

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146794.16 + phase: 0 /pseudo/partial
(1323 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
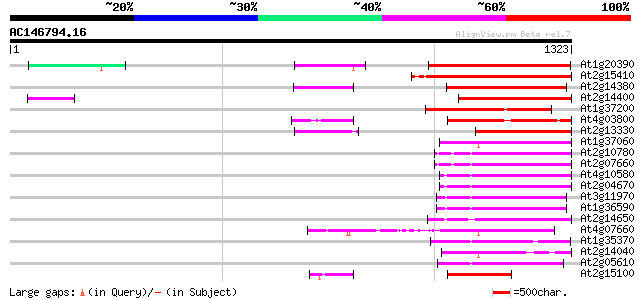
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 342 1e-93
At2g15410 putative retroelement pol polyprotein 338 2e-92
At2g14380 putative retroelement pol polyprotein 291 2e-78
At2g14400 putative retroelement pol polyprotein 285 1e-76
At1g37200 hypothetical protein 279 9e-75
At4g03800 241 2e-63
At2g13330 F14O4.9 234 2e-61
At1g37060 Athila retroelment ORF 1, putative 219 1e-56
At2g10780 pseudogene 217 4e-56
At2g07660 putative retroelement pol polyprotein 213 8e-55
At4g10580 putative reverse-transcriptase -like protein 209 7e-54
At2g04670 putative retroelement pol polyprotein 204 2e-52
At3g11970 hypothetical protein 202 8e-52
At1g36590 hypothetical protein 202 8e-52
At2g14650 putative retroelement pol polyprotein 193 5e-49
At4g07660 putative athila transposon protein 193 6e-49
At1g35370 hypothetical protein 192 1e-48
At2g14040 putative retroelement pol polyprotein 191 3e-48
At2g05610 putative retroelement pol polyprotein 185 1e-46
At2g15100 putative retroelement pol polyprotein 172 2e-42
>At1g20390 hypothetical protein
Length = 1791
Score = 342 bits (876), Expect = 1e-93
Identities = 162/336 (48%), Positives = 231/336 (68%)
Query: 987 ELINIGTEENKREIKIGAALEEGVKKKIIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPT 1046
E++NI + R + +GA + ++ ++I LL+ FAWS EDM G+DP I H +
Sbjct: 712 EMVNIDESDPTRCVGVGAEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNV 771
Query: 1047 KPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMC 1106
P PV+QK R+ P+ A + EV+K + AG ++ V+YPEW+AN V V KK+GK R+C
Sbjct: 772 DPTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVC 831
Query: 1107 VDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITP 1166
VD+ DLNKA KD++PLPHID LV+ T+ + + SFMD FSGYNQI M +D+EKTSF+T
Sbjct: 832 VDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTD 891
Query: 1167 WGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKM 1226
GT+CYKVM FGL NAGATYQR + + D I + VEVY+DDM+VKS E HVE+L+K
Sbjct: 892 RGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKC 951
Query: 1227 FERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGF 1286
F+ L Y ++LNP KCTFGV SG+ LG++V+++GIE +P ++RAI E+P+P+ ++V+
Sbjct: 952 FDVLNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRL 1011
Query: 1287 LGRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDE 1322
GR+ ++RFIS T C P + LL++ W+ +
Sbjct: 1012 TGRIAALNRFISRSTDKCLPFYNLLKRRAQFDWDKD 1047
Score = 73.6 bits (179), Expect = 7e-13
Identities = 47/176 (26%), Positives = 81/176 (45%), Gaps = 7/176 (3%)
Query: 671 FSEDELPEAGKHHNLALHISVNCKSDMISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRS 730
F++ +L HN L + + ++ VL+DTGSS++++ K L ++ ++
Sbjct: 555 FTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIKPV 614
Query: 731 TFLVKAFDGTRKSVLGEIDLPITIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTST 790
+ + FDG +G I LPI +G + F V+ A Y+ +LG PWIH A+ ST
Sbjct: 615 SKPLAGFDGDFVMTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPST 674
Query: 791 LHQKLKFAKSGKLVTIHG-------EEAYLVSQLSSFSCIEAGSAEGTAFQGLTVE 839
HQ +KF + T+ +Y S+L + ++ T G+ E
Sbjct: 675 YHQCVKFPTHNGIFTLRAPKEAKTPSRSYEESELCRTEMVNIDESDPTRCVGVGAE 730
Score = 45.1 bits (105), Expect = 3e-04
Identities = 44/239 (18%), Positives = 89/239 (36%), Gaps = 10/239 (4%)
Query: 45 DRYNGLTCPQNHIIKY---VRKMGNYKDN-DSLMIHCFQDSLMEDAAEWYTSLSKNDIHT 100
+ YNG P+ + + + + DN D+ F + L A W++ L N I +
Sbjct: 175 ESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIEHLTGPAHNWFSRLKPNSIDS 234
Query: 101 FDELAVAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRGAAARITPALDEEEMT 160
F +L +F HY + + L S+SQ +ES R + R++ IT + +
Sbjct: 235 FHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFVDRFKLVVTNITVPDEAAIVA 294
Query: 161 QTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFEKAESSVNAS------ 214
+ + + + AP+ + + +R E + +I+ K S+ +
Sbjct: 295 LRNAVWYDSRFRDDITLHAPSTLEDALHRASRFIELEEEKLILARKHNSTKTPACKDAVV 354
Query: 215 KRYGNGHHKKKETEIGMVSAGAGQPMATVALINAAQMPPSYPYVPYSQHPFFPPFYHQY 273
+ G + + + +P + + P Y+ + P P Y +Y
Sbjct: 355 IKVGPDDSNEPRQHLDRNPSAGRKPTSFLVSTETPDAKPWNKYIRDADSPAAGPMYCEY 413
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 338 bits (866), Expect = 2e-92
Identities = 165/375 (44%), Positives = 244/375 (65%), Gaps = 7/375 (1%)
Query: 949 FEFPVYEAEDEEGDDIPYEITRLLEQEKKAIQPH*EEIELINIGTEENKREIKIGAALEE 1008
FEF V + I Y + + Q+++ P + +NI + R + I L
Sbjct: 686 FEFAVVDKP------IIYNVILVPTQDERP-NPQKGTVVQVNIDESDPSRCVGIRIDLPS 738
Query: 1009 GVKKKIIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKI 1068
++ +++ LR+ FAWS EDMPG+D + H + P P++QK R+ PD +
Sbjct: 739 ELQNELVNFLRQNAATFAWSVEDMPGIDSAVTCHELNVDPTYKPLKQKRRKLGPDRTKDV 798
Query: 1069 KSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLPHIDV 1128
EV+K +DAG ++ V YP+W+ N V V KK+GK R+C+DF DLNKA KD+FPLPHID
Sbjct: 799 NEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKNGKWRVCIDFTDLNKACPKDSFPLPHIDR 858
Query: 1129 LVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQR 1188
LV+ TA +++ SFMD FSGYNQI M DREKT FIT GT+CYKVMPFGL NAGATY R
Sbjct: 859 LVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQGTYCYKVMPFGLKNAGATYPR 918
Query: 1189 GMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRS 1248
+ +F D + +EVY+DDM+VKS E+H+ +L + F+ L +Y ++LNP+KCTFGV S
Sbjct: 919 LVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCFQVLNRYNMKLNPSKCTFGVTS 978
Query: 1249 GKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFISHMTATCGPIF 1308
G+ LG++V+++GIE +P ++ AI ++P+P+ ++V+ +GR+ ++RFIS T C P +
Sbjct: 979 GEFLGYLVTRRGIEANPKQISAIIDLPSPRNTREVQRLIGRIAALNRFISRSTDKCLPFY 1038
Query: 1309 KLLRKNQPIVWNDEC 1323
+LLR N+ W+++C
Sbjct: 1039 QLLRANKRFEWDEKC 1053
Score = 34.3 bits (77), Expect = 0.49
Identities = 24/102 (23%), Positives = 42/102 (40%), Gaps = 4/102 (3%)
Query: 47 YNGLTCPQNHIIKYVRKMGNY----KDNDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFD 102
Y GL P+ + +G +D D+ F L A W++ L N I +
Sbjct: 258 YEGLVDPRPFLTSVSIAIGRAHFSDEDRDAGSCQLFVKHLSGAALTWFSRLEANSIDSVH 317
Query: 103 ELAVAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWR 144
L +F +YG + L +++Q +ES R + R++
Sbjct: 318 ALTTSFLKNYGVFMEKGASNVDLWTMAQTAKESLRSFIGRFK 359
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 291 bits (744), Expect = 2e-78
Identities = 139/282 (49%), Positives = 196/282 (69%)
Query: 1031 DMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWV 1090
DM G+DP + H + P V+QK R+ P+ + + EV K +DAG ++ V+YPEW+
Sbjct: 476 DMVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEWL 535
Query: 1091 ANIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQ 1150
AN V V KK+ K R+C+DF DLNKA KD+FPLPHID +V+ T +++ SFMD FSGYNQ
Sbjct: 536 ANPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQ 595
Query: 1151 IKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMI 1210
I M +D+EKTSFI GT+CYKVMPFGL N GA YQR + +F + K +EVY+DDM+
Sbjct: 596 IPMHKDDQEKTSFIIDRGTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDDML 655
Query: 1211 VKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRA 1270
VKST H+++L FE L KY ++LNP KC FGV SG+ LG+IV+++GIE +P ++RA
Sbjct: 656 VKSTRSADHIDHLKACFETLNKYNMKLNPAKCLFGVTSGEFLGYIVTKRGIEANPKQIRA 715
Query: 1271 IREMPAPQTQKQVRGFLGRLNYISRFISHMTATCGPIFKLLR 1312
I ++ +P+ +K+V+ GR+ ++RFI+ T P ++LLR
Sbjct: 716 ILDLQSPRNKKEVQRLTGRIAGLNRFIARSTDKSLPFYQLLR 757
Score = 81.6 bits (200), Expect = 3e-15
Identities = 47/142 (33%), Positives = 72/142 (50%)
Query: 669 LWFSEDELPEAGKHHNLALHISVNCKSDMISNVLVDTGSSLNVMPKSTLDQLSYRGTPLR 728
L F+ ++L HN L I ++ ++ +L+DTGSS+NV+ K L ++ ++
Sbjct: 330 LTFTSEDLFGVDLPHNDPLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIK 389
Query: 729 RSTFLVKAFDGTRKSVLGEIDLPITIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVT 788
S + FDG G I LPI +G F V+D Y+ +LG PWIHD A+
Sbjct: 390 PSVRPLTGFDGNTMMTNGTIKLPIYLGGAATWHKFVVVDKPTIYNIILGTPWIHDMQAIP 449
Query: 789 STLHQKLKFAKSGKLVTIHGEE 810
S+ HQ +K S + TI G +
Sbjct: 450 SSYHQCIKIPTSIGIETIRGNQ 471
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 285 bits (729), Expect = 1e-76
Identities = 132/266 (49%), Positives = 193/266 (71%)
Query: 1058 RRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASL 1117
R+ + A + EV K + G + V+YP+W+AN V V KK+GK R+C+DF DLNKA
Sbjct: 470 RKLGVERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKACP 529
Query: 1118 KDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPF 1177
KD+FPLPHID LV++TA +++ SFMD FSGYNQI M+PED+EKT FIT G +CYKVMPF
Sbjct: 530 KDSFPLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPF 589
Query: 1178 GLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRL 1237
GL NAGATY R + +F + + K +EVY+DDM++KS +E HV++L + F L +Y+++L
Sbjct: 590 GLRNAGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKL 649
Query: 1238 NPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFI 1297
NP KCTFGV SG+ LG+IV+++GIE +P+++ A MP+P+ K+V+ GR+ ++RFI
Sbjct: 650 NPAKCTFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRFI 709
Query: 1298 SHMTATCGPIFKLLRKNQPIVWNDEC 1323
S T P +++L+ N+ +W+++C
Sbjct: 710 SRSTDKSLPFYQILKGNKEFLWDEKC 735
Score = 47.0 bits (110), Expect = 7e-05
Identities = 28/113 (24%), Positives = 51/113 (44%), Gaps = 4/113 (3%)
Query: 43 DFDRYNGLTCPQNHIIKYVRKMG----NYKDNDSLMIHCFQDSLMEDAAEWYTSLSKNDI 98
+ + YNGL P+ ++ ++ G N D D F ++L A W+T L I
Sbjct: 161 NLESYNGLEDPKGYLAAFLIAAGRVDLNEADEDVRYCKLFSENLCGQALMWFTQLEPGSI 220
Query: 99 HTFDELAVAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRGAAARIT 151
F+EL+V F Y + L +LSQ E+ R + +++ ++++
Sbjct: 221 SNFNELSVVFLKQYSILMDKSISDTDLWNLSQGPNETLRAFITKFKYVLSKLS 273
Score = 39.3 bits (90), Expect = 0.015
Identities = 24/89 (26%), Positives = 49/89 (54%), Gaps = 4/89 (4%)
Query: 667 SNLWFSEDELPEAGKHHNLALHISVNCKSDMISNVLVDTGSSLNVMPKSTLDQLSYRGTP 726
+++ F E E + H+ AL ++++ + +S +L+DTGSS++++ STL+++
Sbjct: 407 TSILFDEKETQHLERSHDDALVVTLDVANFEVSRILIDTGSSVDLIFLSTLERMGISRAD 466
Query: 727 LRRSTFLVKAFDGTRKSVLGEIDLPITIG 755
+ R V+ K+V E+D + IG
Sbjct: 467 VNRRKLGVE----RAKAVNDEVDKLLKIG 491
>At1g37200 hypothetical protein
Length = 1564
Score = 279 bits (713), Expect = 9e-75
Identities = 134/298 (44%), Positives = 197/298 (65%), Gaps = 4/298 (1%)
Query: 981 PH*EEIELINIGTEENKREIKIGAALEEGVKKKIIQLLREYPDIFAWSYEDMPGLDPMIV 1040
P I + I + K+ + +G L+ +++ I L+E D FAWS ++ G+ +
Sbjct: 614 PQKSSITQVYIDESDPKQCVGVGQDLDPAIREDFITFLKENKDSFAWSSANLQGISLEVT 673
Query: 1041 EHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKD 1100
H + P P++QK R+ P+ A ++ EV + + G + V+YP+W+AN V V K+
Sbjct: 674 SHELNVDPTYRPIKQKRRKLGPERAKAVQDEVDRLLKIGSIREVKYPDWLANPVVVKNKN 733
Query: 1101 GKVRMCVDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREK 1160
GK R+C+DF DLNKA KD+FPLPHID LV TA ++ SFMD FSGYNQI M P+D+EK
Sbjct: 734 GKWRVCIDFMDLNKACPKDSFPLPHIDRLVKATAGHELLSFMDAFSGYNQILMRPDDQEK 793
Query: 1161 TSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHV 1220
T+FIT CYKVMPFGL N GATYQR + +F D + K +E+Y+DDM+VKS E+ H+
Sbjct: 794 TAFITD----CYKVMPFGLKNTGATYQRLVNRMFADQLGKTMELYIDDMLVKSAHEKDHL 849
Query: 1221 EYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQ 1278
L + F+ L K++++LNP KC+FGV SG+ LG++V+Q+GIE +P ++ A +MP+P+
Sbjct: 850 PQLRECFKILNKFEMKLNPEKCSFGVPSGEFLGYLVTQRGIEANPKQIAAFIDMPSPK 907
Score = 42.0 bits (97), Expect = 0.002
Identities = 22/53 (41%), Positives = 29/53 (54%), Gaps = 6/53 (11%)
Query: 770 ASYSCLLGRPWIHDAGAVTSTLHQKLKFAKSGKLVTIHGEEAYLVSQLSSFSC 822
A ++ +LGRPW+H AV ST HQ +KF + I+G SQ SS C
Sbjct: 530 APFNAILGRPWLHAMKAVPSTYHQCIKFPSEKGIAVIYG------SQRSSRRC 576
Score = 30.4 bits (67), Expect = 7.1
Identities = 15/45 (33%), Positives = 27/45 (59%), Gaps = 2/45 (4%)
Query: 669 LWFSEDELPEAGKHHNLALHISVNCKSDMISNVLVDTGSSLNVMP 713
LW E E + K H+ A+ I ++ + +S +++DTGSS++ P
Sbjct: 489 LW--ESETTDLDKPHDDAIVIRIDVGNYKLSRIMIDTGSSVDKAP 531
>At4g03800
Length = 637
Score = 241 bits (615), Expect = 2e-63
Identities = 121/292 (41%), Positives = 187/292 (63%), Gaps = 21/292 (7%)
Query: 1032 MPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVA 1091
MP +DP I+ H + P P++QK R+ + A + ++ K + G + V+YP+WVA
Sbjct: 321 MPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWVA 380
Query: 1092 NIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQI 1151
V V KK+GK R+C+DF DLNKA KD+FPLPHID LV++TA +++ +FMD F GYNQI
Sbjct: 381 ITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLTFMDAFLGYNQI 440
Query: 1152 KMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIV 1211
M+PED+EKTSFIT + GATYQ + +F++ + K +EV +DD +V
Sbjct: 441 MMNPEDQEKTSFIT---------------DRGATYQWLVNKMFNEHLRKTMEVSIDDTLV 485
Query: 1212 KSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAI 1271
KS +E HV++L + FE L +Y+++LN KCTFGV SG+ LG+IV+++GIE +P+++ A
Sbjct: 486 KSLKKEDHVKHLGECFEILNQYQMKLNLAKCTFGVPSGEFLGYIVTKRGIEANPNQINAF 545
Query: 1272 REMPAPQTQKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDEC 1323
+ P+ + K+V+ GR+ S T P +++L+ N +W+++C
Sbjct: 546 LKTPSLRNFKEVQRLTGRI------ASRSTDKSLPFYQILKGNNGFLWDEKC 591
Score = 78.6 bits (192), Expect = 2e-14
Identities = 51/144 (35%), Positives = 76/144 (52%), Gaps = 14/144 (9%)
Query: 666 CSNLWFSEDELPEAGKHHNLALHISVNCKSDMISNVLVDTGSSLNVMPKSTLDQLSYRGT 725
CS++ F E+E + H+ AL I+++ + IS +LVDTGSS++++ + G
Sbjct: 182 CSSVSFDEEETRHIERPHDDALIITLDVANFKISRILVDTGSSVDLI---------FLGP 232
Query: 726 PLRRSTFLVKAFDGTRKSVLGEIDLPITIGPETFLITFQVMDINASYSCLLGRPWIHDAG 785
P + AF LG I LP+ G + ++ F V D A+Y+ +LG PWI
Sbjct: 233 PSP-----LVAFTSESAMSLGTIKLPVLAGKMSKIVDFVVFDKPATYNIILGTPWIFQMK 287
Query: 786 AVTSTLHQKLKFAKSGKLVTIHGE 809
AV ST HQ LKF S + TI G+
Sbjct: 288 AVPSTYHQWLKFPTSNGVETIWGD 311
>At2g13330 F14O4.9
Length = 889
Score = 234 bits (597), Expect = 2e-61
Identities = 111/226 (49%), Positives = 159/226 (70%)
Query: 1098 KKDGKVRMCVDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPED 1157
+K+GK R+C+DFRDLNKA KD+FPLPHID LV+ T + + SFMD F GYNQI M +
Sbjct: 289 EKNGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDG 348
Query: 1158 REKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEE 1217
+EKT+FIT GT+ YKVMPFGL NAG TYQR + +F D + K +EVY+DDM+VKS E+
Sbjct: 349 QEKTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEK 408
Query: 1218 QHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAP 1277
HV L + F+ L K++++LNP KC+F V+S + L ++V+++GIE +P ++ A EMP+P
Sbjct: 409 DHVPQLRECFKILIKFEMKLNPEKCSFEVQSREFLEYLVTERGIEANPKQIAAFIEMPSP 468
Query: 1278 QTQKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDEC 1323
+ ++V+ GR+ ++ FIS C P ++ LRK + WN +C
Sbjct: 469 KMAREVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKEFDWNKDC 514
Score = 74.3 bits (181), Expect = 4e-13
Identities = 48/152 (31%), Positives = 75/152 (48%), Gaps = 6/152 (3%)
Query: 671 FSEDELPEAGKHHNLALHISVNCKSDMISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRS 730
F E E+ + K H+ AL I ++ + +S ++VDTGSS++V+ + + + L+
Sbjct: 49 FWESEITDLDKPHDDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSKLQGR 108
Query: 731 TFLVKAFDGTRKSVLGEIDLPITIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTST 790
+ F G +G I LP L F V+D A ++ +LGRPW+H AV ST
Sbjct: 109 KTPLTGFAGDTTFSIGTIQLPTIARGVRQLTNFLVVDKKAPFNAILGRPWLHVMKAVPST 168
Query: 791 LHQKLKFAKSGKLVTIHGEEAYLVSQLSSFSC 822
HQ +KF + ++G SQ SS C
Sbjct: 169 YHQCIKFPSYKGIAVVYG------SQRSSRKC 194
Score = 32.3 bits (72), Expect = 1.9
Identities = 17/50 (34%), Positives = 28/50 (56%)
Query: 986 IELINIGTEENKREIKIGAALEEGVKKKIIQLLREYPDIFAWSYEDMPGL 1035
I + I + KR + IG L+ V++ +I L+E D FAWS ++ G+
Sbjct: 237 ITQVCIDESDPKRCVGIGHDLDLTVREDLITFLKENKDSFAWSSANLQGI 286
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 219 bits (557), Expect = 1e-56
Identities = 118/328 (35%), Positives = 182/328 (54%), Gaps = 18/328 (5%)
Query: 1014 IIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQ 1073
+I L +Y +S +D+ G+ P + HRI + E + RR +P++ +K E+
Sbjct: 861 LITELMKYRKAIGYSLDDIKGISPTLCTHRIHLENESYSSIEPQRRLNPNLKEVVKKEIL 920
Query: 1074 KQIDAGFLMTVEYPEWVANIVPVPKKDGKV------------------RMCVDFRDLNKA 1115
K +DAG + + WV+ + VPKK G RMC+++R LN A
Sbjct: 921 KLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRTITGHRMCIEYRKLNVA 980
Query: 1116 SLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVM 1175
S K++FPLP ID +++ A + F+D +SG+ QI + P D+ KT+F P+GTF YK M
Sbjct: 981 SRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRM 1040
Query: 1176 PFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKL 1235
PFGL NA AT+QR MT++F D+I + VEV++DD V + + L ++ +R + L
Sbjct: 1041 PFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFSSCLLNLCRVLKRCEETNL 1100
Query: 1236 RLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISR 1295
LN KC F VR G +LG +S++GIEVD K+ + ++ P+T K +R FLG +
Sbjct: 1101 VLNWEKCHFMVREGIVLGRKISEEGIEVDKAKIDVMMQLQPPKTVKDIRSFLGHAGFYRI 1160
Query: 1296 FISHMTATCGPIFKLLRKNQPIVWNDEC 1323
FI + P+ +LL K ++DEC
Sbjct: 1161 FIKDFSKLARPLTRLLCKETEFAFDDEC 1188
Score = 38.5 bits (88), Expect = 0.026
Identities = 27/107 (25%), Positives = 52/107 (48%), Gaps = 6/107 (5%)
Query: 45 DRYNGLTC--PQNHIIKYVRKMGNYKDN----DSLMIHCFQDSLMEDAAEWYTSLSKNDI 98
++++GL P +H+ ++ R K N D + F SL + A +W SL + I
Sbjct: 52 NKFHGLPMEDPLDHLDEFDRLCSLTKINRVSEDGFKLRLFPFSLGDKAHQWEKSLPQGSI 111
Query: 99 HTFDELAVAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRG 145
++++ AF + + N+R R + +Q E+F E +R++G
Sbjct: 112 TSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNNETFYEAWERFKG 158
>At2g10780 pseudogene
Length = 1611
Score = 217 bits (552), Expect = 4e-56
Identities = 117/324 (36%), Positives = 188/324 (57%), Gaps = 10/324 (3%)
Query: 1001 KIGAALEEGVKKKIIQLLREYPDIFAWSYEDMPGL-DPMIVEHRIPTKPECPPVRQKLRR 1059
++GA+ E K I ++ E+ D+FA P DP +E +P P+ + R
Sbjct: 615 EVGASAE----LKDIPIVNEFSDVFAAVSGVPPDRSDPFTIE----LEPGTTPISKAPYR 666
Query: 1060 THPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASLKD 1119
P K+K ++++ +D GF+ P W A ++ V KKDG R+C+D+R LNK ++K+
Sbjct: 667 MAPAEMAKLKKQLEELLDKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKN 725
Query: 1120 NFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGL 1179
+PLP ID L+D ++ FS +D SGY+QI + P D KT+F T + F + VMPFGL
Sbjct: 726 KYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGL 785
Query: 1180 INAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNP 1239
NA A + + M +F D + + V ++++D++V S E H E+L + ERLR+++L
Sbjct: 786 TNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLREHELFAKL 845
Query: 1240 NKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFISH 1299
+KC+F RS LG ++S +G+ VDP+K+R+I+E P P+ ++R FLG Y RF+
Sbjct: 846 SKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMS 905
Query: 1300 MTATCGPIFKLLRKNQPIVWNDEC 1323
+ P+ +L K+ W+DEC
Sbjct: 906 FASMAQPLTRLTGKDTAFNWSDEC 929
Score = 31.6 bits (70), Expect = 3.2
Identities = 21/70 (30%), Positives = 30/70 (42%), Gaps = 9/70 (12%)
Query: 203 VFEKAESSVNAS--KRYGNGHHKKKETEIGMVSAGAGQPMATVAL-------INAAQMPP 253
V+E +E + +A + G K+ T+ GMV A GQ M L +N MP
Sbjct: 476 VYELSEEANDAGNFRAITGGFRKEPNTDYGMVRAAGGQAMYPTGLVRGISVVVNGVNMPA 535
Query: 254 SYPYVPYSQH 263
VP +H
Sbjct: 536 DLIIVPLKKH 545
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 213 bits (541), Expect = 8e-55
Identities = 116/324 (35%), Positives = 187/324 (56%), Gaps = 10/324 (3%)
Query: 1001 KIGAALEEGVKKKIIQLLREYPDIFAWSYEDMPGL-DPMIVEHRIPTKPECPPVRQKLRR 1059
++GA+ E K I ++ E+ D+FA P DP +E +P P+ + R
Sbjct: 110 EVGASAE----LKDILIVNEFSDVFAAVSGVPPDRSDPFTIE----LEPGTTPISKAPYR 161
Query: 1060 THPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASLKD 1119
P ++K ++++ + GF+ P W A ++ V KKDG R+C+D+R LNK ++K+
Sbjct: 162 MAPAEMAELKKQLEELLAKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKN 220
Query: 1120 NFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGL 1179
+PLP ID L+D ++ FS +D SGY+QI + P D KT+F T +G F + VMPFGL
Sbjct: 221 KYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYGHFEFVVMPFGL 280
Query: 1180 INAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNP 1239
NA A + + M +F D + + V +++DD++V S E H E+L + ERLR+++L
Sbjct: 281 TNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLERLREHELFAKL 340
Query: 1240 NKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFISH 1299
+K +F RS LG ++S +G+ VDP+K+R+I+E P P+ ++R FLG Y RF+
Sbjct: 341 SKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMS 400
Query: 1300 MTATCGPIFKLLRKNQPIVWNDEC 1323
+ P+ +L K+ W+DEC
Sbjct: 401 FASMAQPLTRLTGKDTAFNWSDEC 424
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 209 bits (533), Expect = 7e-54
Identities = 111/310 (35%), Positives = 181/310 (57%), Gaps = 6/310 (1%)
Query: 1015 IQLLREYPDIFAWSYEDMPGLDPMIVE-HRIPTKPECPPVRQKLRRTHPDMALKIKSEVQ 1073
I++++E+ D+F + + GL P + I +P P+ + R P ++K +++
Sbjct: 457 IRVVQEFQDVF----QSLQGLPPSQSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLK 512
Query: 1074 KQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLPHIDVLVDNT 1133
+ GF+ P W A ++ V KKDG R+C+D+R+LN+ ++K+ +PLP ID L+D
Sbjct: 513 DLLGKGFIRPSTSP-WGAPVLFVKKKDGSFRLCIDYRELNRVTVKNRYPLPRIDELLDQL 571
Query: 1134 AQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTL 1193
+ FS +D SGY+QI ++ D KT+F T +G F + VMPFGL NA A + R M ++
Sbjct: 572 RGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSV 631
Query: 1194 FHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLG 1253
F + + + V +++DD++V S E+ +L ++ E+LR+ KL +KC+F R LG
Sbjct: 632 FQEFLDEFVIIFIDDILVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLG 691
Query: 1254 FIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFISHMTATCGPIFKLLRK 1313
IVS +G+ VDP+K+ AIR+ P P ++R FLG Y RF+ + P+ KL K
Sbjct: 692 HIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTGK 751
Query: 1314 NQPIVWNDEC 1323
+ P VW+ EC
Sbjct: 752 DVPFVWSQEC 761
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 204 bits (520), Expect = 2e-52
Identities = 110/310 (35%), Positives = 178/310 (56%), Gaps = 6/310 (1%)
Query: 1015 IQLLREYPDIFAWSYEDMPGLDPMIVE-HRIPTKPECPPVRQKLRRTHPDMALKIKSEVQ 1073
I++++E+ D+F + + GL P + I +P P+ + R P ++K +++
Sbjct: 483 IRVIQEFEDVF----QSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMTELKKQLE 538
Query: 1074 KQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLPHIDVLVDNT 1133
+ GF+ P W A ++ V KKDG R+C+D+R LN ++K+ +PLP ID L+D
Sbjct: 539 DLLGKGFIRPSTSP-WGAPVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQL 597
Query: 1134 AQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTL 1193
+ FS +D SGY+QI ++ D KT+F T +G F + VMPF L NA A + R M ++
Sbjct: 598 RGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNSV 657
Query: 1194 FHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLG 1253
F + + + V +++DD++V S E+H +L ++ E+LR+ KL +KC+F R LG
Sbjct: 658 FQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREIGFLG 717
Query: 1254 FIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFISHMTATCGPIFKLLRK 1313
IVS +G+ VDP+K+ AIR+ P P ++R FL Y RF+ + P+ KL K
Sbjct: 718 HIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLRLTGYYRRFVKGFASMAQPMTKLTGK 777
Query: 1314 NQPIVWNDEC 1323
+ P VW+ EC
Sbjct: 778 DVPFVWSPEC 787
>At3g11970 hypothetical protein
Length = 1499
Score = 202 bits (515), Expect = 8e-52
Identities = 117/308 (37%), Positives = 176/308 (56%), Gaps = 7/308 (2%)
Query: 1007 EEGVKKKIIQLLREYPDIFAWSYEDMPGLDPMIVEH--RIPTKPECPPVRQKLRRTHPDM 1064
E G + + ++L EYPDIF + L P +H +I PV Q+ R
Sbjct: 558 ELGEESVVEEVLNEYPDIFI----EPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQ 613
Query: 1065 ALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLP 1124
+I V+ + G + P + + +V V KKDG R+CVD+R+LN ++KD+FP+P
Sbjct: 614 KNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIP 672
Query: 1125 HIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGA 1184
I+ L+D + +FS +D +GY+Q++M P+D +KT+F T G F Y VMPFGL NA A
Sbjct: 673 LIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPA 732
Query: 1185 TYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTF 1244
T+Q M +F + K V V+ DD++V S+ E+H ++L ++FE +R KL +KC F
Sbjct: 733 TFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAF 792
Query: 1245 GVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFISHMTATC 1304
V + LG +S +GIE DP K++A++E P P T KQ+RGFLG Y RF+
Sbjct: 793 AVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIA 852
Query: 1305 GPIFKLLR 1312
GP+ L +
Sbjct: 853 GPLHALTK 860
>At1g36590 hypothetical protein
Length = 1499
Score = 202 bits (515), Expect = 8e-52
Identities = 117/308 (37%), Positives = 176/308 (56%), Gaps = 7/308 (2%)
Query: 1007 EEGVKKKIIQLLREYPDIFAWSYEDMPGLDPMIVEH--RIPTKPECPPVRQKLRRTHPDM 1064
E G + + ++L EYPDIF + L P +H +I PV Q+ R
Sbjct: 558 ELGEESVVEEVLNEYPDIFI----EPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQ 613
Query: 1065 ALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLP 1124
+I V+ + G + P + + +V V KKDG R+CVD+R+LN ++KD+FP+P
Sbjct: 614 KNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIP 672
Query: 1125 HIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGA 1184
I+ L+D + +FS +D +GY+Q++M P+D +KT+F T G F Y VMPFGL NA A
Sbjct: 673 LIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPA 732
Query: 1185 TYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTF 1244
T+Q M +F + K V V+ DD++V S+ E+H ++L ++FE +R KL +KC F
Sbjct: 733 TFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAF 792
Query: 1245 GVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFISHMTATC 1304
V + LG +S +GIE DP K++A++E P P T KQ+RGFLG Y RF+
Sbjct: 793 AVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIA 852
Query: 1305 GPIFKLLR 1312
GP+ L +
Sbjct: 853 GPLHALTK 860
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 193 bits (491), Expect = 5e-49
Identities = 113/340 (33%), Positives = 180/340 (52%), Gaps = 18/340 (5%)
Query: 985 EIELINIGTEENKREIKIGAALEEGVKKKIIQLLREYPDIFAWSYEDMPGLDPMIVE-HR 1043
E L+ G + I A E + +K + E+ D+F + + GL P +
Sbjct: 414 EGSLVYQGVRPTSGSLVISAVQAEKMIEKGCEAYLEFEDVF----QSLQGLPPSRSDPFT 469
Query: 1044 IPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKV 1103
I +P P+ + R P ++K +++ + A L V KKDG
Sbjct: 470 IELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGAPVLF-------------VKKKDGSF 516
Query: 1104 RMCVDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSF 1163
R+C+D+R LN ++K+ +PLP ID L+D + FS +D SGY+ I ++ D KT+F
Sbjct: 517 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAF 576
Query: 1164 ITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYL 1223
T +G F + VMPFGL NA A + R M ++F +++ + V +++DD++V S E+H +L
Sbjct: 577 RTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHEVHL 636
Query: 1224 TKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQV 1283
++ E+LR+ KL +KC+F R LG IVS +G+ VDP+K+ AIR+ P ++
Sbjct: 637 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTPTNATEI 696
Query: 1284 RGFLGRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDEC 1323
R FLG Y RF+ + P+ KL K+ P VW+ EC
Sbjct: 697 RSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPEC 736
>At4g07660 putative athila transposon protein
Length = 724
Score = 193 bits (490), Expect = 6e-49
Identities = 165/619 (26%), Positives = 272/619 (43%), Gaps = 92/619 (14%)
Query: 702 LVDTGSSLNVMPKSTLDQLSYRGTPLRRSTFLVKAFDGTRKSVLGEIDLPITIGPETFLI 761
L D G+ +++MP S +L + + ++ L E +LPI IG
Sbjct: 35 LCDLGALVSLMPLSVAKRLGFTQYKSCNISLILADRSVRIPHSLFE-NLPIRIGAVDIPT 93
Query: 762 TFQV--MDINASYSCLLGRPWIHDAGAVTSTLHQKL-----KFAK-------SGKLVTIH 807
F V MD +LGRP++ AGA+ K+ K+ + + K TI
Sbjct: 94 DFVVLEMDEEPKDPLILGRPFLATAGAMNDVKKGKIDLNLGKYCRMTFDVKDAMKKPTIK 153
Query: 808 GEEAYL--VSQLSSFSCIEAGSAEGTAFQGLTVEGTEP--KRDGTAMASLKDAQRAVQEG 863
G+ ++ + QL+ +E + E + LT G + + L D+ +A++E
Sbjct: 154 GQLFWIEEIDQLAD-ELLEERAEEDHLYSALTKRGEDGFLHLETLGYQKLLDSHKAMEES 212
Query: 864 QAAGWGRLIQLRENKHKEGLGFSPTSGVSTGAFYSAGF-VNAITEEATGFGPRPVFVIPG 922
+ + E + S +S + N T + G +IP
Sbjct: 213 EP------FEELNGPETEVMVMSEEGSTQVQPAHSRTYSTNNSTSTISNSGE---LIIP- 262
Query: 923 GIARDWDAIDIPSIMHVSDNPAFPPNFEFPVYEAEDEEGDDIPYEITRLLEQEKKAIQPH 982
+ DW + P + D + P + + G + Y +
Sbjct: 263 -TSDDWSELKAPKV----DLKSLPKGLRYVFF------GPNSTYPV-------------- 297
Query: 983 *EEIELINIGTEENKREIKIGAALEEGVKKKIIQLLREYPDIFAWSYEDMPGLDPMIVEH 1042
+IN+ E N E+ + ++ LR+Y +S D+ + P + H
Sbjct: 298 -----IINV--ELNDNEVNL-----------LLSELRKYKRAIGYSLSDIKRISPSLCNH 339
Query: 1043 RIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGK 1102
RI + E + RR + ++ +K E+ K +DAG + + WV + VPKK G
Sbjct: 340 RIHLENESYSSIEPQRRLNLNLKEVVKKEILKLLDAGVIYPISDSTWVFPVHCVPKKGGM 399
Query: 1103 V------------------RMCVDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDG 1144
R+C+D+R LN AS KD+FPLP + +++ A F+DG
Sbjct: 400 TVVKNEKDELIPTRTITGHRVCIDYRKLNAASRKDHFPLPFTNQMLEGLANHLYNCFLDG 459
Query: 1145 FSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEV 1204
+SG+ QI + P D+EKT+F P+GTF YK MPFGL NA T+QR MT++F D+I K VEV
Sbjct: 460 YSGFFQIPIHPNDQEKTTFTCPYGTFAYKRMPFGLCNAPTTFQRCMTSIFSDLIEKMVEV 519
Query: 1205 YVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVD 1264
++DD V + L ++ + + L LN KC F V+ G +LG +S+KGIEVD
Sbjct: 520 FMDDFSVYGPSFSSCLLNLGRVLTKWEETNLVLNWEKCYFMVKEGIVLGHKISEKGIEVD 579
Query: 1265 PDKVRAIREMPAPQTQKQV 1283
+K++ + ++ P+T K +
Sbjct: 580 KEKIKVMMQLQPPKTVKDI 598
>At1g35370 hypothetical protein
Length = 1447
Score = 192 bits (488), Expect = 1e-48
Identities = 120/331 (36%), Positives = 184/331 (55%), Gaps = 13/331 (3%)
Query: 993 TEENKREIKIGAALEEGVKKKIIQ-LLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPECP 1051
++E + I A + V++ ++Q ++ E+PD+FA + P + +H+I
Sbjct: 522 SDEEQEIGSISALTSDVVEESVVQNIVEEFPDVFAEPTDLPPFREKH--DHKIKLLEGAN 579
Query: 1052 PVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRD 1111
PV Q+ R +I VQ I +G + P + + +V V KKDG R+CVD+ +
Sbjct: 580 PVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSP-FASPVVLVKKKDGTWRLCVDYTE 638
Query: 1112 LNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFC 1171
LN ++KD F +P I+ L+D S VFS +D +GY+Q++M P+D +KT+F T G F
Sbjct: 639 LNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFE 698
Query: 1172 YKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLR 1231
Y VM FGL NA AT+Q M ++F D + K V V+ DD+++ S+ E+H E+L +FE +R
Sbjct: 699 YLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMR 758
Query: 1232 KYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLN 1291
+KL F S + LG +S + IE DP K++A++E P P T KQVRGFLG
Sbjct: 759 LHKL--------FAKGSKEHLGHFISAREIETDPAKIQAVKEWPTPTTVKQVRGFLGFAG 810
Query: 1292 YISRFISHMTATCGPIFKLLRKNQPIVWNDE 1322
Y RF+ + GP+ L K W+ E
Sbjct: 811 YYRRFVRNFGVIAGPL-HALTKTDGFCWSLE 840
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 191 bits (484), Expect = 3e-48
Identities = 111/324 (34%), Positives = 166/324 (50%), Gaps = 46/324 (14%)
Query: 1018 LREYPDIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQID 1077
LR+Y +S +D+ G+ P + H I + E + RR + ++ +K E+ K +D
Sbjct: 340 LRKYRKALGYSLDDITGISPTLCMHMIHLEGESITSVEHQRRLNSNLRDVVKKEIMKLLD 399
Query: 1078 AGFLMTVEYPEWVANIVPVPKKDGKV------------------RMCVDFRDLNKASLKD 1119
AG + + WV+ + VPKK G + RMC+D+R LN A+ KD
Sbjct: 400 AGIIYPISDSTWVSPVHVVPKKGGVIVIKNEKNELIPTRTVTGHRMCIDYRKLNSATRKD 459
Query: 1120 NFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGL 1179
NFPL ID +++ + + F+DG+ G+ QI + P+D+EKT+F P+GTF Y+ MPFGL
Sbjct: 460 NFPLSFIDQMLERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTFTCPYGTFAYRRMPFGL 519
Query: 1180 INAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNP 1239
NA AT+Q M +F DMI +EV++DD +V E++H L LN
Sbjct: 520 CNAPATFQHCMKYIFSDMIEDFMEVFIDDFLVLQRCEDKH---------------LVLNW 564
Query: 1240 NKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFISH 1299
K F VR G +LG +S+KG+EVD K+ +R FLG + RFI
Sbjct: 565 EKSHFMVRDGIVLGHKISEKGVEVDRAKIEIMR-------------FLGHAGFYRRFIKD 611
Query: 1300 MTATCGPIFKLLRKNQPIVWNDEC 1323
+ P +LL K Q ++D C
Sbjct: 612 FSKIARPCTQLLCKEQKSEFDDTC 635
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 185 bits (470), Expect = 1e-46
Identities = 107/297 (36%), Positives = 165/297 (55%), Gaps = 3/297 (1%)
Query: 1009 GVKKKIIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKI 1068
G + + +++ E+PDIF + P P +H+I + PV Q+ R +I
Sbjct: 2 GEESFVEKVVTEFPDIFVEPTDLPPFRAPH--DHKIELLEDSNPVNQRPYRYVVHQKNEI 59
Query: 1069 KSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLPHIDV 1128
V + +G + P + + +V V KKDG R+CVD+R+LN ++KD FP+P I+
Sbjct: 60 DKIVDDMLASGTIQASSSP-YASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIED 118
Query: 1129 LVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQR 1188
L+D S V+S +D +GY+Q++M P D KT+F T G + Y VMPFGL NA A++Q
Sbjct: 119 LMDELGGSNVYSKIDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQS 178
Query: 1189 GMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRS 1248
M + F + K V V+ DD+++ ST E+H ++L +FE +R + L +KC F V
Sbjct: 179 LMNSFFKPFLRKFVLVFFDDILIYSTSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPR 238
Query: 1249 GKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFISHMTATCG 1305
+ LG +S +GI DP K++A+++ P P KQ+ GFLG Y RF CG
Sbjct: 239 VEYLGHFISGEGIATDPAKIKAVQDWPVPVNLKQLCGFLGLTGYYRRFFVVEMDACG 295
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 172 bits (435), Expect = 2e-42
Identities = 84/151 (55%), Positives = 106/151 (69%)
Query: 1032 MPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVA 1091
M G+ ++ H + P PV+QK R+ PD A + EV + ++ G + V+YPEW+A
Sbjct: 412 MTGISTEVISHELNVDPTFKPVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREVKYPEWLA 471
Query: 1092 NIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQI 1151
N V V KK+GK R+CVDF DLNKA KD FPLPHID LV++T ++ SFMD FSGYNQI
Sbjct: 472 NPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPHIDRLVESTTGHEMLSFMDAFSGYNQI 531
Query: 1152 KMSPEDREKTSFITPWGTFCYKVMPFGLINA 1182
M+PED+EKTSFIT GT+ YKVMPFGL NA
Sbjct: 532 LMNPEDQEKTSFITECGTYYYKVMPFGLKNA 562
Score = 48.5 bits (114), Expect = 3e-05
Identities = 34/110 (30%), Positives = 53/110 (47%), Gaps = 12/110 (10%)
Query: 707 SSLNVMPKSTLDQLSYRGTPLRR------STFLVKAFDGTRKSVLGEIDLPITIGPETFL 760
S+ N PK+ + + T LR+ +T L + LG + LP+T +
Sbjct: 314 SASNAAPKAPQPATTKKPTELRKPAAGTCTTMLCET-----SMSLGTVVLPVTAQGVVKM 368
Query: 761 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKF-AKSGKLVTIHGE 809
+ F V D A+Y+ +LG PW+++ V ST HQ +KF GK+ I E
Sbjct: 369 VEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFPTPVGKMTGISTE 418
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,117,203
Number of Sequences: 26719
Number of extensions: 1455068
Number of successful extensions: 4847
Number of sequences better than 10.0: 151
Number of HSP's better than 10.0 without gapping: 74
Number of HSP's successfully gapped in prelim test: 77
Number of HSP's that attempted gapping in prelim test: 4426
Number of HSP's gapped (non-prelim): 356
length of query: 1323
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1212
effective length of database: 8,352,787
effective search space: 10123577844
effective search space used: 10123577844
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146794.16


