLHNVFHVPKLRKYVHDASHVIRVDDLEVRDNLTVETWPVRIEDRELKRLRGKEIVLV 759
+ H VFHV LRK +H V+ +++ N+T+E VR+ +R +K L+ K+I L+
Sbjct: 248 MRAFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARQVRVLERRIKELQRKKISLI 307
Query: 760 KVIWVGPTGESATWEPESGMMVSYRSCF 787
KV+W TW+PE+ M ++ F
Sbjct: 308 KVLWDCDGVTEETWQPEARMKARFKKWF 335
>At3g11970 hypothetical protein
Length = 1499
Score = 281 bits (718), Expect = 1e-75
Identities = 174/523 (33%), Positives = 274/523 (52%), Gaps = 43/523 (8%)
Query: 259 GKANVVADVLSR----KTLHMYALMVKELELIEQFGELSLVSELTPDGVRLGMLKLTSNI 314
GK NVVAD LSR + LHM A+ V E +L
Sbjct: 999 GKENVVADALSRVEGSEVLHM-AMTVVECDL----------------------------- 1028
Query: 315 LEEIKNGQKEDLELVDRVTLVNQGKGGD--FRLDENDVLMFRDRVCVPDVLDLKRQILDE 372
L++I+ G D +L D +T + + F +N +L + ++ VP ++K IL
Sbjct: 1029 LKDIQAGYANDSQLQDIITALQRDPDSKKYFSWSQN-ILRRKSKIVVPANDNIKNTILLW 1087
Query: 373 GHRSSLSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLF 432
H S + H G +Q +K LF+W GM K+I ++ +C CQ+ + GL+QPL
Sbjct: 1088 LHGSGVGGHSGRDVTHQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLP 1147
Query: 433 VPEWTWDSISMDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQI 492
+P+ W +SMDF+ LP S I V+VDRL+K+ HF+ + S +A Y++ +
Sbjct: 1148 IPDTIWSEVSMDFIEGLP-VSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNV 1206
Query: 493 VRLHGIPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLR 552
+LHG P+SIVSDRD FTS FW G L L+SAYHPQ+DGQTE + LE LR
Sbjct: 1207 FKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLR 1266
Query: 553 ACVLEQGVSWDECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPE 612
++ W + L L E+ YN ++HSS RM PFE +YG+ L + +
Sbjct: 1267 CMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVAR 1326
Query: 613 VVQETTEKVKMIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKC-R 671
+QE + + ++ + +Q K + D+ R + EF++GD+V++ + P ++ +
Sbjct: 1327 SLQEREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQ 1386
Query: 672 KLTPRFVGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHVIRVDDLEVR 731
KL+P++ GP+ +I++ G VAY++AL PS S +H VFHV +L+ V + S + + + ++
Sbjct: 1387 KLSPKYFGPYKIIDRCGEVAYKLAL-PSYSQVHPVFHVSQLKVLVGNVSTTVHLPSV-MQ 1444
Query: 732 DNLTVETWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWE 774
D E P ++ +R++ +GK + V V W E ATWE
Sbjct: 1445 D--VFEKVPEKVVERKMVNRQGKAVTKVLVKWSNEPLEEATWE 1485
>At1g36590 hypothetical protein
Length = 1499
Score = 276 bits (706), Expect = 3e-74
Identities = 173/523 (33%), Positives = 274/523 (52%), Gaps = 43/523 (8%)
Query: 259 GKANVVADVLSR----KTLHMYALMVKELELIEQFGELSLVSELTPDGVRLGMLKLTSNI 314
GK NVVAD LSR + LHM A+ V E +L
Sbjct: 999 GKENVVADALSRVEGSEVLHM-AMTVVECDL----------------------------- 1028
Query: 315 LEEIKNGQKEDLELVDRVTLVNQGKGGD--FRLDENDVLMFRDRVCVPDVLDLKRQILDE 372
L++I+ G D +L D +T + + F +N +L + ++ VP ++K IL
Sbjct: 1029 LKDIQAGYANDSQLQDIITALQRDPDSKKYFSWSQN-ILRRKSKIVVPANDNIKNTILLW 1087
Query: 373 GHRSSLSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLF 432
H S + H G +Q +K LF+ GM K+I ++ +C CQ+ + GL+QPL
Sbjct: 1088 LHGSGVGGHSGRDVTHQRVKGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLP 1147
Query: 433 VPEWTWDSISMDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQI 492
+P+ W +SMDF+ LP S I V+VDRL+K+ HF+ + S +A+ Y++ +
Sbjct: 1148 IPDTIWSEVSMDFIEGLP-VSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNV 1206
Query: 493 VRLHGIPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLR 552
+LHG P+SIVSDRD FTS FW G L L+SAYHPQ+DGQTE + LE LR
Sbjct: 1207 FKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLR 1266
Query: 553 ACVLEQGVSWDECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPE 612
++ W + L L E+ YN ++HSS RM PFE +YG+ L + +
Sbjct: 1267 CMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVAR 1326
Query: 613 VVQETTEKVKMIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKC-R 671
+QE + + ++ + +Q K + D+ R + EF++GD+V++ + P ++ +
Sbjct: 1327 SLQEREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQ 1386
Query: 672 KLTPRFVGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHVIRVDDLEVR 731
KL+P++ GP+ +I++ G VAY++AL PS S +H VFHV +L+ V + S + + + ++
Sbjct: 1387 KLSPKYFGPYKIIDRCGEVAYKLAL-PSYSQVHPVFHVSQLKVLVGNVSTTVHLPSV-MQ 1444
Query: 732 DNLTVETWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWE 774
D E P ++ +R++ +GK + V V W E ATWE
Sbjct: 1445 D--VFEKVPEKVVERKMVNRQGKAVTKVLVKWSNEPLEEATWE 1485
>At4g03840 putative transposon protein
Length = 973
Score = 251 bits (642), Expect = 9e-67
Identities = 128/277 (46%), Positives = 177/277 (63%), Gaps = 19/277 (6%)
Query: 514 FWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLEQGVSWDECLPLIEFTY 573
FW++ Q ALGT+++LS+AYHPQTDGQ+ERTIQ+LED+LRAC L+ G +W++ L L
Sbjct: 690 FWKAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACALDWGGNWEKYLRL----- 744
Query: 574 NNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPEVVQETTEKVKMIQEKMKASQS 633
ALYGR CRTPLCW GE + P +V ETTE++K ++ K+K +Q
Sbjct: 745 --------------ALYGRACRTPLCWTPVGERRLFGPIIVDETTERMKFLKIKLKEAQD 790
Query: 634 T*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKLTPRFVGPFDVIEKVGVVAYR 693
KSY +KRRK++EFQV D V+L G G +KL+PR+VGP+ VIE+VG VAY+
Sbjct: 791 RQKSYANKRRKELEFQVEDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYK 850
Query: 694 IALPPSLSNLHNVFHVPKLRKYVHDASHVIRVDDLEVRDNLTVETWPVRIEDRELKRLRG 753
+ LPP L+ HNVFHV +LRK + + + +++N+TVE WPV+I DR K RG
Sbjct: 851 LDLPPKLNAFHNVFHVSQLRKCLSNQEESVEDVPPGLKENMTVEAWPVQIMDRMTKGTRG 910
Query: 754 KEIVLVKVIWVGPTGESATWEPESGMMVSYRSCFLQV 790
K L+KV+W E TWE E+ M ++ F ++
Sbjct: 911 KSRDLLKVLWNCGGREQYTWETENKMKANFSEWFKEM 947
Score = 134 bits (336), Expect = 3e-31
Identities = 80/190 (42%), Positives = 112/190 (58%), Gaps = 11/190 (5%)
Query: 259 GKANVVADVLSRKTLHMYALMVKELELIEQFGEL---SLVSELTPDGVRLGMLKLTSNIL 315
GKAN VAD LSR+ + A +++L+ G L +L E+ P G LG +N+L
Sbjct: 508 GKANQVADALSRRRSEVEAER-SQVDLVNMMGTLHVNALSKEVEPLG--LGAAD-QANLL 563
Query: 316 EEIKNGQKEDLELVDRVTLVNQGKGGDFRLDENDVLMFRDRVCVPDVLDLKRQILDEGHR 375
I+ Q+ D E + L N+ +++ N ++ RVCVP+ LK +IL E H+
Sbjct: 564 SRIRLAQERDEE-IKGWALNNKT---EYQTSNNGTIVVNGRVCVPNNRALKEEILREAHQ 619
Query: 376 SSLSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVPE 435
S SIHPG+ K+Y+DLKR + W GMKK++A +V C CQ EHQ PSGL+Q L +PE
Sbjct: 620 SKFSIHPGSNKIYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPIPE 679
Query: 436 WTWDSISMDF 445
W WD I+MDF
Sbjct: 680 WKWDHITMDF 689
>At1g35370 hypothetical protein
Length = 1447
Score = 242 bits (618), Expect = 5e-64
Identities = 155/522 (29%), Positives = 256/522 (48%), Gaps = 64/522 (12%)
Query: 259 GKANVVADVLSR----KTLHMYALMVKELELIEQFGELSLVSELTPDGVRLGMLKLTSNI 314
GK N+VAD LSR + LHM AL + E +
Sbjct: 970 GKENLVADALSRVEGSEVLHM-ALSIVECDF----------------------------- 999
Query: 315 LEEIKNGQKEDLELVDRVTLVNQGKGGDFRLD-ENDVLMFRDRVCVPDVLDLKRQILDEG 373
L+EI+ + D L D ++ + Q D+L + ++ VP+ +++ ++L
Sbjct: 1000 LKEIQVAYESDGVLKDIISALQQHPDAKKHYSWSQDILRRKSKIVVPNDVEITNKLLQWL 1059
Query: 374 HRSSLSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFV 433
H S + G +Q +K LF+W GM K+I F+ +C CQ+ ++ GL+QPL +
Sbjct: 1060 HCSGMGGRSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPLPI 1119
Query: 434 PEWTWDSISMDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIV 493
P+ W +SMDF+ LP + I V+VDRL+K+ HFV + S +A+ +++ +
Sbjct: 1120 PDKIWCDVSMDFIEGLPNSGGK-SVIMVVVDRLSKAAHFVALAHPYSALTVAQAFLDNVY 1178
Query: 494 RLHGIPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRA 553
+ HG P+SIVSDRD FTS+FW+ G +L +SSAYHPQ+DGQTE + LE+ LR
Sbjct: 1179 KHHGCPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCLENYLRC 1238
Query: 554 CVLEQGVSWDECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPEV 613
+ W++ LPL E+ YN ++HSS +M PFE +YG+ L + +
Sbjct: 1239 MCHARPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLPYLPGKSKVAVVARS 1298
Query: 614 VQETTEKVKMIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKC-RK 672
+QE + ++ + +Q K + D+ R + F +GD V++ + P L+ +K
Sbjct: 1299 LQERENMLLFLKFHLMRAQHRMKQFADQHRTERTFDIGDFVYVKLQPYRQQSVVLRVNQK 1358
Query: 673 LTPRFVGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHVIRVDDLEVRD 732
L+P++ GP+ +IEK G V + N+ +P + + + +
Sbjct: 1359 LSPKYFGPYKIIEKCGEV--------MVGNVTTSTQLPSVLPDIFEKA------------ 1398
Query: 733 NLTVETWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWE 774
P I +R+L + +G+ +V V W+G E ATW+
Sbjct: 1399 -------PEYILERKLVKRQGRAATMVLVKWIGEPVEEATWK 1433
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 227 bits (578), Expect = 2e-59
Identities = 127/288 (44%), Positives = 180/288 (62%), Gaps = 31/288 (10%)
Query: 466 LTKSTHFVPIKTSMSIAR-LAEIYIEQIVRL-HGIPSSIVSD------RDPRFTSNFWES 517
L + H+V +K +AR +A+ Q+V+ H +PS ++ + + T +F +
Sbjct: 635 LKRYYHWVGMKKD--VARWVAKCPTCQLVKAEHQVPSGLLQNLPIPEWKWDHITMDF--A 690
Query: 518 LQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLEQGVSWDECLPLIEFTYNNSF 577
Q ALGT+++LS+AYHPQTDGQ+ERTIQ+LED+LRACVL+ G +W++ L L
Sbjct: 691 FQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLTL--------- 741
Query: 578 HSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPEVVQETTEKVKMIQEKMKASQST*KS 637
ALYGR CRTPLCW GE + P +V ETTE++K ++ K+K + KS
Sbjct: 742 ----------ALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAHDRQKS 791
Query: 638 YHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKLTPRFVGPFDVIEKVGVVAYRIALP 697
Y +KRRK++EFQVGD V+L G G +KL+PR+VGP+ VIE+VG VAY++ LP
Sbjct: 792 YANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLP 851
Query: 698 PSLSNLHNVFHVPKLRKYVHDASHVIRVDDLEVRDNLTVETWPVRIED 745
P L+ HNVFHV +LRK + D + +++N+TVE WPVRI D
Sbjct: 852 PKLNAFHNVFHVSQLRKCLSDQEESVEDVPPGLKENMTVEAWPVRIMD 899
Score = 136 bits (342), Expect = 5e-32
Identities = 79/189 (41%), Positives = 113/189 (58%), Gaps = 7/189 (3%)
Query: 258 PGKANVVADVLSRKTLHMYALMVKELELIEQFGELSLVSELTPDGVRLGMLKLT-SNILE 316
PGKAN VAD LSR+ + A +++L+ G L V+ L+ + LG+ +++L
Sbjct: 507 PGKANQVADALSRRRSEVEAER-SQVDLVNMMGTLH-VNALSKEVESLGLGAADQADLLS 564
Query: 317 EIKNGQKEDLELVDRVTLVNQGKGGDFRLDENDVLMFRDRVCVPDVLDLKRQILDEGHRS 376
I+ Q+ D E + L N+ +++ N ++ RVCVP+ LK +IL E H+S
Sbjct: 565 RIRLAQERDEE-IKGWALNNKT---EYQTSNNGTIVVNGRVCVPNDRALKEEILREAHQS 620
Query: 377 SLSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVPEW 436
SIHPG+ KMY+DLKR + W GMKK++A +V C CQ EHQ PSGL+Q L +PEW
Sbjct: 621 KFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPIPEW 680
Query: 437 TWDSISMDF 445
WD I+MDF
Sbjct: 681 KWDHITMDF 689
>At4g07850 putative polyprotein
Length = 1138
Score = 208 bits (529), Expect = 1e-53
Identities = 101/248 (40%), Positives = 154/248 (61%), Gaps = 2/248 (0%)
Query: 351 LMFRDRVCVPDVLDLKRQILDEGHRSSLSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYA 410
L + +R+C+P+ L+ + E H L H G +K + ++ F WP MK+++
Sbjct: 745 LFYDNRLCIPNS-SLRELFIREAHGGGLMGHFGVSKTLKVMQDHFHWPHMKRDVERMCER 803
Query: 411 CLVCQKSMIEHQRPSGLMQPLFVPEWTWDSISMDFVGALPKTSKSFDTIWVIVDRLTKST 470
C C+++ + Q P GL PL +P W+ ISMDFV LP+T D+I+V+VDR +K
Sbjct: 804 CTTCKQAKAKSQ-PHGLCTPLPIPLHPWNDISMDFVVGLPRTRTGKDSIFVVVDRFSKMA 862
Query: 471 HFVPIKTSMSIARLAEIYIEQIVRLHGIPSSIVSDRDPRFTSNFWESLQAALGTKLSLSS 530
HF+P + +A ++ ++VRLHG+P +IVSDRD +F S FW++L + LGTKL S+
Sbjct: 863 HFIPCHKTDDAMHIANLFFREVVRLHGMPKTIVSDRDTKFLSYFWKTLWSKLGTKLLFST 922
Query: 531 AYHPQTDGQTERTIQSLEDLLRACVLEQGVSWDECLPLIEFTYNNSFHSSIRMAPFEALY 590
HPQTDGQTE ++L LLRA + + +W++CLP +EF YN+S HS+ + +PF+ +Y
Sbjct: 923 TCHPQTDGQTEVVNRTLSTLLRALIKKNLKTWEDCLPHVEFAYNHSVHSATKFSPFQIVY 982
Query: 591 GRRCRTPL 598
G TPL
Sbjct: 983 GFNPITPL 990
>At3g31480 hypothetical protein
Length = 338
Score = 187 bits (475), Expect = 2e-47
Identities = 100/251 (39%), Positives = 149/251 (58%), Gaps = 43/251 (17%)
Query: 443 MDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLHGIPSSI 502
MDFV LP S++ D IWVI DRLT S F+ I+ + +A LA Y+ +IV+LHG+ SI
Sbjct: 1 MDFVVGLPM-SRTKDAIWVIGDRLTNSADFLAIRKTDGVAVLANKYVSEIVKLHGVHVSI 59
Query: 503 VSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLEQGVSW 562
VSD+D +FTS FW + QA +GTK+ +S+AYHP+TDG+ E+TIQ+LED+LR
Sbjct: 60 VSDKDSKFTSAFWIAFQAEMGTKVQISTAYHPKTDGKFEKTIQTLEDMLR---------- 109
Query: 563 DECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPEVVQETTEKVK 622
TPLCW + GE ++ V ETTE+++
Sbjct: 110 --------------------------------MTPLCWTQVGERSIYGANYVHETTERIQ 137
Query: 623 MIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKLTPRFVGPFD 682
+++ MK + +SY D RR+++EF+VGD V+L ++ + + KL+P+++GPF
Sbjct: 138 VLKLNMKEAHDRQRSYADMRRREVEFEVGDRVYLKMDMLQSPKRFILETKLSPKYMGPFR 197
Query: 683 VIEKVGVVAYR 693
++E+VG VAYR
Sbjct: 198 IVERVGPVAYR 208
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 167 bits (424), Expect = 2e-41
Identities = 108/308 (35%), Positives = 161/308 (52%), Gaps = 17/308 (5%)
Query: 484 LAEIYIEQIVRLHGIPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERT 543
+A ++ +V+LHGIP SIVSD DP F S FW+ TKL +S+AYHPQTDGQTE
Sbjct: 634 VAAKFVSHVVKLHGIPRSIVSDCDPIFMSLFWQEFWKLSRTKLWMSTAYHPQTDGQTEVV 693
Query: 544 IQSLEDLLRACVLEQGVSWDECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFES 603
+ +E LR V W +P E+ YN +FH+S M PF+ALYGR +P+ +E
Sbjct: 694 NRCIEQFLRCFVHYHPKQWSSFIPWAEYWYNTTFHASTGMTPFQALYGRP-PSPIPAYEL 752
Query: 604 GESAMLAPEVVQETTEKVKMIQE---KMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNP 660
G +++ E+ ++ + +++ E + + + K D R +D+ FQVGD V L + P
Sbjct: 753 G--SVVCGELNEQMAARDELLAELKQHLVTANNCMKQQADSRLRDVSFQVGDWVLLRIQP 810
Query: 661 V-TGVGHALKCRKLTPRFVGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDA 719
+KL+ RF GPF V K G VAYR+ LP + +H VFHV L+ +V D
Sbjct: 811 YRQKTLFRRSSQKLSHRFYGPFQVASKHGEVAYRLTLPEG-TRIHPVFHVSLLKPWVGDG 869
Query: 720 SHVIRVDDL----EVRDNLTVETWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWEP 775
D+ +R+N ++ P + + + K + + V W G E ATWE
Sbjct: 870 E-----PDMGQLPPLRNNGELKLQPTAVLEVRWRSQDKKRVADLLVQWEGLHIEDATWEE 924
Query: 776 ESGMMVSY 783
+ S+
Sbjct: 925 YDQLAASF 932
>At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein
Length = 280
Score = 162 bits (411), Expect = 5e-40
Identities = 101/276 (36%), Positives = 150/276 (53%), Gaps = 20/276 (7%)
Query: 443 MDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLHGIPSSI 502
MDFV LP+T + D+++V+VDR +K THF+ K + + +A+++ +++VRLHG+P SI
Sbjct: 1 MDFVLGLPRTQRGVDSVFVVVDRFSKMTHFIACKKTADASNIAKLFFKEVVRLHGVPKSI 60
Query: 503 VSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLEQGVSW 562
SDRD +F S+FW +L GT L+ SS H Q DGQTE T ++L +++R+ + W
Sbjct: 61 TSDRDTKFLSHFWSTLWRMFGTALNRSSTPHTQIDGQTEVTNRTLGNMVRSICGDNPKQW 120
Query: 563 DECLPLIEFTYNNSFHSSIRMAPFEALYGRRCRTPLCWFESGESAMLAPEVVQETTEKVK 622
D LP IEF YN+ H + +AL G SA E + E VK
Sbjct: 121 DLALPQIEFAYNSVVHVVDLVKLPKAL--------------GASAETMAEEILVVKEVVK 166
Query: 623 MIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHALKCRKLTPRFVGPFD 682
K++A+ K DKRR+ F+ GD V + + G K+ PR GPF
Sbjct: 167 ---AKLEATGKKNKVAADKRRRFKVFKEGDDVMVLLR--KGRFAVGTYNKVKPRKYGPFK 221
Query: 683 VIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHD 718
V+ K+ AY +ALP S+ N+ N F+V + +Y D
Sbjct: 222 VLRKINDNAYVVALPKSM-NISNTFNVADIHEYHAD 256
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 135 bits (341), Expect = 7e-32
Identities = 99/276 (35%), Positives = 139/276 (49%), Gaps = 60/276 (21%)
Query: 258 PGKANVVADVLSRKTLHMYALMVKELELIEQFGELSLVSELTPDGVRLGMLKLTSNILEE 317
PGKANVV D LSRK + A + + +E+ LVSE+ G L+L + E
Sbjct: 889 PGKANVVVDALSRKRVG--AALGQSVEV--------LVSEI-------GALRLCAVAREP 931
Query: 318 IKNGQKEDLELVDRVTLVNQGKGGDFRLDENDVLMFRDRVCVPDVLDLKRQILDEGHRSS 377
+ LE VDR L+ + + Q DEG R
Sbjct: 932 L------GLEAVDRADLLTRVR--------------------------LAQKKDEGLR-- 957
Query: 378 LSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVPEWT 437
ATKMY+DLKR + W GMK ++A +V C VCQ EHQ G++Q L +PEW
Sbjct: 958 ------ATKMYRDLKRYYQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWK 1011
Query: 438 WDSISMDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLHG 497
WD I+MD V L + S++ D IWVIVDRLTKS HF+ I+ + A LA+ ++ +IV+LHG
Sbjct: 1012 WDFITMDLVVGL-RVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHG 1070
Query: 498 IPSSI--VSDRDPRFTSNFWESLQAALGTKLSLSSA 531
+P ++ DR + L+ +G ++ L A
Sbjct: 1071 VPLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMA 1106
Score = 135 bits (340), Expect = 9e-32
Identities = 70/182 (38%), Positives = 110/182 (59%), Gaps = 1/182 (0%)
Query: 606 SAMLAPEVVQETTEKVKMIQEKMKASQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVG 665
+A+LA + V E K+ + MK +Q +SY DKRR+++EF+VGD V+L + + G
Sbjct: 1054 AAVLAKKFVSEIV-KLHGVPLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPN 1112
Query: 666 HALKCRKLTPRFVGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHVIRV 725
++ KL+PR++GPF ++E+V VAYR+ LP + H VFHV LRK +H +
Sbjct: 1113 RSISETKLSPRYMGPFKIVERVEPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEALAK 1172
Query: 726 DDLEVRDNLTVETWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWEPESGMMVSYRS 785
+++ N+T+E PVR+ +R +K LR K+I L+KV+W TWEPE+ M ++
Sbjct: 1173 IPEDLQPNMTLEARPVRVLERRIKELRQKKIPLIKVLWDCDGVTEETWEPEARMKARFKK 1232
Query: 786 CF 787
F
Sbjct: 1233 WF 1234
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 120 bits (301), Expect = 3e-27
Identities = 81/250 (32%), Positives = 123/250 (48%), Gaps = 36/250 (14%)
Query: 259 GKANVVADVLSR----KTLHMYALMVKELELIEQFGELSLVSELTPDGVRLGMLKLTSNI 314
GK N+VAD LSR + LHM AL V E +L
Sbjct: 390 GKENLVADALSRVEGSEVLHM-ALSVVECDL----------------------------- 419
Query: 315 LEEIKNGQKEDLELVDRVTLVNQGKGGDFRLD-ENDVLMFRDRVCVPDVLDLKRQILDEG 373
L+EI+ G D ++ +T++ Q VL ++++ VP+ +K IL
Sbjct: 420 LKEIQAGYVTDGDIQGIITILQQQADSKKHYTWSQGVLRRKNKIVVPNNSGIKDTILRWL 479
Query: 374 HRSSLSIHPGATKMYQDLKRLFWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFV 433
H S + H G +Q +K LF+W M K+I F+ +C CQ+ ++ GL+QPL +
Sbjct: 480 HCSGMGGHSGKEVTHQRVKGLFYWKSMVKDIQAFIRSCGTCQQCKSDNAASPGLLQPLPI 539
Query: 434 PEWTWDSISMDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIV 493
P+ W +SMDF+ LP S I V+VDRL+K+ HF+ + S +A+ Y++ +
Sbjct: 540 PDRIWSDVSMDFIDGLP-LSNGKTVIMVVVDRLSKAAHFIALAHPYSAMTVAQAYLDNVF 598
Query: 494 RLHGIPSSIV 503
+LHG PSSIV
Sbjct: 599 KLHGCPSSIV 608
Score = 78.6 bits (192), Expect = 1e-14
Identities = 47/156 (30%), Positives = 85/156 (54%), Gaps = 5/156 (3%)
Query: 583 MAPFEALYGRRCRTPLCWFESGESAMLAPEVVQETTEKVKMIQEKMKASQST*KSYHDKR 642
M P+EA+YG+ L + + +QE + ++ + +Q K D+
Sbjct: 625 MTPYEAVYGQPPPLHLPYLPGESKVAVVARSMQERESMILFLKFHLMRAQHRMKQLADQH 684
Query: 643 RKDIEFQVGDHVFLWVNPVTGVGHALK-CRKLTPRFVGPFDVIEKVGVVAYRIALPPSLS 701
+ EF+VGD+VF+ + P ++ +KL+P++ GP+ VI++ G VAY++ LP + S
Sbjct: 685 ITEREFEVGDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQLPAN-S 743
Query: 702 NLHNVFHVPKLRKY---VHDASHVIRVDDLEVRDNL 734
+H VFHV +LR V ++H++R + +R+NL
Sbjct: 744 QVHPVFHVSQLRVLVGTVTTSTHLLRCYLMSLRENL 779
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.340 0.151 0.500
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,780,065
Number of Sequences: 26719
Number of extensions: 632057
Number of successful extensions: 2343
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 2209
Number of HSP's gapped (non-prelim): 87
length of query: 796
length of database: 11,318,596
effective HSP length: 107
effective length of query: 689
effective length of database: 8,459,663
effective search space: 5828707807
effective search space used: 5828707807
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146806.4


