

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146793.2 - phase: 0
(438 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
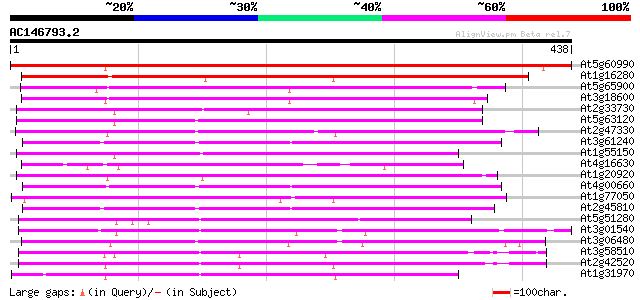
Score E
Sequences producing significant alignments: (bits) Value
At5g60990 replication protein A1 - like 647 0.0
At1g16280 putative ATP-dependent RNA helicase 341 4e-94
At5g65900 ATP-dependent RNA helicase-like 252 3e-67
At3g18600 DEAD box helicase protein, putative 250 1e-66
At2g33730 putative U5 small nuclear ribonucleoprotein, an RNA he... 232 3e-61
At5g63120 ATP-dependent RNA helicase-like protein 231 6e-61
At2g47330 putative ATP-dependent RNA helicase 226 2e-59
At3g61240 DEAD box RNA helicase RH12 225 4e-59
At1g55150 unknown protein 223 1e-58
At4g16630 RNA helicase like protein 221 6e-58
At1g20920 putative RNA helicase 220 1e-57
At4g00660 RNA helicase like protein 219 2e-57
At1g77050 217 9e-57
At2g45810 putative ATP-dependent RNA helicase 216 2e-56
At5g51280 DEAD-box protein abstrakt 212 4e-55
At3g01540 AT3g01540/F4P13_9 211 5e-55
At3g06480 putative RNA helicase 211 6e-55
At3g58510 ATP-dependent RNA helicase-like protein 211 8e-55
At2g42520 putative ATP-dependent RNA helicase 209 2e-54
At1g31970 unknown protein 209 2e-54
>At5g60990 replication protein A1 - like
Length = 456
Score = 647 bits (1670), Expect = 0.0
Identities = 332/456 (72%), Positives = 386/456 (83%), Gaps = 18/456 (3%)
Query: 1 MEEENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKT 60
MEEEN+ +K+F +LG+ E LV+ACE++GWK P +IQ EA+P ALEGKD+IGLA+TGSGKT
Sbjct: 1 MEEENEVVKTFAELGVREELVKACERLGWKNPSKIQAEALPFALEGKDVIGLAQTGSGKT 60
Query: 61 GAFALPILHALLE---------APRPNH-FFACVMSPTRELAIQISEQFEALGSEIGVKC 110
GAFA+PIL ALLE RP+ FFACV+SPTRELAIQI+EQFEALG++I ++C
Sbjct: 61 GAFAIPILQALLEYVYDSEPKKGRRPDPAFFACVLSPTRELAIQIAEQFEALGADISLRC 120
Query: 111 AVLVGGIDMVQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNED 170
AVLVGGID +QQ++ + K PH+IV TPGR+ DH+ +TKGFSL LKYLVLDEADRLLNED
Sbjct: 121 AVLVGGIDRMQQTIALGKRPHVIVATPGRLWDHMSDTKGFSLKSLKYLVLDEADRLLNED 180
Query: 171 FEESLNEILGMIPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYR 230
FE+SLN+IL IP ER+TFLFSATMT KV KLQR CLRNPVKIE +SKYSTVDTLKQQYR
Sbjct: 181 FEKSLNQILEEIPLERKTFLFSATMTKKVRKLQRACLRNPVKIEAASKYSTVDTLKQQYR 240
Query: 231 FLPAKHKDCYLVYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRL 290
F+ AK+KDCYLVYILSEM STSM+FTRTCD TR LAL+LR+LG +AIPI+G M+Q KRL
Sbjct: 241 FVAAKYKDCYLVYILSEMPESTSMIFTRTCDGTRFLALVLRSLGFRAIPISGQMTQSKRL 300
Query: 291 GALNKFKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVA 350
GALNKFK+G+CNIL+CTDVASRGLDIP+VD+VINYDIPTNSKDYIHRVGRTARAGRSGV
Sbjct: 301 GALNKFKAGECNILVCTDVASRGLDIPSVDVVINYDIPTNSKDYIHRVGRTARAGRSGVG 360
Query: 351 ISLVNQYELEWYVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRR 410
ISLVNQYELEWY+QIEKLIGKKLPEYPA E+EVL L ERV EAK+L+A MKESGG+KRR
Sbjct: 361 ISLVNQYELEWYIQIEKLIGKKLPEYPAEEDEVLSLLERVAEAKKLSAMNMKESGGRKRR 420
Query: 411 GEGDI-------GEEDDVDK-YFGLKDRKSSKKFRR 438
GE D G +D +K G KD+KSSKKF+R
Sbjct: 421 GEDDEESERFLGGNKDRGNKERGGNKDKKSSKKFKR 456
>At1g16280 putative ATP-dependent RNA helicase
Length = 491
Score = 341 bits (875), Expect = 4e-94
Identities = 185/401 (46%), Positives = 265/401 (65%), Gaps = 7/401 (1%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
+F+ LGL E VE C+++G + P +Q +P L G+D++GLA+TGSGKT AFALPILH
Sbjct: 59 NFEGLGLAEWAVETCKELGMRKPTPVQTHCVPKILAGRDVLGLAQTGSGKTAAFALPILH 118
Query: 70 ALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIAKL 129
L E P FA V++PTRELA Q++EQF+ALGS + ++C+V+VGG+DM+ Q++ +
Sbjct: 119 RLAEDPYG--VFALVVTPTRELAFQLAEQFKALGSCLNLRCSVIVGGMDMLTQTMSLVSR 176
Query: 130 PHIIVGTPGRVLDHLKNTKGFS--LARLKYLVLDEADRLLNEDFEESLNEILGMIPRERR 187
PHI++ TPGR+ L+N +R K+LVLDEADR+L+ F++ L I +P+ R+
Sbjct: 177 PHIVITTPGRIKVLLENNPDVPPVFSRTKFLVLDEADRVLDVGFQDELRTIFQCLPKSRQ 236
Query: 188 TFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYILSE 247
T LFSATMT+ ++ L E TVDTL QQ+ F K+ YLV+ILS+
Sbjct: 237 TLLFSATMTSNLQALLEHSSNKAYFYEAYEGLKTVDTLTQQFIFEDKDAKELYLVHILSQ 296
Query: 248 MAGS---TSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNIL 304
M ++M+F TC + + L+L+L L ++ I ++ SQ RL AL+KFKSG IL
Sbjct: 297 MEDKGIRSAMIFVSTCRTCQRLSLMLDELEVENIAMHSLNSQSMRLSALSKFKSGKVPIL 356
Query: 305 LCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQ 364
L TDVASRGLDIP VD+VINYDIP + +DY+HRVGRTARAGR G+A+S++ + +++ +
Sbjct: 357 LATDVASRGLDIPTVDLVINYDIPRDPRDYVHRVGRTARAGRGGLAVSIITETDVKLIHK 416
Query: 365 IEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESG 405
IE+ +GKK+ Y L +V +AKR+A KM ++G
Sbjct: 417 IEEEVGKKMEPYNKKVITDSLEVTKVSKAKRVAMMKMLDNG 457
>At5g65900 ATP-dependent RNA helicase-like
Length = 633
Score = 252 bits (643), Expect = 3e-67
Identities = 155/386 (40%), Positives = 227/386 (58%), Gaps = 11/386 (2%)
Query: 9 KSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALP-- 66
K+F+ L L ++ ++ +++G+ +IQ +AIPP + G+D++G A+TGSGKT AF +P
Sbjct: 154 KTFESLSLSDNTYKSIKEMGFARMTQIQAKAIPPLMMGEDVLGAARTGSGKTLAFLIPAV 213
Query: 67 -ILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVK 125
+L+ + PR N V+ PTRELAIQ + L ++GG ++
Sbjct: 214 ELLYRVKFTPR-NGTGVLVICPTRELAIQSYGVAKELLKYHSQTVGKVIGGEKRKTEAEI 272
Query: 126 IAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRE 185
+AK +++V TPGR+LDHL+NT GF LK+LV+DEADR+L ++FEE L +IL ++P+
Sbjct: 273 LAKGVNLLVATPGRLLDHLENTNGFIFKNLKFLVMDEADRILEQNFEEDLKKILNLLPKT 332
Query: 186 RRTFLFSATMTNKVEKLQRVCLRNPVKIETSS--KYSTVDTLKQQYRFLPAKHKDCYLVY 243
R+T LFSAT + KVE L RV L +PV I+ K T + L+Q Y +P+ + +L+
Sbjct: 333 RQTSLFSATQSAKVEDLARVSLTSPVYIDVDEGRKEVTNEGLEQGYCVVPSAMRLLFLLT 392
Query: 244 ILSEMAGSTS-MVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCN 302
L G MVF TC ST+ A + R + + I G + Q KR +F +
Sbjct: 393 FLKRFQGKKKIMVFFSTCKSTKFHAELFRYIKFDCLEIRGGIDQNKRTPTFLQFIKAETG 452
Query: 303 ILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARA-GRSGVAISLVNQYELEW 361
ILLCT+VA+RGLD P VD ++ YD P N DYIHRVGRTAR G G A+ ++ EL++
Sbjct: 453 ILLCTNVAARGLDFPHVDWIVQYDPPDNPTDYIHRVGRTARGEGAKGKALLVLTPQELKF 512
Query: 362 YVQIEKLIGKKLPEYPANEEEVLLLE 387
I+ L K+P EE LL+
Sbjct: 513 ---IQYLKAAKIPVEEHEFEEKKLLD 535
Score = 30.4 bits (67), Expect = 2.0
Identities = 25/73 (34%), Positives = 38/73 (51%), Gaps = 9/73 (12%)
Query: 371 KKLPEYPANEEEVLLLEERVGEAKRLAAT-----KMKESGGKKRRGEGDIGEEDDVDKYF 425
KKL + PA EEE + G+AK A K K+ K+RG+ D GE++ V +
Sbjct: 30 KKLKQ-PAMEEEP---DHEDGDAKENNALIDEEPKKKKKKKNKKRGDTDDGEDEAVAEEE 85
Query: 426 GLKDRKSSKKFRR 438
K +K +KK ++
Sbjct: 86 PKKKKKKNKKLQQ 98
>At3g18600 DEAD box helicase protein, putative
Length = 568
Score = 250 bits (638), Expect = 1e-66
Identities = 151/372 (40%), Positives = 222/372 (59%), Gaps = 9/372 (2%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
+F L L E A +++G++ +IQ +I P LEGKD++G A+TGSGKT AF +P +
Sbjct: 90 TFDSLDLSEQTSIAIKEMGFQYMTQIQAGSIQPLLEGKDVLGAARTGSGKTLAFLIPAVE 149
Query: 70 ALLE---APRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKI 126
L + +PR N V+ PTRELAIQ E L ++++GG + ++ +I
Sbjct: 150 LLFKERFSPR-NGTGVIVICPTRELAIQTKNVAEELLKHHSQTVSMVIGGNNRRSEAQRI 208
Query: 127 AKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRER 186
A ++++ TPGR+LDHL+NTK F LK LV+DEADR+L E+FEE +N+IL ++P+ R
Sbjct: 209 ASGSNLVIATPGRLLDHLQNTKAFIYKHLKCLVIDEADRILEENFEEDMNKILKILPKTR 268
Query: 187 RTFLFSATMTNKVEKLQRVCLRNPVKIETSS--KYSTVDTLKQQYRFLPAKHKDCYLVYI 244
+T LFSAT T+KV+ L RV L +PV ++ + T + L+Q Y +P+K + L+
Sbjct: 269 QTALFSATQTSKVKDLARVSLTSPVHVDVDDGRRKVTNEGLEQGYCVVPSKQRLILLISF 328
Query: 245 LSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNIL 304
L + MVF TC S + I++ + I+G M Q +R F IL
Sbjct: 329 LKKNLNKKIMVFFSTCKSVQFHTEIMKISDVDVSDIHGGMDQNRRTKTFFDFMKAKKGIL 388
Query: 305 LCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARA-GRSGVAISLVNQYELEW-- 361
LCTDVA+RGLDIP+VD +I YD P +YIHRVGRTAR G G A+ ++ EL++
Sbjct: 389 LCTDVAARGLDIPSVDWIIQYDPPDKPTEYIHRVGRTARGEGAKGKALLVLIPEELQFIR 448
Query: 362 YVQIEKLIGKKL 373
Y++ K+ K+L
Sbjct: 449 YLKAAKVPVKEL 460
>At2g33730 putative U5 small nuclear ribonucleoprotein, an RNA
helicase
Length = 733
Score = 232 bits (591), Expect = 3e-61
Identities = 138/387 (35%), Positives = 216/387 (55%), Gaps = 24/387 (6%)
Query: 6 KEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFAL 65
+ M+S+++ L L++A E+ G+K P IQ+ AIP L+ +D+IG+A+TGSGKT AF L
Sbjct: 310 RPMRSWEESKLTSELLKAVERAGYKKPSPIQMAAIPLGLQQRDVIGIAETGSGKTAAFVL 369
Query: 66 PILHALLEAPRPNHF------FACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDM 119
P+L + P + +A VM+PTRELA QI E+ +G + +VGG +
Sbjct: 370 PMLAYISRLPPMSEENETEGPYAVVMAPTRELAQQIEEETVKFAHYLGFRVTSIVGGQSI 429
Query: 120 VQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEIL 179
+Q +KI + I++ TPGR++D L+ L + Y+VLDEADR+++ FE + +L
Sbjct: 430 EEQGLKITQGCEIVIATPGRLIDCLERRYAV-LNQCNYVVLDEADRMIDMGFEPQVAGVL 488
Query: 180 GMIPRE-----------------RRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTV 222
+P R T++FSATM VE+L R LRNPV + + T
Sbjct: 489 DAMPSSNLKPENEEEELDEKKIYRTTYMFSATMPPGVERLARKYLRNPVVVTIGTAGKTT 548
Query: 223 DTLKQQYRFLPAKHKDCYLVYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPING 282
D + Q + K L +L E+ T++VF T + +A L G + ++G
Sbjct: 549 DLISQHVIMMKESEKFFRLQKLLDELGEKTAIVFVNTKKNCDSIAKNLDKAGYRVTTLHG 608
Query: 283 HMSQPKRLGALNKFKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTA 342
SQ +R +L F++ N+L+ TDV RG+DIP V VINYD+P + + Y HR+GRT
Sbjct: 609 GKSQEQREISLEGFRAKRYNVLVATDVVGRGIDIPDVAHVINYDMPKHIEMYTHRIGRTG 668
Query: 343 RAGRSGVAISLVNQYELEWYVQIEKLI 369
RAG+SGVA S + ++ E + +++++
Sbjct: 669 RAGKSGVATSFLTLHDTEVFYDLKQML 695
>At5g63120 ATP-dependent RNA helicase-like protein
Length = 591
Score = 231 bits (589), Expect = 6e-61
Identities = 139/369 (37%), Positives = 210/369 (56%), Gaps = 6/369 (1%)
Query: 6 KEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFAL 65
K MK F+D P++++EA K+G+ P IQ + P AL+G+DLIG+A+TGSGKT A+ L
Sbjct: 162 KPMKMFQDANFPDNILEAIAKLGFTEPTPIQAQGWPMALKGRDLIGIAETGSGKTLAYLL 221
Query: 66 PILHALLEAPRPNHF---FACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQ 122
P L + PR +++PTRELA+QI E+ G GV+ + GG Q
Sbjct: 222 PALVHVSAQPRLGQDDGPIVLILAPTRELAVQIQEESRKFGLRSGVRSTCIYGGAPKGPQ 281
Query: 123 SVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMI 182
+ + I++ TPGR++D L+ + +L R+ YLVLDEADR+L+ FE + +I+ I
Sbjct: 282 IRDLRRGVEIVIATPGRLIDMLE-CQHTNLKRVTYLVLDEADRMLDMGFEPQIRKIVSQI 340
Query: 183 PRERRTFLFSATMTNKVEKLQRVCLRNPVK-IETSSKYSTVDTLKQQYRFLPAKHKDCYL 241
+R+T L+SAT +VE L R LR+P K I S+ ++ Q +P K L
Sbjct: 341 RPDRQTLLWSATWPREVETLARQFLRDPYKAIIGSTDLKANQSINQVIEIVPTPEKYNRL 400
Query: 242 VYILSE-MAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGD 300
+ +L + M GS ++F T + LR G A+ I+G +Q +R L +FKSG
Sbjct: 401 LTLLKQLMDGSKILIFVETKRGCDQVTRQLRMDGWPALAIHGDKTQSERDRVLAEFKSGR 460
Query: 301 CNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELE 360
I+ TDVA+RGLD+ + V+NYD P +DYIHR+GRT RAG G+A + +
Sbjct: 461 SPIMTATDVAARGLDVKDIKCVVNYDFPNTLEDYIHRIGRTGRAGAKGMAFTFFTHDNAK 520
Query: 361 WYVQIEKLI 369
+ ++ K++
Sbjct: 521 FARELVKIL 529
>At2g47330 putative ATP-dependent RNA helicase
Length = 760
Score = 226 bits (576), Expect = 2e-59
Identities = 141/417 (33%), Positives = 225/417 (53%), Gaps = 17/417 (4%)
Query: 5 NKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFA 64
++ +K+F+D G ++ A +K ++ P IQ +A+P L G+D+IG+AKTGSGKT AF
Sbjct: 224 HRPVKTFEDCGFSSQIMSAIKKQAYEKPTAIQCQALPIVLSGRDVIGIAKTGSGKTAAFV 283
Query: 65 LPILHALLEAP---RPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQ 121
LP++ +++ P R + +PTRELA QI + + G++ + + GG+ +
Sbjct: 284 LPMIVHIMDQPELQRDEGPIGVICAPTRELAHQIFLEAKKFSKAYGLRVSAVYGGMSKHE 343
Query: 122 QSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGM 181
Q ++ I+V TPGR++D LK K ++ R YLVLDEADR+ + FE + I+G
Sbjct: 344 QFKELKAGCEIVVATPGRLIDMLK-MKALTMMRASYLVLDEADRMFDLGFEPQVRSIVGQ 402
Query: 182 IPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYL 241
I +R+T LFSATM KVEKL R L +P+++ + + Q +P+ + L
Sbjct: 403 IRPDRQTLLFSATMPWKVEKLAREILSDPIRVTVGEVGMANEDITQVVNVIPSDAEK--L 460
Query: 242 VYILSEMAGSTS----MVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFK 297
++L ++ G +VF + + L K ++G Q R+ L KFK
Sbjct: 461 PWLLEKLPGMIDEGDVLVFASKKATVDEIEAQLTLNSFKVAALHGDKDQASRMETLQKFK 520
Query: 298 SGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAG-RSGVAISLVNQ 356
SG ++L+ TDVA+RGLDI ++ V+NYDI + ++HR+GRT RAG R GVA +LV Q
Sbjct: 521 SGVHHVLIATDVAARGLDIKSLKTVVNYDIAKDMDMHVHRIGRTGRAGDRDGVAYTLVTQ 580
Query: 357 YELEWYVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEG 413
E + ++ + P ++ + + R + + GGKK RG G
Sbjct: 581 REARFAGELVNSLVAAGQNVPPELTDLAMKD------GRFKSKRDGRKGGKKGRGGG 631
>At3g61240 DEAD box RNA helicase RH12
Length = 498
Score = 225 bits (573), Expect = 4e-59
Identities = 130/375 (34%), Positives = 215/375 (56%), Gaps = 6/375 (1%)
Query: 11 FKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILHA 70
F+D L L++ + G++ P IQ E+IP AL G D++ AK G+GKTGAF +P+L
Sbjct: 126 FEDYFLKRDLLKGIYEKGFEKPSPIQEESIPIALTGSDILARAKNGTGKTGAFCIPVLEK 185
Query: 71 LLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIAKLP 130
+ P N A ++ PTRELA+Q S+ + L + ++ V GG + +++ +
Sbjct: 186 I--DPNNNVIQAMILVPTRELALQTSQVCKELSKYLNIQVMVTTGGTSLRDDIMRLHQPV 243
Query: 131 HIIVGTPGRVLDHLKNTKGFSLAR-LKYLVLDEADRLLNEDFEESLNEILGMIPRERRTF 189
H++VGTPGR+LD K KG + + LV+DEAD+LL+ +F+ SL E++ +P+ R+
Sbjct: 244 HLLVGTPGRILDLTK--KGVCVLKDCAMLVMDEADKLLSAEFQPSLEELIQFLPQNRQFL 301
Query: 190 LFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYILSEMA 249
+FSAT V+ + LR P I + T+ + Q Y F+ + K L + S++
Sbjct: 302 MFSATFPVTVKAFKDRHLRKPYVINLMDQL-TLMGVTQYYAFVEERQKVHCLNTLFSKLQ 360
Query: 250 GSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILLCTDV 309
+ S++F + + LLA + LG I+ M Q R ++F++G C L+CTD+
Sbjct: 361 INQSIIFCNSVNRVELLAKKITELGYSCFYIHAKMVQDHRNRVFHEFRNGACRNLVCTDL 420
Query: 310 ASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQIEKLI 369
+RG+DI AV++VIN+D P S+ Y+HRVGR+ R G G+A++LV + Q E+ +
Sbjct: 421 FTRGIDIQAVNVVINFDFPRTSESYLHRVGRSGRFGHLGLAVNLVTYEDRFKMYQTEQEL 480
Query: 370 GKKLPEYPANEEEVL 384
G ++ P+N ++ +
Sbjct: 481 GTEIKPIPSNIDQAI 495
>At1g55150 unknown protein
Length = 501
Score = 223 bits (569), Expect = 1e-58
Identities = 134/350 (38%), Positives = 203/350 (57%), Gaps = 6/350 (1%)
Query: 6 KEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFAL 65
K +KSF+D+G P+ ++E +K G+ P IQ + P A++G+DLIG+A+TGSGKT ++ L
Sbjct: 96 KPVKSFRDVGFPDYVLEEVKKAGFTEPTPIQSQGWPMAMKGRDLIGIAETGSGKTLSYLL 155
Query: 66 PILHALLEAPRPNHF---FACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQ 122
P + + P H V++PTRELA+QI ++ GS +K + GG+ Q
Sbjct: 156 PAIVHVNAQPMLAHGDGPIVLVLAPTRELAVQIQQEASKFGSSSKIKTTCIYGGVPKGPQ 215
Query: 123 SVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMI 182
+ K I++ TPGR++D +++ +L R+ YLVLDEADR+L+ F+ + +I+ I
Sbjct: 216 VRDLQKGVEIVIATPGRLIDMMESNNT-NLRRVTYLVLDEADRMLDMGFDPQIRKIVSHI 274
Query: 183 PRERRTFLFSATMTNKVEKLQRVCLRNPVKIET-SSKYSTVDTLKQQYRFLPAKHKDCYL 241
+R+T +SAT +VE+L + L NP K+ SS ++Q + K L
Sbjct: 275 RPDRQTLYWSATWPKEVEQLSKKFLYNPYKVIIGSSDLKANRAIRQIVDVISESQKYNKL 334
Query: 242 VYILSE-MAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGD 300
V +L + M GS +VF T + LR G A+ I+G SQ +R L++F+SG
Sbjct: 335 VKLLEDIMDGSRILVFLDTKKGCDQITRQLRMDGWPALSIHGDKSQAERDWVLSEFRSGK 394
Query: 301 CNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVA 350
I+ TDVA+RGLD+ V VINYD P + +DY+HR+GRT RAG G A
Sbjct: 395 SPIMTATDVAARGLDVKDVKYVINYDFPGSLEDYVHRIGRTGRAGAKGTA 444
>At4g16630 RNA helicase like protein
Length = 683
Score = 221 bits (563), Expect = 6e-58
Identities = 137/378 (36%), Positives = 199/378 (52%), Gaps = 54/378 (14%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGK---------- 59
+F +L L L+ ACE +G+K P IQ++ IP E +DL A TGSGK
Sbjct: 168 TFMELNLSRPLLRACETLGYKKPTPIQVQFIPQ--EWRDLCASAITGSGKVSTRLMVLCN 225
Query: 60 ----------TGAFALPILHALLEAPRPNHFFAC---VMSPTRELAIQISEQFEALGSEI 106
T AFALP L LL RP FA +++PTRELA+QI + L
Sbjct: 226 TEILFRYLTNTAAFALPTLERLLF--RPKRVFATRVLILTPTRELAVQIHSMIQNLAQFT 283
Query: 107 GVKCAVLVGGIDMVQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRL 166
+KC ++VGG+ + +Q V + +P I+V TPGR++DHL+N+ L L L+LDEADRL
Sbjct: 284 DIKCGLIVGGLSVREQEVVLRSMPDIVVATPGRMIDHLRNSMSVDLDDLAVLILDEADRL 343
Query: 167 LNEDFEESLNEILGMIPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLK 226
L F + E++ + P+ R+T LFSATMT +V++L ++ L P+++ L
Sbjct: 344 LQTGFATEITELVRLCPKRRQTMLFSATMTEEVKELVKLSLNKPLRLSADPSARRPPGLT 403
Query: 227 QQYRFLPAKHKDCYLVYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQ 286
++Y + + A T R L ++ GLKA ++G+++Q
Sbjct: 404 EEY----------LREFFVPSCASGTKQAAHR-------LKILFGLAGLKAAELHGNLTQ 446
Query: 287 PKRLG----------ALNKFKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIH 336
+RL +L F+ + + L+ TDVA+RGLDI V VINY P Y+H
Sbjct: 447 AQRLDVRNLGTSALFSLELFRKQEVDFLIATDVAARGLDIIGVQTVINYACPREIDSYVH 506
Query: 337 RVGRTARAGRSGVAISLV 354
RVGRTARAGR G A++ V
Sbjct: 507 RVGRTARAGREGYAVTFV 524
>At1g20920 putative RNA helicase
Length = 1166
Score = 220 bits (561), Expect = 1e-57
Identities = 124/382 (32%), Positives = 211/382 (54%), Gaps = 8/382 (2%)
Query: 6 KEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFAL 65
+ +K + GL +++ +K+ ++ P+ IQ +A+P + G+D IG+AKTGSGKT F L
Sbjct: 526 RPIKFWHQTGLTSKILDTMKKLNYEKPMPIQTQALPIIMSGRDCIGVAKTGSGKTLGFVL 585
Query: 66 PILHALLEAP---RPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQ 122
P+L + + P + VM+PTREL QI +G++C + GG + QQ
Sbjct: 586 PMLRHIKDQPPVEAGDGPIGLVMAPTRELVQQIHSDIRKFSKPLGIRCVPVYGGSGVAQQ 645
Query: 123 SVKIAKLPHIIVGTPGRVLDHLKNTKG--FSLARLKYLVLDEADRLLNEDFEESLNEILG 180
++ + I+V TPGR++D L + G +L R+ +LV+DEADR+ + FE + I+
Sbjct: 646 ISELKRGTEIVVCTPGRMIDILCTSSGKITNLRRVTFLVMDEADRMFDMGFEPQITRIIQ 705
Query: 181 MIPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCY 240
I ER+T LFSAT +VE L R L PV+I+ + + Q P +
Sbjct: 706 NIRPERQTVLFSATFPRQVETLARKVLNKPVEIQVGGRSVVNKDITQLVEVRPESDRFLR 765
Query: 241 LVYILSEMAGSTS-MVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSG 299
L+ +L E + +VF ++ + L + + ++G Q R ++ FK+
Sbjct: 766 LLELLGEWSEKGKILVFVQSQEKCDALYRDMIKSSYPCLSLHGGKDQTDRESTISDFKND 825
Query: 300 DCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYEL 359
CN+L+ T VA+RGLD+ +++V+N+D P + +DY+HRVGRT RAGR G A++ +++ +
Sbjct: 826 VCNLLIATSVAARGLDVKELELVVNFDAPNHYEDYVHRVGRTGRAGRKGCAVTFISEDDA 885
Query: 360 EWYVQIEKLIGKKLPEYPANEE 381
++ + K + +L E P ++
Sbjct: 886 KYAPDLVKAL--ELSEQPVPDD 905
>At4g00660 RNA helicase like protein
Length = 505
Score = 219 bits (558), Expect = 2e-57
Identities = 125/375 (33%), Positives = 215/375 (57%), Gaps = 6/375 (1%)
Query: 11 FKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILHA 70
F+D L L+ + G++ P IQ E+IP AL G+D++ AK G+GKT AF +P+L
Sbjct: 133 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGRDILARAKNGTGKTAAFCIPVLEK 192
Query: 71 LLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIAKLP 130
+ + N A ++ PTRELA+Q S+ + LG + ++ V GG + +++ +
Sbjct: 193 IDQDN--NVIQAVIIVPTRELALQTSQVCKELGKHLKIQVMVTTGGTSLKDDIMRLYQPV 250
Query: 131 HIIVGTPGRVLDHLKNTKGFSLAR-LKYLVLDEADRLLNEDFEESLNEILGMIPRERRTF 189
H++VGTPGR+LD K KG + + LV+DEAD+LL+++F+ S+ ++ +P R+
Sbjct: 251 HLLVGTPGRILDLTK--KGVCVLKDCSVLVMDEADKLLSQEFQPSVEHLISFLPESRQIL 308
Query: 190 LFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYILSEMA 249
+FSAT V+ + L NP I + T+ + Q Y F+ + K L + S++
Sbjct: 309 MFSATFPVTVKDFKDRFLTNPYVINLMDEL-TLKGITQFYAFVEERQKIHCLNTLFSKLQ 367
Query: 250 GSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILLCTDV 309
+ S++F + + LLA + LG I+ M Q R + F++G C L+CTD+
Sbjct: 368 INQSIIFCNSVNRVELLAKKITELGYSCFYIHAKMLQDHRNRVFHDFRNGACRNLVCTDL 427
Query: 310 ASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQIEKLI 369
+RG+DI AV++VIN+D P N++ Y+HRVGR+ R G G+A++L+ + +IE+ +
Sbjct: 428 FTRGIDIQAVNVVINFDFPKNAETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQEL 487
Query: 370 GKKLPEYPANEEEVL 384
G ++ + P + ++ +
Sbjct: 488 GTEIKQIPPHIDQAI 502
>At1g77050
Length = 513
Score = 217 bits (553), Expect = 9e-57
Identities = 129/393 (32%), Positives = 217/393 (54%), Gaps = 8/393 (2%)
Query: 2 EEENKEMKS--FKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGK 59
E++ K+ KS F+ L L ++ A +K G+K P IQ + +P L G D++ +A+TGSGK
Sbjct: 19 EKQKKKGKSGGFESLNLGPNVFNAIKKKGYKVPTPIQRKTMPLILSGVDVVAMARTGSGK 78
Query: 60 TGAFALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDM 119
T AF +P+L L + A ++SPTR+LA Q + + LG ++ ++LVGG M
Sbjct: 79 TAAFLIPMLEKLKQHVPQGGVRALILSPTRDLAEQTLKFTKELGKFTDLRVSLLVGGDSM 138
Query: 120 VQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEIL 179
Q ++ K P +I+ TPGR++ L +L ++Y+V DEAD L F E L++IL
Sbjct: 139 EDQFEELTKGPDVIIATPGRLMHLLSEVDDMTLRTVEYVVFDEADSLFGMGFAEQLHQIL 198
Query: 180 GMIPRERRTFLFSATMTNKVEKLQRVCLRNP--VKIETSSKYSTVDTLKQQYRFLPAKHK 237
+ R+T LFSAT+ + + + + LR P V+++ +K S LK + + + K
Sbjct: 199 TQLSENRQTLLFSATLPSALAEFAKAGLREPQLVRLDVENKIS--PDLKLSFLTVRPEEK 256
Query: 238 DCYLVYILSEMAGS--TSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNK 295
L+Y++ E S +++F T + + + ++ G M Q R +++
Sbjct: 257 YSALLYLVREHISSDQQTLIFVSTKHHVEFVNSLFKLENIEPSVCYGDMDQDARKIHVSR 316
Query: 296 FKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVN 355
F++ +L+ TD+A+RG+DIP +D VIN+D P K ++HRVGR ARAGR+G A S V
Sbjct: 317 FRARKTMLLIVTDIAARGIDIPLLDNVINWDFPPRPKIFVHRVGRAARAGRTGCAYSFVT 376
Query: 356 QYELEWYVQIEKLIGKKLPEYPANEEEVLLLEE 388
++ + + + + K + P +E + +EE
Sbjct: 377 PEDMPYMLDLHLFLSKPVRPAPTEDEVLKNMEE 409
>At2g45810 putative ATP-dependent RNA helicase
Length = 528
Score = 216 bits (550), Expect = 2e-56
Identities = 127/369 (34%), Positives = 207/369 (55%), Gaps = 6/369 (1%)
Query: 11 FKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILHA 70
F+D L L+ + G++ P IQ E+IP AL G D++ AK G+GKTGAF +P L
Sbjct: 156 FEDYFLKRDLLRGIYEKGFEKPSPIQEESIPIALTGSDILARAKNGTGKTGAFCIPTLEK 215
Query: 71 LLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIAKLP 130
+ P N A ++ PTRELA+Q S+ + L + ++ V GG + +++ +
Sbjct: 216 I--DPENNVIQAVILVPTRELALQTSQVCKELSKYLKIEVMVTTGGTSLRDDIMRLYQPV 273
Query: 131 HIIVGTPGRVLDHLKNTKGFSLAR-LKYLVLDEADRLLNEDFEESLNEILGMIPRERRTF 189
H++VGTPGR+LD K KG + + LV+DEAD+LL+ +F+ S+ E++ +P R+
Sbjct: 274 HLLVGTPGRILDLAK--KGVCVLKDCAMLVMDEADKLLSVEFQPSIEELIQFLPESRQIL 331
Query: 190 LFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYILSEMA 249
+FSAT V+ + L+ P I + T+ + Q Y F+ + K L + S++
Sbjct: 332 MFSATFPVTVKSFKDRYLKKPYIINLMDQL-TLMGVTQYYAFVEERQKVHCLNTLFSKLQ 390
Query: 250 GSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILLCTDV 309
+ S++F + + LLA + LG I+ M Q R + F++G C L+CTD+
Sbjct: 391 INQSIIFCNSVNRVELLAKKITELGYSCFYIHAKMVQDHRNRVFHDFRNGACRNLVCTDL 450
Query: 310 ASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQIEKLI 369
+RG+DI AV++VIN+D P S+ Y+HRVGR+ R G G+A++LV + Q E+ +
Sbjct: 451 FTRGIDIQAVNVVINFDFPRTSESYLHRVGRSGRFGHLGLAVNLVTYEDRFKMYQTEQEL 510
Query: 370 GKKLPEYPA 378
G ++ P+
Sbjct: 511 GTEIKPIPS 519
>At5g51280 DEAD-box protein abstrakt
Length = 591
Score = 212 bits (539), Expect = 4e-55
Identities = 127/365 (34%), Positives = 203/365 (54%), Gaps = 14/365 (3%)
Query: 8 MKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPI 67
+K+FKD+ P +++ ++ G P IQ++ +P L G+D+IG+A TGSGKT F LP+
Sbjct: 145 IKNFKDMKFPRPVLDTLKEKGIVQPTPIQVQGLPVILAGRDMIGIAFTGSGKTLVFVLPM 204
Query: 68 LHALLEAPRPNHFFA------CVMSPTRELAIQ---ISEQFEALGSEIG---VKCAVLVG 115
+ L+ A ++ P+RELA Q + EQF A E G ++ + +G
Sbjct: 205 IMIALQEEMMMPIAAGEGPIGLIVCPSRELARQTYEVVEQFVAPLVEAGYPPLRSLLCIG 264
Query: 116 GIDMVQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESL 175
GIDM Q + + HI+V TPGR+ D L K SL +YL LDEADRL++ FE+ +
Sbjct: 265 GIDMRSQLEVVKRGVHIVVATPGRLKDMLAKKK-MSLDACRYLTLDEADRLVDLGFEDDI 323
Query: 176 NEILGMIPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAK 235
E+ +R+T LFSATM K++ R L PV + + + Q+ ++ +
Sbjct: 324 REVFDHFKSQRQTLLFSATMPTKIQIFARSALVKPVTVNVGRAGAANLDVIQEVEYVKQE 383
Query: 236 HKDCYLVYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNK 295
K YL+ L + + + D + +L G++A+ I+G Q R A++
Sbjct: 384 AKIVYLLECLQKTSPPVLIFCENKADVDDIHEYLLLK-GVEAVAIHGGKDQEDREYAISS 442
Query: 296 FKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVN 355
FK+G ++L+ TDVAS+GLD P + VINYD+P ++Y+HR+GRT R G++G+A + +N
Sbjct: 443 FKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYVHRIGRTGRCGKTGIATTFIN 502
Query: 356 QYELE 360
+ + E
Sbjct: 503 KNQSE 507
>At3g01540 AT3g01540/F4P13_9
Length = 619
Score = 211 bits (538), Expect = 5e-55
Identities = 146/440 (33%), Positives = 224/440 (50%), Gaps = 22/440 (5%)
Query: 8 MKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALP- 66
+ SF+ G P L+ G+ P IQ ++ P A++G+D++ +AKTGSGKT + +P
Sbjct: 157 LMSFEATGFPPELLREVLSAGFSAPTPIQAQSWPIAMQGRDIVAIAKTGSGKTLGYLIPG 216
Query: 67 ILHALLEAPRPNHFFA---CVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
LH L+ R + V+SPTRELA QI E+ G + C L GG Q
Sbjct: 217 FLH--LQRIRNDSRMGPTILVLSPTRELATQIQEEAVKFGRSSRISCTCLYGGAPKGPQL 274
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
+ + I+V TPGR+ D L+ + SL ++ YLVLDEADR+L+ FE + +I+ IP
Sbjct: 275 RDLERGADIVVATPGRLNDILEMRR-ISLRQISYLVLDEADRMLDMGFEPQIRKIVKEIP 333
Query: 184 RERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTV--DTLKQQYRFLPAKHKDCYL 241
+R+T +++AT V K+ L NP ++ + V ++ Q + K L
Sbjct: 334 TKRQTLMYTATWPKGVRKIAADLLVNPAQVNIGNVDELVANKSITQHIEVVAPMEKQRRL 393
Query: 242 VYIL-SEMAGSTSMVFTRTCDSTRLLALILRNLGLK--AIPINGHMSQPKRLGALNKFKS 298
IL S+ GS ++F C + R+ + RNL + A I+G SQP+R LN+F+S
Sbjct: 394 EQILRSQEPGSKVIIF---CSTKRMCDQLTRNLTRQFGAAAIHGDKSQPERDNVLNQFRS 450
Query: 299 GDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYE 358
G +L+ TDVA+RGLD+ + V+NYD P +DY+HR+GRT RAG +G A + +
Sbjct: 451 GRTPVLVATDVAARGLDVKDIRAVVNYDFPNGVEDYVHRIGRTGRAGATGQAFTFFGDQD 510
Query: 359 LEWYVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEE 418
+ + K++ P E + R G + SGG+ R G+ G
Sbjct: 511 SKHASDLIKILEGANQRVPPQIRE---MATRGGGGMNKFSRWGPPSGGRGRGGDSGYGGR 567
Query: 419 DDVDKYFGLKDRKSSKKFRR 438
F +D +SS + R
Sbjct: 568 GS----FASRDSRSSNGWGR 583
>At3g06480 putative RNA helicase
Length = 1088
Score = 211 bits (537), Expect = 6e-55
Identities = 138/430 (32%), Positives = 221/430 (51%), Gaps = 25/430 (5%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
+F+ GLP ++ G+ +P IQ + P AL+ +D++ +AKTGSGKT + +P
Sbjct: 436 TFESSGLPPEILRELLSAGFPSPTPIQAQTWPIALQSRDIVAIAKTGSGKTLGYLIPAFI 495
Query: 70 ALLEAPRP--NHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIA 127
L N +++PTRELA QI ++ G + C L GG Q ++
Sbjct: 496 LLRHCRNDSRNGPTVLILAPTRELATQIQDEALRFGRSSRISCTCLYGGAPKGPQLKELE 555
Query: 128 KLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRERR 187
+ I+V TPGR+ D L+ K ++ LVLDEADR+L+ FE + +I+ IP R+
Sbjct: 556 RGADIVVATPGRLNDILE-MKMIDFQQVSLLVLDEADRMLDMGFEPQIRKIVNEIPPRRQ 614
Query: 188 TFLFSATMTNKVEKLQRVCLRNPVKIETS--SKYSTVDTLKQQYRFLPAKHKDCYLVYIL 245
T +++AT +V K+ L NPV++ + + + Q +P K+ L IL
Sbjct: 615 TLMYTATWPKEVRKIASDLLVNPVQVNIGRVDELAANKAITQYVEVVPQMEKERRLEQIL 674
Query: 246 -SEMAGSTSMVFTRTCDSTRLLALILRNLG--LKAIPINGHMSQPKRLGALNKFKSGDCN 302
S+ GS ++F C + RL + R++G A+ I+G +Q +R LN+F+SG
Sbjct: 675 RSQERGSKVIIF---CSTKRLCDHLARSVGRHFGAVVIHGDKTQGERDWVLNQFRSGKSC 731
Query: 303 ILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWY 362
+L+ TDVA+RGLDI + +VINYD PT +DY+HR+GRT RAG +GVA + + + ++
Sbjct: 732 VLIATDVAARGLDIKDIRVVINYDFPTGVEDYVHRIGRTGRAGATGVAFTFFTEQDWKYA 791
Query: 363 VQIEKLIGKKLPEYPANEEEVLL----------LEERVGEAKRL----AATKMKESGGKK 408
+ K++ + P ++ + ++R G R T+ GG
Sbjct: 792 PDLIKVLEGANQQVPPQVRDIAMRGGGGGGPGYSQDRRGMVNRFDSGGGGTRWDSGGGFG 851
Query: 409 RRGEGDIGEE 418
RG G G E
Sbjct: 852 GRGGGFSGRE 861
>At3g58510 ATP-dependent RNA helicase-like protein
Length = 612
Score = 211 bits (536), Expect = 8e-55
Identities = 148/429 (34%), Positives = 230/429 (53%), Gaps = 33/429 (7%)
Query: 8 MKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPI 67
+ +F D+ L ++L + + P +Q AIP L +DL+ A+TGSGKT AF PI
Sbjct: 150 VNTFADIDLGDALNLNIRRCKYVRPTPVQRHAIPILLAERDLMACAQTGSGKTAAFCFPI 209
Query: 68 LHALL-----EAPRPNHF---FACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDM 119
+ ++ E PR + FA ++SPTRELA QI ++ + + GVK V GG +
Sbjct: 210 ISGIMKDQHVERPRGSRAVYPFAVILSPTRELACQIHDEAKKFSYQTGVKVVVAYGGTPI 269
Query: 120 VQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEI- 178
QQ ++ + I+V TPGR+ D L+ + S+ +++L LDEADR+L+ FE + +I
Sbjct: 270 HQQLRELERGCDILVATPGRLNDLLERAR-VSMQMIRFLALDEADRMLDMGFEPQIRKIV 328
Query: 179 --LGMIPRE-RRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAK 235
+ M PR R+T LFSAT +++++L + N + + S+ D + Q+ F+
Sbjct: 329 EQMDMPPRGVRQTMLFSATFPSQIQRLAADFMSNYIFLAVGRVGSSTDLITQRVEFVQES 388
Query: 236 HKDCYLVYIL-----SEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRL 290
K +L+ +L ++ S ++VF T L L A I+G +Q +R
Sbjct: 389 DKRSHLMDLLHAQRETQDKQSLTLVFVETKRGADTLENWLCMNEFPATSIHGDRTQQERE 448
Query: 291 GALNKFKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVA 350
AL FK+G IL+ TDVA+RGLDIP V V+N+D+P + DY+HR+GRT RAG+SG+A
Sbjct: 449 VALRSFKTGRTPILVATDVAARGLDIPHVAHVVNFDLPNDIDDYVHRIGRTGRAGKSGIA 508
Query: 351 ISLVNQYELEWYVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRR 410
+ N+ Q+ + + + + E AN+E V E A++ GGKKR
Sbjct: 509 TAFFNENN----AQLARSLAELMQE--ANQE--------VPEWLTRYASRASFGGGKKRS 554
Query: 411 GEGDIGEED 419
G G G D
Sbjct: 555 G-GRFGGRD 562
>At2g42520 putative ATP-dependent RNA helicase
Length = 633
Score = 209 bits (533), Expect = 2e-54
Identities = 147/431 (34%), Positives = 224/431 (51%), Gaps = 34/431 (7%)
Query: 8 MKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPI 67
+ +F ++ L E+L + + P +Q AIP LEG+DL+ A+TGSGKT AF PI
Sbjct: 158 VNTFAEIDLGEALNLNIRRCKYVKPTPVQRHAIPILLEGRDLMACAQTGSGKTAAFCFPI 217
Query: 68 LHALLE--------APRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDM 119
+ +++ R + A ++SPTRELA QI ++ + + GVK V GG +
Sbjct: 218 ISGIMKDQHVQRPRGSRTVYPLAVILSPTRELASQIHDEAKKFSYQTGVKVVVAYGGTPI 277
Query: 120 VQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEI- 178
QQ ++ + I+V TPGR+ D L+ + S+ +++L LDEADR+L+ FE + +I
Sbjct: 278 NQQLRELERGVDILVATPGRLNDLLERAR-VSMQMIRFLALDEADRMLDMGFEPQIRKIV 336
Query: 179 --LGMIPRE-RRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAK 235
+ M PR R+T LFSAT ++++L L N + + S+ D + Q+ F+
Sbjct: 337 EQMDMPPRGVRQTLLFSATFPREIQRLAADFLANYIFLAVGRVGSSTDLIVQRVEFVLDS 396
Query: 236 HKDCYLVYILSEMAGS-------TSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPK 288
K +L+ +L + ++VF T L L G A I+G +Q +
Sbjct: 397 DKRSHLMDLLHAQRENGIQGKQALTLVFVETKRGADSLENWLCINGFPATSIHGDRTQQE 456
Query: 289 RLGALNKFKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSG 348
R AL FKSG IL+ TDVA+RGLDIP V V+N+D+P + DY+HR+GRT RAG+SG
Sbjct: 457 REVALKAFKSGRTPILVATDVAARGLDIPHVAHVVNFDLPNDIDDYVHRIGRTGRAGKSG 516
Query: 349 VAISLVNQYELEWYVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKK 408
+A + N + +L+ + AN+E V E A++ GGK
Sbjct: 517 LATAFFNDGNTSLARPLAELMQE------ANQE--------VPEWLTRYASRSSFGGGKN 562
Query: 409 RRGEGDIGEED 419
RR G G D
Sbjct: 563 RRSGGRFGGRD 573
>At1g31970 unknown protein
Length = 537
Score = 209 bits (533), Expect = 2e-54
Identities = 135/361 (37%), Positives = 203/361 (55%), Gaps = 15/361 (4%)
Query: 2 EEENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTG 61
E + +K+F + LPE++++ C+ ++ P IQ P L+G+DLIG+AKTGSGKT
Sbjct: 109 EAKYAALKTFAESNLPENVLDCCKT--FEKPSPIQSHTWPFLLDGRDLIGIAKTGSGKTL 166
Query: 62 AFALP-ILHALLE------APRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLV 114
AF +P I+H L + + + V+SPTRELA+QIS+ G G+K +
Sbjct: 167 AFGIPAIMHVLKKNKKIGGGSKKVNPTCLVLSPTRELAVQISDVLREAGEPCGLKSICVY 226
Query: 115 GGIDMVQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEES 174
GG Q I I++GTPGR+ D +++ L+ + ++VLDEADR+L+ FEE
Sbjct: 227 GGSSKGPQISAIRSGVDIVIGTPGRLRDLIESNV-LRLSDVSFVVLDEADRMLDMGFEEP 285
Query: 175 LNEILGMIPRERRTFLFSATMTNKVEKL-QRVCLRNPVKIETSS-KYSTVDTLKQQYRFL 232
+ IL + R+ +FSAT V KL Q NP+K+ S + + Q L
Sbjct: 286 VRFILSNTNKVRQMVMFSATWPLDVHKLAQEFMDPNPIKVIIGSVDLAANHDVMQIIEVL 345
Query: 233 PAKHKDCYLVYILSEMAGSTS---MVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKR 289
+ +D L+ +L + S +VF L L+ G KA+ I+G+ +Q +R
Sbjct: 346 DERARDQRLIALLEKYHKSQKNRVLVFALYKVEAERLERFLQQRGWKAVSIHGNKAQSER 405
Query: 290 LGALNKFKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGV 349
+L+ FK G C +L+ TDVA+RGLDIP V++VINY P ++DY+HR+GRT RAG+ GV
Sbjct: 406 TRSLSLFKEGSCPLLVATDVAARGLDIPDVEVVINYTFPLTTEDYVHRIGRTGRAGKKGV 465
Query: 350 A 350
A
Sbjct: 466 A 466
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,620,944
Number of Sequences: 26719
Number of extensions: 419351
Number of successful extensions: 1518
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 73
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 1230
Number of HSP's gapped (non-prelim): 135
length of query: 438
length of database: 11,318,596
effective HSP length: 102
effective length of query: 336
effective length of database: 8,593,258
effective search space: 2887334688
effective search space used: 2887334688
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146793.2


