

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146806.1 - phase: 0
(790 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
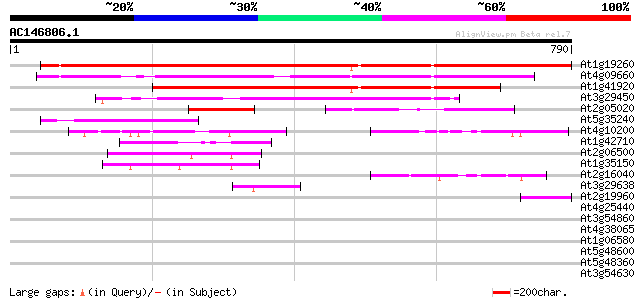
Score E
Sequences producing significant alignments: (bits) Value
At1g19260 hypothetical protein 632 0.0
At4g09660 putative protein 508 e-144
At1g41920 hypothetical protein 409 e-114
At3g29450 hypothetical protein 362 e-100
At2g05020 hypothetical protein 132 8e-31
At5g35240 hypothetical protein 125 9e-29
At4g10200 putative protein 95 1e-19
At1g42710 unknown protein 92 9e-19
At2g06500 Ac-like transposase 66 7e-11
At1g35150 hypothetical protein 60 4e-09
At2g16040 Ac-like transposase 55 2e-07
At3g29638 putative protein 51 2e-06
At2g19960 Ac-like transposase 47 4e-05
At4g25440 putative protein 32 1.1
At3g54860 vacuolar protein sorting protein 33 (VPS33) 31 3.1
At4g38065 hypothetical protein 30 5.3
At1g06580 hypothetical protein 30 5.3
At5g48600 chromosome condensation protein 30 6.9
At5g48360 unknown protein 30 6.9
At3g54630 unknown protein 29 9.0
>At1g19260 hypothetical protein
Length = 769
Score = 632 bits (1630), Expect = 0.0
Identities = 345/756 (45%), Positives = 491/756 (64%), Gaps = 15/756 (1%)
Query: 44 LETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTRRRFNKNWFNLY 103
L +DP R I SYHP+ +DEVR+ YL IR P + RRFN WF+LY
Sbjct: 20 LPSDPAKRKSILSYHPNQRDEVRREYL-IRGPCQPRGHKFKQIAIGKVLRRFNPKWFDLY 78
Query: 104 -DWLEYSESKNLAFCLPCFLFKN-VSNYGG-DHFVGDGFGDWKNPRKLANHA--TSNNSH 158
DWLEYS K AFCL C+LF++ N GG D F+ GF W +L H N+ H
Sbjct: 79 GDWLEYSVEKEKAFCLYCYLFRDQTGNQGGSDSFLSTGFCSWNKADRLDQHVGLDVNSFH 138
Query: 159 VDCVHMGYALMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSDEAD 218
+ LM QSIK A QT + +Y + + S+ +++LL GL FRG DE++
Sbjct: 139 NNAKRKCEDLMRQGQSIKHALHKQTDVVKNDYRIWLNASIDVSRHLLHQGLPFRGHDESE 198
Query: 219 DSLYKGPFLELLD-TLKENNSDVATILDSAPGNSLMTCPKIQKDLASACACEITREIVCD 277
+S KG F+ELL T +N +L +AP N+ MT P IQKD+ + E+TR I+ +
Sbjct: 199 ESTNKGNFVELLKYTAGQNEVVKKVVLKNAPKNNQMTSPPIQKDIVHCFSKEVTRSIIEE 258
Query: 278 IADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDALETM 337
+ +DVF +L+DES D S +EQMAVV R+VD G+VKERF+G+ V+ETS+ SLK A++++
Sbjct: 259 MDNDVFGLLVDESADASDKEQMAVVFRFVDKYGVVKERFIGVIHVQETSSLSLKSAIDSL 318
Query: 338 LSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACAKT 397
+ GLS +RGQGYDGASNM+G F GL++LI E+ SA+YVHCFAHQLQL ++A AK
Sbjct: 319 FAKYGLSLKKLRGQGYDGASNMKGEFNGLRSLILKESSSAYYVHCFAHQLQLVVMAVAKK 378
Query: 398 HKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARA 457
H V FF +++L+N + AS KR++ +R+ + + I GEI+TG+GLNQ+ S+ R
Sbjct: 379 HVEVGEFFYMISVLLNVVGASCKRKDKIREIHRQKVEEKISNGEIKTGTGLNQELSLQRP 438
Query: 458 GDTRWGSHFRTLTSLMTLYGAIV---EVIVEVGNDPSFDKFGETVLLLDVLQSFDFIFML 514
G+TRWGSH++TL L L+ +IV E I + G D + K + +L +FDF+F L
Sbjct: 439 GNTRWGSHYKTLLRLEELFSSIVIVLEYIQDEGTDTT--KRQQAYGILKYFHTFDFVFYL 496
Query: 515 YMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKH 574
+M+ ++G+T+ LS ALQR+DQD+LNAISLV TK QLQ++R++GW+ +++V + K+
Sbjct: 497 ELMLLVMGLTDSLSKALQRKDQDILNAISLVKTTKCQLQKVRDDGWDAFMAKVSSFSEKN 556
Query: 575 EIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTELL 634
+ M+ +++ ++PR+ + ++NLHHYK DC ++VLD+QLQE N RFDE N+ELL
Sbjct: 557 NTGMLKMEEEFVDSRRPRK---KTGITNLHHYKVDCFYTVLDMQLQEFNDRFDEVNSELL 613
Query: 635 QCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAKLK 694
C+S LSP SF FD + L+R+ E YP+DF V L QL Y+ NVK+D +F LK
Sbjct: 614 ICMSSLSPIDSFRQFDKSMLVRLTEFYPDDFSFVERRSLDHQLEIYLDNVKNDERFTDLK 673
Query: 695 GLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKSHLRNKMGDQW 754
L DL ++VET K + LV++LLK++L+LPVATA+VER FSAM VK+ LRN++GD +
Sbjct: 674 CLGDLARVMVETRKHLSHPLVYRLLKVSLILPVATATVERCFSAMNFVKTTLRNRIGDMF 733
Query: 755 LNDRLVTFIERDVLFTISTDVILAHFQQMDDRRFSL 790
L+D LV FIE+ L T++ + ++ FQ M +RR L
Sbjct: 734 LSDCLVCFIEKQALNTVTNESVIKRFQDMSERRVHL 769
>At4g09660 putative protein
Length = 664
Score = 508 bits (1307), Expect = e-144
Identities = 295/707 (41%), Positives = 422/707 (58%), Gaps = 65/707 (9%)
Query: 39 VDYKLLETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTRRRFNKN 98
+++ L +DP R I SYH + +DEVR+ YL IR P + RRFN
Sbjct: 15 INFNKLPSDPAKRKSILSYHLNQRDEVRREYL-IRGPCQPRGHKFKQIVIGKVLRRFNPK 73
Query: 99 WFNLY-DWLEYSESKNLAFCLPCFLFKN-VSNYGG-DHFVGDGFGDWKNPRKLANHATSN 155
WF+LY DWLEYS K AFCL C+LF++ N GG D F+ GF W +L H +
Sbjct: 74 WFDLYGDWLEYSVEKEKAFCLYCYLFRDQAGNQGGSDSFLSTGFCSWNKADRLDQHVGLD 133
Query: 156 NSHVDCVHMGYALMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSD 215
+ +F N K+ + L+R GL+FRG D
Sbjct: 134 VN--------------------SFHNNAKRKCED--------------LMRQGLSFRGHD 159
Query: 216 EADDSLYKGPFLELLD-TLKENNSDVATILDSAPGNSLMTCPKIQKDLASACACEITREI 274
E+++S KG FLELL T +N +L +AP N+ MT P IQKD+ + E+TR I
Sbjct: 160 ESEESTNKGNFLELLKYTAGQNEVVKKVVLKNAPKNNQMTSPPIQKDIVHCFSEEVTRSI 219
Query: 275 VCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDAL 334
+ ++ +DVF +L+DES D S +EQM VV R+VD G+VKERF+G+ VKETS+ SLK A+
Sbjct: 220 IEEMDNDVFGLLVDESADASNKEQMTVVFRFVDKYGVVKERFIGVIHVKETSSLSLKSAI 279
Query: 335 ETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVAC 394
+++ + GLS +RGQGYDGASNM+G F GL++LI E
Sbjct: 280 DSLFAKYGLSLKKLRGQGYDGASNMKGEFNGLRSLILKE--------------------I 319
Query: 395 AKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSI 454
AK H V FF +++L+N + AS R++ +R+ + + I GEI+TG+ LNQ+ S+
Sbjct: 320 AKKHVEVGEFFDMISVLLNVVGASCTRKDKIREIHRQKVEEKISNGEIKTGTRLNQELSL 379
Query: 455 ARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFD--KFGETVLLLDVLQSFDFIF 512
R G+TRWGSH++TL L L+ +IV +++E D D K + +L +FDF+F
Sbjct: 380 QRPGNTRWGSHYKTLLRLEELFSSIV-IVLEYIQDEGTDTTKRQQAYGILKYFHTFDFVF 438
Query: 513 MLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICT 572
L +M+ ++G+T+ LS ALQR+DQD+LN ISLV TK QLQ++R++GW+ ++ V +
Sbjct: 439 YLELMLLVMGLTDSLSKALQRKDQDILNVISLVKTTKCQLQKVRDDGWDAFMAEVSSFSE 498
Query: 573 KHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTE 632
K+ + M+ +++ ++PR+ S ++NLHHYK DC ++VLD+QLQE N RFDE N+E
Sbjct: 499 KNNTAMLKMEEEFVDSRRPRKK---SGITNLHHYKVDCFYTVLDMQLQEFNDRFDEVNSE 555
Query: 633 LLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAK 692
LL C+S LSP SF FD + L+R+ E YP++F V L QL Y+ NVK+D +F
Sbjct: 556 LLICMSSLSPIDSFCQFDKSMLVRLTEFYPDEFSFVERRSLDHQLEIYLDNVKNDERFTD 615
Query: 693 LKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAM 739
LK DL ++VET K + LV++LLKL+L+LPVATA+VER FSAM
Sbjct: 616 LKCFGDLARVMVETRKHLSHPLVYRLLKLSLILPVATATVERCFSAM 662
>At1g41920 hypothetical protein
Length = 496
Score = 409 bits (1050), Expect = e-114
Identities = 217/496 (43%), Positives = 322/496 (64%), Gaps = 11/496 (2%)
Query: 202 KYLLRCGLAFRGSDEADDSLYKGPFLELLD-TLKENNSDVATILDSAPGNSLMTCPKIQK 260
+YL+R L FRG DE+D+S +G FLEL+ T+ +N +L++AP N+ M ++ +
Sbjct: 5 RYLVRQRLPFRGHDESDESANRGNFLELVKYTVGQNEVISKVVLENAPKNNQMVFDEMSE 64
Query: 261 DLASACACEITRE-IVCDIADDVFCVLIDE-SGDVSGREQMAVVLRYVDGDGLVKERFLG 318
L + ++ D+ D V +VS +EQMAVV R+VD G VKERF+G
Sbjct: 65 PLIPETRDPVPSSFLLLDLILDYPVVWQQTVQSNVSDKEQMAVVFRFVDKHGTVKERFIG 124
Query: 319 ITSVKETSAKSLKDALETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAH 378
+ VKETS+ SLK A++++ + GLS +RGQGYDGA+NM+G F L++LI EN SA+
Sbjct: 125 LIHVKETSSASLKCAIDSLFAKRGLSMKQLRGQGYDGANNMKGEFNWLRSLILRENSSAY 184
Query: 379 YVHCFAHQLQLALVACAKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIE 438
Y+HCFAHQLQL +VA AK V FF ++ L+N + AS K ++ +R++ L + + I
Sbjct: 185 YIHCFAHQLQLVVVAVAKKQFEVGDFFDMISALLNVVGASCKGKDRIREEYLKEIEEGIN 244
Query: 439 EGEIETGSGLNQDSSIARAGDTRWGSHFRTLTSLMTLYGAIV---EVIVEVGNDPSFDKF 495
+GEI+TG GLNQ+ S+ R G+TRWG+H+ TL L L+ I+ E + E G D + K
Sbjct: 245 QGEIKTGKGLNQELSLQRPGNTRWGTHYTTLHRLAHLFSVIIKLLEFVEEEGTDST--KR 302
Query: 496 GETVLLLDVLQSFDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEM 555
+ LL +FDF F L +M+ +LG+TN LS+ALQ++DQD+LN +SLV TK+QL ++
Sbjct: 303 RQANGLLKYFNTFDFAFYLQLMLLLLGLTNSLSVALQKKDQDILNPMSLVKSTKQQLCKL 362
Query: 556 RNEGWEELISRVVTICTKHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVL 615
R++GW+ L++ V + C KH+I++ MD ++ + PR+ S+++N HHY+ +C +++L
Sbjct: 363 RDDGWDSLVNEVFSFCKKHDIELVIMDGEFVNPRNPRK---RSNMTNFHHYQVECFYTIL 419
Query: 616 DLQLQELNARFDEENTELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRR 675
D+Q+QE N RFDE NTELL CV+ LSP SF FD K+LR++E YP DF V L
Sbjct: 420 DMQIQEFNDRFDEVNTELLSCVASLSPIDSFHEFDQLKVLRLSEFYPQDFTHVDRRSLEH 479
Query: 676 QLHNYVRNVKSDPKFA 691
QL Y+ N++ D +FA
Sbjct: 480 QLGLYIDNIREDERFA 495
>At3g29450 hypothetical protein
Length = 522
Score = 362 bits (928), Expect = e-100
Identities = 207/521 (39%), Positives = 307/521 (58%), Gaps = 72/521 (13%)
Query: 122 LFKNVSNY-----GGDHFVGDGFGDWKNPRKLANHATSNNSHVDCVHMGYALMNPNQSIK 176
L K + +Y G D FV GF WKNP+ L H NS N ++K
Sbjct: 35 LMKEMRDYTENKGGSDTFVTKGFDTWKNPQSLREHVGLVNSF------------HNNALK 82
Query: 177 AA--FVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSDEADDSLYKGPFLELLD-TL 233
A + Q +Q GL FRG DE+ DS + FLEL+ T
Sbjct: 83 RADCLMRQVRQ----------------------GLPFRGHDESVDSANRENFLELVKYTA 120
Query: 234 KENNSDVATILDSAPGNSLMTCPKIQKDLASACACEITREIVCDIADDVFCVLIDESGDV 293
+N + +L++AP N+ M CPKIQKD+ A E+ R I+ ++ DVF +++DES D+
Sbjct: 121 GQNEAVSKIVLENAPKNNQMACPKIQKDIVHCFAEEVIRSIIQEVDHDVFWLMVDESADI 180
Query: 294 SGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDALETMLSINGLSFSSIRGQGY 353
S +EQMAV +T SLK A++++ + GLS +RGQGY
Sbjct: 181 SDKEQMAV----------------------KTFFASLKCAIDSLFAKLGLSIKQLRGQGY 218
Query: 354 DGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACAKTHKPVSGFFGKVNMLVN 413
DGASNM+G F GL++LI EN SA+Y+HCFAHQLQL +VA AK H + FF +++L+N
Sbjct: 219 DGASNMKGEFNGLRSLILRENSSAYYIHCFAHQLQLVVVAVAKKHFEIGDFFDMISVLIN 278
Query: 414 FIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARAGDTRWGSHFRTLTSLM 473
+ AS KR++ +RD+ + + I +GEI+TG GLNQ S+ R G+TRWG+H+ TL L+
Sbjct: 279 VVGASCKRKDRVRDEFRKKLEERINQGEIKTGKGLNQKLSLQRPGNTRWGTHYTTLLRLV 338
Query: 474 TLYGAIVEVIVEVGND-PSFDKFGETVLLLDVLQSFDFIFMLYMMVEILGITNDLSLALQ 532
L+ I++V+ + +D K + LL +FDF+F L +M+ ILG+TN LS+ALQ
Sbjct: 339 DLFSVIIKVLEWIEDDGTDSTKRRQANGLLKYFNTFDFVFYLQLMLLILGLTNSLSVALQ 398
Query: 533 RRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKHEIDVPDMDAPYMEGKKPR 592
R+DQD+LNA+SLV TK+QL ++R++GW+ ++ V + C H+I+ MD +++ +KPR
Sbjct: 399 RKDQDILNAMSLVKSTKQQLFKLRDDGWDSFLNEVFSFCKDHDIEFVIMDGEFVDPRKPR 458
Query: 593 RVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTEL 633
+ S+++NLHHY+ +C +VLD+Q+QE F++ NT L
Sbjct: 459 K---KSNMTNLHHYQVECFNTVLDMQIQE----FNDPNTSL 492
>At2g05020 hypothetical protein
Length = 370
Score = 132 bits (332), Expect = 8e-31
Identities = 89/268 (33%), Positives = 128/268 (47%), Gaps = 59/268 (22%)
Query: 445 GSGLNQDSSIARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFD--KFGETVLLL 502
G L ++ S+ + +TRWG+H++TL ++ + I+E I E D D K + LL
Sbjct: 119 GLSLKKELSLQKPANTRWGTHYKTLLRIIEFFSCIIEYI-EYIQDEDVDNIKRRQANGLL 177
Query: 503 DVLQSFDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEE 562
+FDF F L +M+ ILG + LS ALQRRDQD+LN +SLV T
Sbjct: 178 KYFHTFDFAFYLQLMLHILGFADILSQALQRRDQDILNVMSLVAST-------------- 223
Query: 563 LISRVVTICTKHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQEL 622
K E+D ++VLD+QLQ
Sbjct: 224 ----------KREVDY--------------------------------YYNVLDMQLQAF 241
Query: 623 NARFDEENTELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVR 682
N FDE N+E+L ++ LSP SFS F + L+R+ ELY +DF V + L QL Y+
Sbjct: 242 NDLFDEVNSEILFWIASLSPMDSFSQFKKSMLVRLTELYLDDFSFVERISLDHQLDIYLD 301
Query: 683 NVKSDPKFAKLKGLSDLCAILVETNKCK 710
NV+ D +F K L DL ++ ++ CK
Sbjct: 302 NVQRDERFTNFKSLGDLSRVMRKSAPCK 329
Score = 102 bits (254), Expect = 8e-22
Identities = 48/92 (52%), Positives = 70/92 (75%)
Query: 253 MTCPKIQKDLASACACEITREIVCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLV 312
MT P IQKD+ A E+T+ ++ +I +DVF +L+DES D+ +EQM VV R+VD DG+V
Sbjct: 30 MTSPPIQKDIMHCFAMEVTKSVIQEINNDVFALLVDESADIPDKEQMTVVFRFVDKDGIV 89
Query: 313 KERFLGITSVKETSAKSLKDALETMLSINGLS 344
KERF+GI+ VKETS+ SLK+A++ + + +GLS
Sbjct: 90 KERFVGISHVKETSSLSLKNAIDPLFTKDGLS 121
>At5g35240 hypothetical protein
Length = 234
Score = 125 bits (314), Expect = 9e-29
Identities = 87/229 (37%), Positives = 118/229 (50%), Gaps = 29/229 (12%)
Query: 44 LETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTR-RRFNKNWFNL 102
L +D R I YHP+ +D+ R GT RRFN +WF
Sbjct: 26 LPSDLAQRKRILDYHPNERDKKRI----------------------GTALRRFNASWFGK 63
Query: 103 Y-DWLEYSESKNLAFCLPCFLFKN-VSNYGGDH-FVGDGFGDWKNPRKLANHA--TSNNS 157
Y WLEYS S N A+CL C+LFK+ + G ++ FV +GF W NP L H T N
Sbjct: 64 YPSWLEYSTSTNKAYCLYCYLFKDDIPKRGNNYAFVEEGFSLWNNPETLREHVGVTPNTF 123
Query: 158 HVDCVHMGYALMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSDEA 217
H LMN QSI + Q + EY VR+ S+ A++YLLR GL FRG DE+
Sbjct: 124 HNISAKKCDDLMNQAQSIVHSLHKQDDVVKKEYRVRLNASIDASRYLLRQGLPFRGHDES 183
Query: 218 DDSLYKGPFLELLD-TLKENNSDVATILDSAPGNSLMTCPKIQKDLASA 265
+S +G FLEL+ T ++N++ IL++AP N + P IQKD ++
Sbjct: 184 KESTNRGNFLELIKYTSEQNDATSKVILENAPKNKQVISPVIQKDTVNS 232
>At4g10200 putative protein
Length = 733
Score = 95.1 bits (235), Expect = 1e-19
Identities = 89/327 (27%), Positives = 142/327 (43%), Gaps = 46/327 (14%)
Query: 83 YPWSDFRGTRRRFNKNWFNLY---------DWLEYSESKNLAFCLPCFLFKNVSNYGGDH 133
Y + G RR F+ ++ +WL YS+ + +C C LF +
Sbjct: 92 YAFPKTAGIRRHFSHRYYKREMKNGDKQDRNWLLYSKVSDKVYCFCCKLFGR--DQDAMQ 149
Query: 134 FVGDGFGDWKNPR-KLANHATSNNSHVDCVHMGYAL---MNPNQSIKAAF---VNQTKQM 186
GF DW+N R +L+ H TS+ H+ C+ L + + +I +N K
Sbjct: 150 LSSTGFNDWRNIRIRLSQHETSHR-HIVCMSKWMELELRLRKHLTIDKCLQKDINIEKNH 208
Query: 187 NVEYCVRVKTSLLATKYLLRCGLAFRGSDEADDSLYKGPFLELLDTLKENNSDVATILDS 246
E +R+ + + K L + LAFRG +E G F LL+ +I D
Sbjct: 209 WREVLLRIFSLV---KNLAKQNLAFRGENEKIGQKNNGNFRVLLNRSATLIQSYESISDE 265
Query: 247 APGNSLMTCPKIQKDLASACACEITREIVCDIADDVFCVLIDESGDVSGREQMAVVLRYV 306
LM KIQ+ F V++D + D S +EQM++++R V
Sbjct: 266 LKKVQLMIIKKIQEA-------------------KYFSVILDCTPDKSHKEQMSLIIRCV 306
Query: 307 D---GDGLVKERFLGITSVKETSAKSLKDAL-ETMLSINGLSFSSIRGQGYDGASNMRGR 362
D V E FL V + S + L + L +T++++N L+ + +RGQGYD NM+G+
Sbjct: 307 DVSMASTQVSEFFLTFVEVSDKSGEGLFELLCDTLVALN-LNINDVRGQGYDNGCNMKGK 365
Query: 363 FGGLKTLIQNENPSAHYVHCFAHQLQL 389
G++ + + N A Y C H L L
Sbjct: 366 HKGVQKKLLDINSRAFYTPCGCHSLNL 392
Score = 55.8 bits (133), Expect = 9e-08
Identities = 68/292 (23%), Positives = 122/292 (41%), Gaps = 36/292 (12%)
Query: 508 FDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRV 567
F+F+F + + +L N +S ALQ + D+ A+ + LQ R G++E +
Sbjct: 456 FEFLFGMVIWYNLLFTMNTVSKALQSENIDIELALVQLKGLVSYLQNYRETGFQEAKAEA 515
Query: 568 VTICTKHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFD 627
I +I+ P K+ R + +ND +L + + RF+
Sbjct: 516 TLIAESMDIE------PKFPVKRKRIIKRKRHFDE--EMENDVETELLSEE-ENFKVRFE 566
Query: 628 --EENTELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQL-HNYVRNV 684
EE + + + FD+ KL ++ D + + + L L H +V
Sbjct: 567 QFEEYGRIFRFL-----------FDLRKLKSASD----DGLKAACINLETSLKHGDSSDV 611
Query: 685 KSDPKFAKLKGLSDLCAILVET-----NKCKTFALVFK----LLKLALLLPVATASVERV 735
+ F +LK L +L + N K F + ++ L +PV+ AS ER
Sbjct: 612 DGNHLFLELKVLKELLPTEITKAIEVLNFLKKFEGCYPNTWIAFRVMLTVPVSVASAERS 671
Query: 736 FSAMKIVKSHLRNKMGDQWLNDRLVTFIERDVLFTISTDVILAHFQQMDDRR 787
FS +K++KS+ R+ M ++ LN + IERD++ + ++ F RR
Sbjct: 672 FSKLKLIKSYSRSTMSEERLNALAILSIERDLVGELDYISLMNDFAAKTARR 723
>At1g42710 unknown protein
Length = 206
Score = 92.4 bits (228), Expect = 9e-19
Identities = 69/214 (32%), Positives = 98/214 (45%), Gaps = 56/214 (26%)
Query: 155 NNSHVDCVHMGYALMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGS 214
N+SH + LMN QSI A Q M EY + + TS+ A++YLLR G+AF G
Sbjct: 34 NSSHNYAIEKCVNLMNQGQSIVHALFKQDDVMKREYHIPLNTSIDASRYLLRQGIAFHGH 93
Query: 215 DEADDSLYKGPFLELLDTLKENNSDVATILDSAPGNSLMTCPKIQKDLASACACEITREI 274
DE+++S KG FLEL+ E N + T ++
Sbjct: 94 DESEESANKGNFLELVKYTGEQN-------------------------------DATEQM 122
Query: 275 VCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDAL 334
V +D+SG VKERF+ + VKETS+ S+K A+
Sbjct: 123 A------VVFRFVDKSG-------------------TVKERFIEVVHVKETSSASVKSAI 157
Query: 335 ETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKT 368
+ + + GLS ++RGQGYDGAS M+ G K+
Sbjct: 158 DDLFAKYGLSLKTVRGQGYDGASIMKVAKGVAKS 191
>At2g06500 Ac-like transposase
Length = 582
Score = 66.2 bits (160), Expect = 7e-11
Identities = 57/226 (25%), Positives = 101/226 (44%), Gaps = 10/226 (4%)
Query: 139 FGDWKNPRKLANHATSNNSHVDCVHMGYALMN--PNQSIKAAFVNQTKQMNVEYCVRVKT 196
+ DW+N K ++ H+ C+ L + N++ +V Q + V
Sbjct: 85 YNDWRNLSKRLEEHEGSHDHIICMTRWTELESRLQNKTTIDKYVQQEINKEKIHWREVLV 144
Query: 197 SLLA-TKYLLRCGLAFRGSDEADDSLYKGPFLELLDTLKENNSDVATILDSAPGNSLMT- 254
++A K L + LAFRG ++ G FL ++ + E + + + + +
Sbjct: 145 RIIALVKTLAKNNLAFRGENKKIGEDRNGNFLSFIEMIAEFDVVMREHIRKIGAGEIYSH 204
Query: 255 --CPKIQKDLASACACEITREIVCDI-ADDVFCVLIDESGDVSGREQMAVVLRYVDGDGL 311
PKIQ +L S A EI I+ I A F +++D + D+S +EQM +++R VD
Sbjct: 205 YLSPKIQNELISMLAQEIRLMIMKTIRASKYFSIILDCTPDISHKEQMTILIRCVDISST 264
Query: 312 ---VKERFLGITSVKETSAKSLKDALETMLSINGLSFSSIRGQGYD 354
V+E FL V + + + L ++ +L L +RG G+D
Sbjct: 265 PIKVEEFFLKFLEVNDKTGEGLFSTIQEVLIDMELEIDDVRGHGWD 310
Score = 40.4 bits (93), Expect = 0.004
Identities = 21/61 (34%), Positives = 35/61 (56%)
Query: 720 KLALLLPVATASVERVFSAMKIVKSHLRNKMGDQWLNDRLVTFIERDVLFTISTDVILAH 779
++ L + V+ AS ER FS +K++K++LR+ M LN + IER +L I ++
Sbjct: 510 RILLTILVSVASAERSFSKLKLIKNYLRSTMSQDRLNGLAILSIERAMLEKIDYATVMDD 569
Query: 780 F 780
F
Sbjct: 570 F 570
>At1g35150 hypothetical protein
Length = 459
Score = 60.5 bits (145), Expect = 4e-09
Identities = 54/231 (23%), Positives = 99/231 (42%), Gaps = 10/231 (4%)
Query: 131 GDHFVGDGFGDWKNPRKLANHATSNNSHVDCVHMGYAL---MNPNQSIKAAFVNQTKQMN 187
G + G+ DW+N K ++ H+ C+ L + N++I + N
Sbjct: 94 GGYLATSGYNDWRNLSKRLKEHKGSHDHITCMTRRAELESRLQKNKTIDKHAQEAINKDN 153
Query: 188 VEYCVRVKTSLLATKYLLRCGLAFRGSDEADDSLYKGPFLELLDTLKENN---SDVATIL 244
+ + + + K + LAFRG +E G FL ++ + E + + +
Sbjct: 154 IHWREVLLRIIALVKTHAKNNLAFRGKNEKVGQDRNGNFLSFIEMIAEFDVVMREHIRRI 213
Query: 245 DSAPGNSLMTCPKIQKDLASACACEITREIVCDIADDVFC-VLIDESGDVSGREQMAVVL 303
+A S KIQ +L EI I+ I +C +++D + D+S +EQM +++
Sbjct: 214 GAAEIYSHYLSHKIQNELIGILTGEIRLMIMKTIHASKYCSIILDCTPDISHKEQMTMII 273
Query: 304 RYVDGDGL---VKERFLGITSVKETSAKSLKDALETMLSINGLSFSSIRGQ 351
R V+ V+E +L VK+ S++ L ++ L L +RGQ
Sbjct: 274 RCVNISSTLTKVEEFYLTFLEVKDKSSEGLFSKIKEALVDMELEIDDVRGQ 324
>At2g16040 Ac-like transposase
Length = 382
Score = 55.1 bits (131), Expect = 2e-07
Identities = 61/265 (23%), Positives = 116/265 (43%), Gaps = 33/265 (12%)
Query: 508 FDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRV 567
F+F+F + + ++L N +S +LQ D DL AIS + L+ + G+E+ ++V
Sbjct: 128 FEFLFGMIIWYDLLAAVNIVSKSLQFEDMDLEVAISQLGGLVTYLKNYKETGFEK--AKV 185
Query: 568 VTICTKHEIDVPDMDAPYMEGKKPRRVPPVSSVSNL----HHYKNDCLFSVLDLQLQELN 623
+ E+ + + KK + V V + ++ D +++D + +
Sbjct: 186 ESTQIAIEMKIAPVFPKKSVKKKKQFVEDVEKIDESKIAEESFRIDYFINIMDQAIMCIE 245
Query: 624 ARFDEENTELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQL-HNYVR 682
RF++ + F K L++AE +D + S ++L L H+
Sbjct: 246 IRFEQFQVY----------EQIFGFLFGVKRLKVAE---DDELRTSCMKLEASLKHDVHS 292
Query: 683 NVKSDPKFAKLKGLSDLCAILVETNKCKTFALVFKLL-----------KLALLLPVATAS 731
+V + F +LK L D+ + E K K++ ++ L +PV+ A
Sbjct: 293 DVDGEDLFMELKLLKDV--LPKEITKPVEVLKFLKIMDSCYPNTWIAYRILLTIPVSVAL 350
Query: 732 VERVFSAMKIVKSHLRNKMGDQWLN 756
ER FS +K++K +LR+ M + LN
Sbjct: 351 AERTFSKLKLIKKYLRSTMSQERLN 375
Score = 30.4 bits (67), Expect = 4.0
Identities = 23/72 (31%), Positives = 35/72 (47%), Gaps = 4/72 (5%)
Query: 257 KIQKDLASACACEITREIVCDIAD-DVFCVLIDESGDVSGREQMAVVLRYVDGDGL---V 312
K Q +L A + I+ I D F V++D DVS +EQM ++R VD +
Sbjct: 29 KSQNELTELLANDTKMMILKKIKDAKYFSVILDSIPDVSRKEQMTFLIRCVDVSTCSPKI 88
Query: 313 KERFLGITSVKE 324
+E FL +K+
Sbjct: 89 EEFFLTFLHIKD 100
>At3g29638 putative protein
Length = 412
Score = 51.2 bits (121), Expect = 2e-06
Identities = 32/100 (32%), Positives = 46/100 (46%), Gaps = 4/100 (4%)
Query: 314 ERFLGITSVKETSAKSLKDALETMLSIN----GLSFSSIRGQGYDGASNMRGRFGGLKTL 369
E F S+ + K +K+ L +L L +RGQGYD SN++G+ ++
Sbjct: 124 ETFEEFDSLTQEHIKRVKEGLFELLCYTFGDLKLKIDDVRGQGYDNGSNIKGKHKRVQKR 183
Query: 370 IQNENPSAHYVHCFAHQLQLALVACAKTHKPVSGFFGKVN 409
+ + N A Y C H L LAL AK+ FFG N
Sbjct: 184 LLDINSIAFYASCGCHSLNLALADMAKSSSKAILFFGISN 223
>At2g19960 Ac-like transposase
Length = 173
Score = 47.0 bits (110), Expect = 4e-05
Identities = 24/71 (33%), Positives = 42/71 (58%)
Query: 720 KLALLLPVATASVERVFSAMKIVKSHLRNKMGDQWLNDRLVTFIERDVLFTISTDVILAH 779
++ L +PV+ AS ER FS +K++KS+LR+ M + L+D + IER ++ + + +
Sbjct: 103 QVLLTIPVSVASAERSFSKLKLIKSYLRSSMSQERLSDLAILSIERALVREVDFERLAND 162
Query: 780 FQQMDDRRFSL 790
F RR +L
Sbjct: 163 FVGKKGRRITL 173
>At4g25440 putative protein
Length = 668
Score = 32.3 bits (72), Expect = 1.1
Identities = 18/66 (27%), Positives = 29/66 (43%), Gaps = 13/66 (19%)
Query: 349 RGQGYDGASNMRGRFGGLKTLIQNE-------------NPSAHYVHCFAHQLQLALVACA 395
RG G+ G +N GRFGG +T+ + E Y+HC++ +L+
Sbjct: 81 RGPGFSGTANNWGRFGGNRTVTKTEKLCKFWVDGNCPYGDKCRYLHCWSKGDSFSLLTQL 140
Query: 396 KTHKPV 401
H+ V
Sbjct: 141 DGHQKV 146
>At3g54860 vacuolar protein sorting protein 33 (VPS33)
Length = 592
Score = 30.8 bits (68), Expect = 3.1
Identities = 25/97 (25%), Positives = 43/97 (43%), Gaps = 9/97 (9%)
Query: 675 RQLHNYVRNVKSDPKFAKLKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVER 734
R+L N +++V+ LS ++++ T+K K L + L PV T +
Sbjct: 23 RELVNLLKDVRGTKCLVIDPKLSGSVSLIIPTSKLKELGLELRHLTAE---PVQTECTKV 79
Query: 735 VF------SAMKIVKSHLRNKMGDQWLNDRLVTFIER 765
V+ S MK + SH++N + D V F+ R
Sbjct: 80 VYLVRSQLSFMKFIASHIQNDIAKAIQRDYYVYFVPR 116
>At4g38065 hypothetical protein
Length = 1050
Score = 30.0 bits (66), Expect = 5.3
Identities = 37/145 (25%), Positives = 59/145 (40%), Gaps = 39/145 (26%)
Query: 530 ALQRRDQDL----------LNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKHEIDVP 579
ALQ++DQDL L ++SL+ K+ M + WE+L +R I T E +
Sbjct: 841 ALQQKDQDLCEVKHELEGSLKSVSLLLQQKQNEVNMLRKTWEKLTAR--QILTAVETESK 898
Query: 580 DMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCL--------------------FSVLDLQL 619
M +EG+ +SS+S N+ + + Q+
Sbjct: 899 KMMIIELEGE-------ISSLSQKLETSNESVSCFRQEATKSRAELETKQTELKEVTTQM 951
Query: 620 QELNARFDEENTELLQCVSCLSPAK 644
QE + E TEL++ V+ LS K
Sbjct: 952 QEKLRTSEAEKTELVKEVASLSTEK 976
>At1g06580 hypothetical protein
Length = 500
Score = 30.0 bits (66), Expect = 5.3
Identities = 25/98 (25%), Positives = 45/98 (45%), Gaps = 10/98 (10%)
Query: 436 LIEEGEIETGSGLNQDSSIARAGDT------RWGSHFRTLTSLMTLYGAIVEVIVEVGND 489
L+E+G + T L +S A +G + R G H +TL+ + E
Sbjct: 21 LLEKGNLVTALSLRICNSRAFSGRSDYRERLRSGLHSIKFNDALTLFCDMAESHPL---- 76
Query: 490 PSFDKFGETVLLLDVLQSFDFIFMLYMMVEILGITNDL 527
PS F ++ + L ++ + L+ +E+LGI++DL
Sbjct: 77 PSIVDFSRLLIAIAKLNKYEAVISLFRHLEMLGISHDL 114
>At5g48600 chromosome condensation protein
Length = 1241
Score = 29.6 bits (65), Expect = 6.9
Identities = 32/131 (24%), Positives = 56/131 (42%), Gaps = 38/131 (29%)
Query: 537 DLLNAISLV-NDTKKQLQEMRNEGWEELIS-------------RVVTICTKHEIDVPDMD 582
D LN+++ +DT+KQ E+R +E ++ +++T+ E+++ D
Sbjct: 1059 DELNSVTQERDDTRKQYDELRKRRLDEFMAGFNTISLKLKEMYQMITLGGDAELELVDSL 1118
Query: 583 APYMEGKKPRRVPPVSSVSN-------------------LHHYKNDCLFSVLDLQLQELN 623
P+ EG PP S N LHHYK L+ V+D E++
Sbjct: 1119 DPFSEGVVFSVRPPKKSWKNIANLSGGEKTLSSLALVFALHHYKPTPLY-VMD----EID 1173
Query: 624 ARFDEENTELL 634
A D +N ++
Sbjct: 1174 AALDFKNVSIV 1184
>At5g48360 unknown protein
Length = 782
Score = 29.6 bits (65), Expect = 6.9
Identities = 47/198 (23%), Positives = 83/198 (41%), Gaps = 20/198 (10%)
Query: 546 NDTKKQL---QEMRNEGWEELISRVVTICTKHEIDVPDMDAPYMEGKKPRRVPPVSSVSN 602
ND KKQ Q + WE L S + +K ++ + PR +P + V +
Sbjct: 417 NDVKKQSFSDQPPKQLHWERLRSSSSKL-SKEMVETMFI----ANSSNPRDLPIQNQVLD 471
Query: 603 LHHYKNDC-LFSVLDLQLQE-----LNARFDEENTELLQCVSCLSPAK----SFSAFDVN 652
+N L +L+L ++ L+ D ELL+C+S L+P+K +F
Sbjct: 472 PRKAQNIATLLQLLNLSTKDVCQALLDGDCDVLGAELLECLSRLAPSKEEERKLKSFSDG 531
Query: 653 KLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAKLKGLSDLCAILVETNKCKTF 712
+ AE + + + V V R +V N S+ K + K S + E + F
Sbjct: 532 SEIGPAERFLKELLHVPFVFKRVDALLFVANFHSEIKRLR-KSFSVVQVACEELRNSRMF 590
Query: 713 ALVFK-LLKLALLLPVAT 729
+++ + +LK ++ V T
Sbjct: 591 SILLEAILKTGNMMSVRT 608
>At3g54630 unknown protein
Length = 560
Score = 29.3 bits (64), Expect = 9.0
Identities = 24/95 (25%), Positives = 42/95 (43%), Gaps = 19/95 (20%)
Query: 607 KNDCLFSVLDLQLQELNARFDE------ENTELLQCVSCLSPAKSFSAFDVNKLLRMAEL 660
+N + V++ + +EL A+ +E EN EL + V ++FSA DVN++ R +
Sbjct: 268 RNPAMEKVVEEKAKELKAKEEERERISVENKELKKSVEL----QNFSAADVNRMRRELQA 323
Query: 661 YPNDFVDVSEV---------ELRRQLHNYVRNVKS 686
D D EL Q+ N +++
Sbjct: 324 VERDVADAEVARDGWDQKAWELNSQIRNQFHQIQT 358
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,461,337
Number of Sequences: 26719
Number of extensions: 759204
Number of successful extensions: 1935
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1870
Number of HSP's gapped (non-prelim): 35
length of query: 790
length of database: 11,318,596
effective HSP length: 107
effective length of query: 683
effective length of database: 8,459,663
effective search space: 5777949829
effective search space used: 5777949829
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146806.1


