

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146792.2 + phase: 0
(577 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
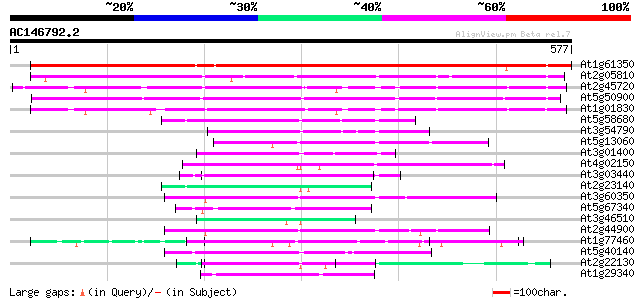
Score E
Sequences producing significant alignments: (bits) Value
At1g61350 unknown 640 0.0
At2g05810 hypothetical protein 246 2e-65
At2g45720 unknown protein 243 2e-64
At5g50900 unknown protein 231 8e-61
At1g01830 unknown protein 219 2e-57
At5g58680 unknown protein 70 2e-12
At3g54790 unknown protein 65 1e-10
At5g13060 unknown protein 65 1e-10
At3g01400 unknown protein 62 1e-09
At4g02150 AtKAP alpha 60 3e-09
At3g03440 unknown protein 60 4e-09
At2g23140 hypothetical protein 57 3e-08
At3g60350 Arm repeat containing protein - like 57 4e-08
At5g67340 unknown proteins 54 3e-07
At3g46510 arm repeat containing protein homolog 51 2e-06
At2g44900 hypothetical protein 50 3e-06
At1g77460 unknown protein 50 3e-06
At5g40140 putative protein 50 3e-06
At2g22130 unknown protein 49 6e-06
At1g29340 arm repeat-containing protein, putative 49 8e-06
>At1g61350 unknown
Length = 573
Score = 640 bits (1650), Expect = 0.0
Identities = 341/563 (60%), Positives = 441/563 (77%), Gaps = 10/563 (1%)
Query: 22 LRRALELICSLLSLTLSIRVFAGKWQLIRNKLEELHSGLIAAENSDSGENPSLSRLVTSI 81
+ +A+E I SL+SL+ SI+ F KWQLIR KL+EL+SGL + N +SG +PSLS L+++I
Sbjct: 14 IEKAIEAISSLISLSHSIKSFNIKWQLIRTKLQELYSGLDSLRNLNSGFDPSLSSLISAI 73
Query: 82 VATVKECHDLGQRCVDFSYSGKLLMQSDLDVAFSKLDGLAKKLSEIYRTGILTNGFALVV 141
+ ++K+ +DL RCV+ S+SGKLLMQSDLDV K DG + LS IY GIL++GFA+VV
Sbjct: 74 LISLKDTYDLATRCVNVSFSGKLLMQSDLDVMAGKFDGHTRNLSRIYSAGILSHGFAIVV 133
Query: 142 SKPCLGACKEDMRFYVRDILTRMKIGELGMKKQALRNLLEVVVEDEKYVKVIVVDVSDVV 201
KP ACK+DMRFY+RD+LTRMKIG+L MKKQAL L E + ED++YVK I++++SD+V
Sbjct: 134 LKPNGNACKDDMRFYIRDLLTRMKIGDLEMKKQALVKLNEAMEEDDRYVK-ILIEISDMV 192
Query: 202 HVLVGFLGSNEVEIQEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAA 261
+VLVGFL S E+ IQEESAK V ++GF SY+ VLI +GVI PL+RVL+ G+ +G+ A+A
Sbjct: 193 NVLVGFLDS-EIGIQEESAKAVFFISGFGSYRDVLIRSGVIGPLVRVLENGNGVGREASA 251
Query: 262 RCLMKLTENSDNAWAVSAHGGVTALLNICGNDDCKGDLVGPACGVLRNLVGVEEVKRFMV 321
RCLMKLTENS+NAW+VSAHGGV+ALL IC D G+L+G +CGVLRNLVGVEE+KRFM+
Sbjct: 252 RCLMKLTENSENAWSVSAHGGVSALLKICSCSDFGGELIGTSCGVLRNLVGVEEIKRFMI 311
Query: 322 EED-AVSTFIRLVKSKEEAIQVNSIGFIQNIAFGDELVRQMVIREGGIRALLRVL-DPKW 379
EED V+TFI+L+ SKEE +QVNSI + ++ DE R +++REGGI+ L+ VL DP
Sbjct: 312 EEDHTVATFIKLIGSKEEIVQVNSIDLLLSMCCKDEQTRDILVREGGIQELVSVLSDPNS 371
Query: 380 SYSSKTKEITMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGEVSIQELALKVAFRLS 439
SSK+KEI +RAI++LCF S+ ++ LM F+D LL +R+GE+S+QE ALKV RL
Sbjct: 372 LSSSKSKEIALRAIDNLCFGSAGCLNALMGCKFLDHLLNLLRNGEISVQESALKVTSRLC 431
Query: 440 GTSEEAKKAMGDAGFMVEFVKFLNAKSFEVREMAAEALSGMVTVPRNRKRFVQDDHNIAL 499
EE K+ MG+AGFM E VKFL+AKS +VREMA+ AL +++VPRNRK+F QDD NI+
Sbjct: 432 SLQEEVKRIMGEAGFMPELVKFLDAKSIDVREMASVALYCLISVPRNRKKFAQDDFNISY 491
Query: 500 LLQLLDPEEG-----NSGNKKFLISILMSLTSCNSARKKIVSSGYAKNIDKLADAEVSCD 554
+LQLLD E+G +SGN KFLISILMSLTSCNSAR+KI SSGY K+I+KLA+ E S D
Sbjct: 492 ILQLLDHEDGSNVSSDSGNTKFLISILMSLTSCNSARRKIASSGYLKSIEKLAETEGS-D 550
Query: 555 AKKLVKKLSTNRFRSMLNGIWHS 577
AKKLVKKLS NRFRS+L+GIWHS
Sbjct: 551 AKKLVKKLSMNRFRSILSGIWHS 573
>At2g05810 hypothetical protein
Length = 580
Score = 246 bits (628), Expect = 2e-65
Identities = 173/563 (30%), Positives = 310/563 (54%), Gaps = 25/563 (4%)
Query: 22 LRRALELICSLLSL----TLSIRVFAGKWQLIRNKLEELHSGLIA-AENSDSGENPSLSR 76
L+ ++LI ++LSL +L++R F G+WQ++R+KL L+S L + +E+ +NP L
Sbjct: 20 LQPLVDLITNVLSLLLLSSLTVRSFIGRWQILRSKLFTLNSSLSSLSESPHWSQNPLLHT 79
Query: 77 LVTSIVATVKECHDLGQRCVDFSYSG-KLLMQSDLDVAFSKLDGLAKKLSEIYRTGILTN 135
L+ S+++ ++ L +C S+SG KLLMQSDLD+A S L L + R+G+L
Sbjct: 80 LLPSLLSNLQRLSSLSDQCSSASFSGGKLLMQSDLDIASSSLSTHISDLDLLLRSGVLHQ 139
Query: 136 GFALVVSKPCLGACKEDMRFYVRDILTRMKIGELGMKKQALRNLLEVVVEDEKYVKVIVV 195
A+V+S P + K+D+ F++RD+ TR++IG KK++L +LL+++ ++EK ++I
Sbjct: 140 QNAIVLSLPPPTSDKDDIAFFIRDLFTRLQIGGAEFKKKSLESLLQLLTDNEKSARIIAK 199
Query: 196 DVSDVVHVLVGFLGSNEVEIQEESAKVVCVL---AGFDSYKGVLISAGVIAPLIRVLDCG 252
+ + V LV L + + E A L + DS K V G + PL+R+L+ G
Sbjct: 200 EGN--VGYLVTLLDLHHHPLIREHALAAVSLLTSSSADSRKTVFEQGG-LGPLLRLLETG 256
Query: 253 SELGKVAAARCLMKLTENSDNAWAVSAHGGVTALLNICGNDDCKGDLVGPACGVLRNLVG 312
S K AA + +T + AWA+SA+GGVT L+ C + + G + N+
Sbjct: 257 SSPFKTRAAIAIEAITADPATAWAISAYGGVTVLIEACRSG--SKQVQEHIAGAISNIAA 314
Query: 313 VEEVKRFMVEEDAVSTFIRLVKSKEEAIQVNSIGFIQNIAFGDELVRQMVIRE-GGIRAL 371
VEE++ + EE A+ I+L+ S ++Q + FI I+ E R +++RE GG++ L
Sbjct: 315 VEEIRTTLAEEGAIPVLIQLLISGSSSVQEKTANFISLISSSGEYYRDLIVRERGGLQIL 374
Query: 372 LRVLDPKWSYSSKTKEITMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGEVSIQELA 431
+ ++ S + T E + A+ + + S + S F+ +L ++HG V +Q+++
Sbjct: 375 IHLVQE--SSNPDTIEHCLLALSQISAMETVSRVLSSSTRFIIRLGELIKHGNVILQQIS 432
Query: 432 LKVAFRLSGTSEEAKKAMGDAGFMVEFVKFLNA-KSFEVREMAAEALSGMVTVPRNRKRF 490
+ L+ S+ K+A+ D + ++ + + K ++E A EA ++TV NRK
Sbjct: 433 TSLLSNLT-ISDGNKRAVADC--LSSLIRLMESPKPAGLQEAATEAAKSLLTVRSNRKEL 489
Query: 491 VQDDHNIALLLQLLDPEEGNSGNKKFLISILMSLTSCNS--ARKKIVSSGYAKNIDKLAD 548
++D+ ++ L+Q+LDP NK+ + ++ ++ S S AR K++ G + + L +
Sbjct: 490 MRDEKSVIRLVQMLDPRNERMNNKELPVMVVTAILSGGSYAARTKLIGLGADRYLQSLEE 549
Query: 549 AEVSCDAKKLVKKLST-NRFRSM 570
EV AKK V++L+ NR +S+
Sbjct: 550 MEVP-GAKKAVQRLAAGNRLKSI 571
>At2g45720 unknown protein
Length = 553
Score = 243 bits (621), Expect = 2e-64
Identities = 181/581 (31%), Positives = 305/581 (52%), Gaps = 44/581 (7%)
Query: 4 EELTGESQLAGVPTGACRLRRALELICSLLSLTLSIRVFAGKWQLIRNKLEELHSGLIAA 63
EE TG L T L +A EL+ LS +++ F+ +W++I ++LE++ + L
Sbjct: 3 EEKTGNVTLLD-QTVEDLLLQAQELVPIALSKARTVKGFSSRWRVIISRLEKIPTCL--- 58
Query: 64 ENSDSGENPSLSR------LVTSIVATVKECHDLGQRCVDFSYSGKLLMQSDLDVAFSKL 117
SD +P S+ + +++ T+KE +L CV GKL MQSDLD +K+
Sbjct: 59 --SDLSSHPCFSKHTLCKEQLQAVLETLKETIELANVCVSEKQEGKLKMQSDLDSLSAKI 116
Query: 118 DGLAKKLSEIYRTGILTNGFALVVSKPCLGACKEDMRFYVRDILTRMKIGELGMKKQALR 177
D K + +TG+L V+KP + ++ F VR++L R++IG L K++AL
Sbjct: 117 DLSLKDCGLLMKTGVLGE-----VTKPLSSSTQDLETFSVRELLARLQIGHLESKRKALE 171
Query: 178 NLLEVVVEDEKYVKVIVVDVSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGFDSYKGVLI 237
L+EV+ EDEK V + + V LV L + ++E + V+C LA + LI
Sbjct: 172 QLVEVMKEDEKAVITALGRTN--VASLVQLLTATSPSVRENAVTVICSLAESGGCENWLI 229
Query: 238 SAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNAWAVSAHGGVTALLNICGNDDCKG 297
S + LIR+L+ GS + K A L +++ +S+ + ++ HGGV L+ IC D
Sbjct: 230 SENALPSLIRLLESGSIVAKEKAVISLQRMSISSETSRSIVGHGGVGPLIEICKTGDSVS 289
Query: 298 DLVGPACGVLRNLVGVEEVKRFMVEEDAVSTFIRLVK------SKEEAIQVNSIGFIQNI 351
AC L+N+ V EV++ + EE V I ++ SKE A + +QN+
Sbjct: 290 QSAS-AC-TLKNISAVPEVRQNLAEEGIVKVMINILNCGILLGSKEYAAEC-----LQNL 342
Query: 352 AFGDELVRQMVIREGGIRALLRVLDPKWSYSSKTKEITMRAIESLCFTSSSSVSILMSYG 411
+E +R+ VI E GI+ LL LD +E + AI +L SVS+ +
Sbjct: 343 TSSNETLRRSVISENGIQTLLAYLD-----GPLPQESGVAAIRNLV----GSVSVETYFK 393
Query: 412 FVDQLLYYVRHGEVSIQELALKVAFRLSGTSEEAKKAMGDAGFMVEFVKFLNAKSFEVRE 471
+ L++ ++ G + Q+ A R++ TS E K+ +G++G + ++ L AK+ RE
Sbjct: 394 IIPSLVHVLKSGSIGAQQAAASTICRIA-TSNETKRMIGESGCIPLLIRMLEAKASGARE 452
Query: 472 MAAEALSGMVTVPRNRKRFVQDDHNIALLLQLLDPEEGNSGNKKFLISILMSLTSCNSAR 531
+AA+A++ +VTVPRN + +D+ ++ L+ LL+P GNS KK+ +S L +L S +
Sbjct: 453 VAAQAIASLVTVPRNCREVKRDEKSVTSLVMLLEPSPGNSA-KKYAVSGLAALCSSRKCK 511
Query: 532 KKIVSSGYAKNIDKLADAEVSCDAKKLVKKLSTNRFRSMLN 572
K +VS G + KL++ EV +KKL++++ + +S +
Sbjct: 512 KLMVSHGAVGYLKKLSELEVP-GSKKLLERIEKGKLKSFFS 551
>At5g50900 unknown protein
Length = 555
Score = 231 bits (589), Expect = 8e-61
Identities = 170/548 (31%), Positives = 289/548 (52%), Gaps = 15/548 (2%)
Query: 23 RRALELICSLLSLTLSIRVFAGKWQLIRNKLEELHSGLIA-AENSDSGENPSLSRLVTSI 81
R E+I SL+ ++ F KW IR KL +L + L ++ + S N L+ S+
Sbjct: 11 RSLTEVITSLIDSIPNLLSFKCKWSSIRAKLADLKTQLSDFSDFAGSSSNKLAVDLLVSV 70
Query: 82 VATVKECHDLGQRCVDFSYS-GKLLMQSDLDVAFSKLDGLAKKLSEIYRTGILTNGFALV 140
T+ + + RC + GKL QS++D ++LD K + ++G+L + +V
Sbjct: 71 RETLNDAVAVAARCEGPDLAEGKLKTQSEVDSVMARLDRHVKDAEVLIKSGLLIDN-GIV 129
Query: 141 VSKPCLGACKEDMRFYVRDILTRMKIGELGMKKQALRNLLEVVVEDEKYVKVIVVDVSDV 200
VS + + KE +R R+++ R++IG + K A+ +L+E++ ED+K V + V V
Sbjct: 130 VSGFSISSKKEAVRLEARNLVIRLQIGGVESKNSAIDSLIELLQEDDKNVMICVAQ--GV 187
Query: 201 VHVLVGFLGSNEVEIQEESAKVVCVLAGFDSYKGVLISAGV--IAPLIRVLDCGSELGKV 258
V VLV L S + ++E++ V+ ++ +S K VLI+ G+ + L+RVL+ GS K
Sbjct: 188 VPVLVRLLDSCSLVMKEKTVAVISRISMVESSKHVLIAEGLSLLNHLLRVLESGSGFAKE 247
Query: 259 AAARCLMKLTENSDNAWAVSAHGGVTALLNICGNDDCKGDLVGPACGVLRNLVGVEEVKR 318
A L L+ + +NA A+ GG+++LL IC A GVLRNL E K
Sbjct: 248 KACVALQALSLSKENARAIGCRGGISSLLEICQGGSPGSQAF--AAGVLRNLALFGETKE 305
Query: 319 FMVEEDAVSTFIRLVKSKEEAIQVNSIGFIQNIAFGDELVRQMVIREGGIRALLRVLDPK 378
VEE+A+ I +V S Q N++G + N+ GDE + V+REGGI+ L D
Sbjct: 306 NFVEENAIFVLISMVSSGTSLAQENAVGCLANLTSGDEDLMISVVREGGIQCLKSFWDSV 365
Query: 379 WSYSSKTKEITMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGEVSIQELALKVAFRL 438
S S + + +LC +++S GF+ +L+ + G + ++ A + L
Sbjct: 366 SSVKSLEVGVVLLKNLALC---PIVREVVISEGFIPRLVPVLSCGVLGVRIAAAEAVSSL 422
Query: 439 SGTSEEAKKAMGDAGFMVEFVKFLNAKSFEVREMAAEALSGMVTVPRNRKRFVQDDHNIA 498
G S +++K MG++G +V + L+ K+ E +E A++ALS ++ NRK F + D +
Sbjct: 423 -GFSSKSRKEMGESGCIVPLIDMLDGKAIEEKEAASKALSTLLVCTSNRKIFKKSDKGVV 481
Query: 499 LLLQLLDPEEGNSGNKKFLISILMSLTSCNSARKKIVSSGYAKNIDKLADAEVSCDAKKL 558
L+QLLDP+ +K++ +S L L + RK++V++G ++ KL D + AKKL
Sbjct: 482 SLVQLLDPKI-KKLDKRYTVSALELLVTSKKCRKQVVAAGACLHLQKLVDMDTE-GAKKL 539
Query: 559 VKKLSTNR 566
+ LS ++
Sbjct: 540 AENLSRSK 547
>At1g01830 unknown protein
Length = 574
Score = 219 bits (559), Expect = 2e-57
Identities = 166/569 (29%), Positives = 290/569 (50%), Gaps = 49/569 (8%)
Query: 22 LRRALELICSLLSLTLSIRVFAGKWQLIRNKLEELHSGLIAAENSDSGENPSLSR----- 76
L R LI S+LS +++ F G+W+ I +K+E++ + L SD +P S+
Sbjct: 36 LSRVNSLIPSVLSKAKTVKKFTGRWKTIISKIEQIPACL-----SDLSSHPCFSKNKLCN 90
Query: 77 -LVTSIVATVKECHDLGQRCVDFSYSGKLLMQSDLDVAFSKLDGLAKKLSEIYRTGILTN 135
+ S+ T+ E +L ++C Y GKL MQSDLD KLD + + +TG+L
Sbjct: 91 EQLQSVAKTLSEVIELAEQCSTDKYEGKLRMQSDLDSLSGKLDLNLRDCGVLIKTGVLGE 150
Query: 136 G-FALVVSK----PCLGACKEDMRFYVRDILTRMKIGELGMKKQALRNLLEVVVEDEKYV 190
L +S P + + KE +L R++IG L K AL +LL + EDEK V
Sbjct: 151 ATLPLYISSSSETPKISSLKE--------LLARLQIGHLESKHNALESLLGAMQEDEKMV 202
Query: 191 KVIVVDVSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLD 250
+ ++ ++V LV L + I+E++ ++ VLA LIS GV+ PL+R+++
Sbjct: 203 LMPLIGRANVA-ALVQLLTATSTRIREKAVNLISVLAESGHCDEWLISEGVLPPLVRLIE 261
Query: 251 CGSELGKVAAARCLMKLTENSDNAWAVSAHGGVTALLNICGNDDCKGDLVGPACGVLRNL 310
GS K AA + +L+ +NA ++ HGG+T L+++C D A L+N+
Sbjct: 262 SGSLETKEKAAIAIQRLSMTEENAREIAGHGGITPLIDLCKTGDSVSQAASAAA--LKNM 319
Query: 311 VGVEEVKRFMVEEDAVSTFIRLVK------SKEEAIQVNSIGFIQNIAFGDELVRQMVIR 364
V E+++ + EE + I L+ S+E + +QN+ + +R+ ++
Sbjct: 320 SAVSELRQLLAEEGIIRVSIDLLNHGILLGSREHMAEC-----LQNLTAASDALREAIVS 374
Query: 365 EGGIRALLRVLDPKWSYSSKTKEITMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGE 424
EGG+ +LL LD ++ + A+ +L S + I ++ + +L + ++ G
Sbjct: 375 EGGVPSLLAYLD-----GPLPQQPAVTALRNLI--PSVNPEIWVALNLLPRLRHVLKSGS 427
Query: 425 VSIQELALKVAFRLSGTSEEAKKAMGDAGFMVEFVKFLNAKSFEVREMAAEALSGMVTVP 484
+ Q+ A R + S E K+ +G++G + E VK L +KS RE AA+A++G+V
Sbjct: 428 LGAQQAAASAICRFA-CSPETKRLVGESGCIPEIVKLLESKSNGCREAAAQAIAGLVAEG 486
Query: 485 RNRKRFVQDDHNIAL-LLQLLDPEEGNSGNKKFLISILMSLTSCNSARKKIVSSGYAKNI 543
R R+ +D ++ L+ LLD GN+ KK+ ++ L+ ++ ++K +VS G +
Sbjct: 487 RIRRELKKDGKSVLTNLVMLLDSNPGNTA-KKYAVAGLLGMSGSEKSKKMMVSYGAIGYL 545
Query: 544 DKLADAEVSCDAKKLVKKLSTNRFRSMLN 572
KL++ EV A KL++KL + RS +
Sbjct: 546 KKLSEMEV-MGADKLLEKLERGKLRSFFH 573
>At5g58680 unknown protein
Length = 357
Score = 70.5 bits (171), Expect = 2e-12
Identities = 58/262 (22%), Positives = 121/262 (46%), Gaps = 9/262 (3%)
Query: 157 VRDILTRMKIGE-LGMKKQALRNLLEVVVEDEKYVKVIVVDVSDVVHVLVGFLGSNEVEI 215
+R+++T ++ + +KQA + ++ K I + + + LV + S+++++
Sbjct: 62 IRNLITHLESSSSIEEQKQAAMEIR--LLSKNKPENRIKLAKAGAIKPLVSLISSSDLQL 119
Query: 216 QEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNAW 275
QE V L+ D K +++S+G + PL+ L G+ K AA L++L++ +N
Sbjct: 120 QEYGVTAVLNLSLCDENKEMIVSSGAVKPLVNALRLGTPTTKENAACALLRLSQVEENKI 179
Query: 276 AVSAHGGVTALLNICGNDDCKGDLVGPACGVLRNLVGVEEVKRFMVEEDAVSTFIRLVKS 335
+ G + L+N+ N + A L +L E K VE + + L+
Sbjct: 180 TIGRSGAIPLLVNLLENGGFRAK--KDASTALYSLCSTNENKTRAVESGIMKPLVELMID 237
Query: 336 KEEAIQVNSIGFIQNIAFGDELVRQMVIREGGIRALLRVLDPKWSYSSKTKEITMRAIES 395
E + V+ F+ N+ + V+ EGG+ L+ +++ + + + KEI++ +
Sbjct: 238 FESDM-VDKSAFVMNLLMSAPESKPAVVEEGGVPVLVEIVE---AGTQRQKEISVSILLQ 293
Query: 396 LCFTSSSSVSILMSYGFVDQLL 417
LC S +++ G V L+
Sbjct: 294 LCEESVVYRTMVAREGAVPPLV 315
>At3g54790 unknown protein
Length = 760
Score = 65.1 bits (157), Expect = 1e-10
Identities = 57/229 (24%), Positives = 103/229 (44%), Gaps = 8/229 (3%)
Query: 204 LVGFLGSNEVEIQEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAARC 263
L+ L S E QE + + L+ + K +++ G I PL+ VL+ G++ K +A
Sbjct: 519 LLSLLYSEEKLTQEHAVTALLNLSISELNKAMIVEVGAIEPLVHVLNTGNDRAKENSAAS 578
Query: 264 LMKLTENSDNAWAVS-AHGGVTALLNICGNDDCKGDLVGPACGVLRNLVGVEEVKRFMVE 322
L L+ N + ++ + AL+N+ G +G A L NL + K +V+
Sbjct: 579 LFSLSVLQVNRERIGQSNAAIQALVNLLGKGTFRGKK--DAASALFNLSITHDNKARIVQ 636
Query: 323 EDAVSTFIRLVKSKEEAIQVNSIGFIQNIAFGDELVRQMVIREGGIRALLRVLDPKWSYS 382
AV + L+ E + ++ + N++ E RQ ++REGGI L+ +D S
Sbjct: 637 AKAVKYLVELLDPDLEMVD-KAVALLANLSAVGE-GRQAIVREGGIPLLVETVD---LGS 691
Query: 383 SKTKEITMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGEVSIQELA 431
+ KE + LC S ++++ G + L+ + G +E A
Sbjct: 692 QRGKENAASVLLQLCLNSPKFCTLVLQEGAIPPLVALSQSGTQRAKEKA 740
>At5g13060 unknown protein
Length = 709
Score = 64.7 bits (156), Expect = 1e-10
Identities = 63/296 (21%), Positives = 132/296 (44%), Gaps = 21/296 (7%)
Query: 210 SNEVEIQEESAKVVCVLAGFD-SYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLT 268
S E +++++ A + ++A Y+ +++ AG I P +++L E G+ A +++
Sbjct: 116 SCEHKLEKDCALALGLIAAIQPGYQQLIVDAGAIVPTVKLLKRRGECGECMFANAVIRRA 175
Query: 269 ---------ENSDNAWAVSAHGGVTALLNICGNDDCKGDLVGPACGVLRNL-VGVEEVKR 318
+N + GG+ L+ + D K + A G LR + +E K
Sbjct: 176 ADIITNIAHDNPRIKTNIRVEGGIAPLVELLNFPDVK--VQRAAAGALRTVSFRNDENKS 233
Query: 319 FMVEEDAVSTFIRLVKSKEEAIQVNSIGFIQNIAFGDELVRQMVIREGGIRALLRVLDPK 378
+VE +A+ T + +++S++ + +IG I N+ +++ VIR G ++ ++ +L
Sbjct: 234 QIVELNALPTLVLMLQSQDSTVHGEAIGAIGNLVHSSPDIKKEVIRAGALQPVIGLLS-- 291
Query: 379 WSYSSKTKEITMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGEVSIQELALKVAFRL 438
S +T+ I S + G + L+ + + + E++ AF L
Sbjct: 292 -STCLETQREAALLIGQFAAPDSDCKVHIAQRGAITPLIKMLESSDEQVVEMS---AFAL 347
Query: 439 SGTSEEAKKAMGDA--GFMVEFVKFLNAKSFEVREMAAEALSGMVTVPRNRKRFVQ 492
+++A G A G ++ + L+ K+ V+ AA AL G+ N F++
Sbjct: 348 GRLAQDAHNQAGIAHRGGIISLLNLLDVKTGSVQHNAAFALYGLADNEENVADFIK 403
>At3g01400 unknown protein
Length = 355
Score = 61.6 bits (148), Expect = 1e-09
Identities = 46/207 (22%), Positives = 96/207 (46%), Gaps = 10/207 (4%)
Query: 193 IVVDVSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCG 252
I + + + L+ + S+++++QE + L+ D K + S+G I PL+R L G
Sbjct: 99 IKIAKAGAIKPLISLISSSDLQLQEYGVTAILNLSLCDENKESIASSGAIKPLVRALKMG 158
Query: 253 SELGKVAAARCLMKLTENSDNAWAVSAHGGVTALLNI--CGNDDCKGDLVGPACGVLRNL 310
+ K AA L++L++ +N A+ G + L+N+ G K D A L +L
Sbjct: 159 TPTAKENAACALLRLSQIEENKVAIGRSGAIPLLVNLLETGGFRAKKD----ASTALYSL 214
Query: 311 VGVEEVKRFMVEEDAVSTFIRLVKSKEEAIQVNSIGFIQNIAFGDELVRQMVIREGGIRA 370
+E K V+ + + L+ + + V+ F+ ++ + ++ EGG+
Sbjct: 215 CSAKENKIRAVQSGIMKPLVELM-ADFGSNMVDKSAFVMSLLMSVPESKPAIVEEGGVPV 273
Query: 371 LLRVLDPKWSYSSKTKEITMRAIESLC 397
L+ +++ + + KE+ + + LC
Sbjct: 274 LVEIVEVG---TQRQKEMAVSILLQLC 297
>At4g02150 AtKAP alpha
Length = 531
Score = 60.1 bits (144), Expect = 3e-09
Identities = 84/377 (22%), Positives = 154/377 (40%), Gaps = 48/377 (12%)
Query: 178 NLLEVVVEDEKYVKVIVVDVSDVVHVLVGFLGSNEV-EIQEESAKVVCVLA-GFDSYKGV 235
NLL ++ E+ + V S VV +V FL ++ ++Q E+A + +A G V
Sbjct: 97 NLLRKLLSIEQNPPINEVVQSGVVPRVVKFLSRDDFPKLQFEAAWALTNIASGTSENTNV 156
Query: 236 LISAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNAW-AVSAHGGVTALLNICGNDD 294
+I +G + I++L SE + A L + +S V ++G +T LL+ +
Sbjct: 157 IIESGAVPIFIQLLSSASEDVREQAVWALGNVAGDSPKCRDLVLSYGAMTPLLSQFNENT 216
Query: 295 ---------------CKG------DLVGPACGVLRNLVGV--EEVK-------------- 317
C+G + PA VL LV EEV
Sbjct: 217 KLSMLRNATWTLSNFCRGKPPPAFEQTQPALPVLERLVQSMDEEVLTDACWALSYLSDNS 276
Query: 318 ----RFMVEEDAVSTFIRLVKSKEEAIQVNSIGFIQNIAFGDELVRQMVIREGGIRALLR 373
+ ++E V I+L+ ++ + ++ I NI GD+L QMV+ + + LL
Sbjct: 277 NDKIQAVIEAGVVPRLIQLLGHSSPSVLIPALRTIGNIVTGDDLQTQMVLDQQALPCLLN 336
Query: 374 VLDPKWSYSSKTKEITMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGEVSI-QELAL 432
+L K +Y K+ I ++ ++ + ++ G + L++ ++ E + +E A
Sbjct: 337 LL--KNNYKKSIKKEACWTISNITAGNADQIQAVIDAGIIQSLVWVLQSAEFEVKKEAAW 394
Query: 433 KVAFRLSGTSEEAKKAMGDAGFMVEFVKFLNAKSFEVREMAAEALSGMVTVPRNRKRFVQ 492
++ SG + + K M G + L +V + EAL ++ V K
Sbjct: 395 GISNATSGGTHDQIKFMVSQGCIKPLCDLLTCPDLKVVTVCLEALENILVVGEAEKNLGH 454
Query: 493 DDHNIALLLQLLDPEEG 509
+ L Q++D EG
Sbjct: 455 TGED-NLYAQMIDEAEG 470
>At3g03440 unknown protein
Length = 408
Score = 59.7 bits (143), Expect = 4e-09
Identities = 50/203 (24%), Positives = 98/203 (47%), Gaps = 5/203 (2%)
Query: 175 ALRNLLEVVVEDEKYVKVIVVDVSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGFDSYKG 234
AL LL + V+DEK KV +++ + ++ FL SN +QE ++ + L+ + K
Sbjct: 125 ALLALLNLAVKDEKN-KVSIIEAG-ALEPIINFLQSNSPTLQEYASASLLTLSASANNKP 182
Query: 235 VLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNAWAVSAHGGVTALLNICGNDD 294
++ + GV+ L++V+ GS K A L L+ DN + A ++ +LN+ +
Sbjct: 183 IIGANGVVPLLVKVIKHGSPQAKADAVMALSNLSTLPDNLSMILATKPLSPILNLLKSSK 242
Query: 295 CKGDLVGPACGVLRNL-VGVEEVKRFMV-EEDAVSTFIRLVKSKEEAIQVNSIGFIQNIA 352
C ++ L V EE + +V +E V + ++++ + +++G + +
Sbjct: 243 KSSKTSEKCCSLIEALMVSGEEARTGLVSDEGGVLAVVEVLENGSLQAREHAVGVLLTLC 302
Query: 353 FGD-ELVRQMVIREGGIRALLRV 374
D R+ ++REG I LL +
Sbjct: 303 QSDRSKYREPILREGVIPGLLEL 325
Score = 47.0 bits (110), Expect = 3e-05
Identities = 53/212 (25%), Positives = 99/212 (46%), Gaps = 14/212 (6%)
Query: 198 SDVVHVLVGFLGSNEVEIQEESAKVVCV-LAGFDSYKGV-LISAGVIAPLIRVLDCGSEL 255
S V LV L + E E+A + + LA D V +I AG + P+I L S
Sbjct: 103 SQAVEPLVSMLRFDSPESHHEAALLALLNLAVKDEKNKVSIIEAGALEPIINFLQSNSPT 162
Query: 256 GKVAAARCLMKLTENSDNAWAVSAHGGVTALLNIC--GNDDCKGDLVGPACGVLRNLVGV 313
+ A+ L+ L+ +++N + A+G V L+ + G+ K D V L NL +
Sbjct: 163 LQEYASASLLTLSASANNKPIIGANGVVPLLVKVIKHGSPQAKADAVM----ALSNLSTL 218
Query: 314 EEVKRFMVEEDAVSTFIRLVKSKEEAIQVNS--IGFIQNIAF-GDELVRQMVIREGGIRA 370
+ ++ +S + L+KS +++ + + I+ + G+E +V EGG+ A
Sbjct: 219 PDNLSMILATKPLSPILNLLKSSKKSSKTSEKCCSLIEALMVSGEEARTGLVSDEGGVLA 278
Query: 371 LLRVLDPKWSYSSKTKEITMRAIESLCFTSSS 402
++ VL+ + S + +E + + +LC + S
Sbjct: 279 VVEVLE---NGSLQAREHAVGVLLTLCQSDRS 307
Score = 33.1 bits (74), Expect = 0.43
Identities = 31/150 (20%), Positives = 64/150 (42%), Gaps = 1/150 (0%)
Query: 387 EITMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGEVSIQELALKVAFRLSGTSEEAK 446
E + A+ +L + ++ G ++ ++ +++ ++QE A LS ++ K
Sbjct: 123 EAALLALLNLAVKDEKNKVSIIEAGALEPIINFLQSNSPTLQEYASASLLTLSASANN-K 181
Query: 447 KAMGDAGFMVEFVKFLNAKSFEVREMAAEALSGMVTVPRNRKRFVQDDHNIALLLQLLDP 506
+G G + VK + S + + A ALS + T+P N + +L L
Sbjct: 182 PIIGANGVVPLLVKVIKHGSPQAKADAVMALSNLSTLPDNLSMILATKPLSPILNLLKSS 241
Query: 507 EEGNSGNKKFLISILMSLTSCNSARKKIVS 536
++ + ++K I + S AR +VS
Sbjct: 242 KKSSKTSEKCCSLIEALMVSGEEARTGLVS 271
>At2g23140 hypothetical protein
Length = 924
Score = 57.0 bits (136), Expect = 3e-08
Identities = 56/254 (22%), Positives = 101/254 (39%), Gaps = 40/254 (15%)
Query: 157 VRDILTRMKIGELGMKKQALRNLLEVVVEDEKYVKVIVVDVSDVVHVLVGFLGSNEVEIQ 216
V+ ++ +K L ++QA L ++ IV+ S + +LV L S + Q
Sbjct: 622 VKKLVEELKSSSLDTQRQATAELR--LLAKHNMDNRIVIGNSGAIVLLVELLYSTDSATQ 679
Query: 217 EESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNAWA 276
E + + L+ D+ K + AG I PLI VL+ GS K +A L L+ +N
Sbjct: 680 ENAVTALLNLSINDNNKKAIADAGAIEPLIHVLENGSSEAKENSAATLFSLSVIEENKIK 739
Query: 277 VSAHGGVTALLNICGNDDCKG-------------------------------DLVGPACG 305
+ G + L+++ GN +G DL+ PA G
Sbjct: 740 IGQSGAIGPLVDLLGNGTPRGKKDAATALFNLSIHQENKAMIVQSGAVRYLIDLMDPAAG 799
Query: 306 -------VLRNLVGVEEVKRFMVEEDAVSTFIRLVKSKEEAIQVNSIGFIQNIAFGDELV 358
VL NL + E + + +E + + +V+ + N+ + ++
Sbjct: 800 MVDKAVAVLANLATIPEGRNAIGQEGGIPLLVEVVELGSARGKENAAAALLQLSTNSGRF 859
Query: 359 RQMVIREGGIRALL 372
MV++EG + L+
Sbjct: 860 CNMVLQEGAVPPLV 873
Score = 36.6 bits (83), Expect = 0.039
Identities = 37/134 (27%), Positives = 59/134 (43%), Gaps = 7/134 (5%)
Query: 415 QLLYYVRHGEVSIQELALKVAFRLSGTSEEAKKAMGDAGFMVEFVKFLNAKSFEVREMAA 474
+LLY + + QE A+ LS ++ KKA+ DAG + + L S E +E +A
Sbjct: 669 ELLYST---DSATQENAVTALLNLS-INDNNKKAIADAGAIEPLIHVLENGSSEAKENSA 724
Query: 475 EALSGMVTVPRNRKRFVQDDHNIALLLQLLDPEEGNSGNKKFLISILMSLTSCNSARKKI 534
L + + N+ + Q I L+ LL G KK + L +L+ + I
Sbjct: 725 ATLFSLSVIEENKIKIGQSG-AIGPLVDLLG--NGTPRGKKDAATALFNLSIHQENKAMI 781
Query: 535 VSSGYAKNIDKLAD 548
V SG + + L D
Sbjct: 782 VQSGAVRYLIDLMD 795
>At3g60350 Arm repeat containing protein - like
Length = 928
Score = 56.6 bits (135), Expect = 4e-08
Identities = 79/354 (22%), Positives = 151/354 (42%), Gaps = 18/354 (5%)
Query: 160 ILTRMKIGELGMKKQALRNLLEVVVEDEKYVKVIVVDVSDV-----VHVLVGFLGSNEVE 214
+L+ M+ + ++++A L +V D++ + V + +L+ S
Sbjct: 385 LLSLMQSAQEDVQERAATGLATFIVVDDENASIDCGRAEAVMRDGGIRLLLELAKSWREG 444
Query: 215 IQEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNA 274
+Q E+AK + L+ + G I+ L + + L AA L L+ ++
Sbjct: 445 LQSEAAKAIANLSVNAKVAKAVAEEGGISVLADLAKSMNRLVAEEAAGGLWNLSVGEEHK 504
Query: 275 WAVSAHGGVTALLNICGN--DDCKGDLVGPACGVLRNLVGVEEVKRFMVEEDAVSTFIRL 332
A++ GGV AL+++ C G ++ A G L NL ++ + V + L
Sbjct: 505 NAIAQAGGVNALVDLIFRWPHGCDG-VLERAAGALANLAADDKCSMEVARAGGVHALVML 563
Query: 333 VKS-KEEAIQVNSIGFIQNIA-FGDELVRQMVIRE--GGIRALLRVLDPKWSYSSKTKEI 388
++ K E Q + + N+A GD + + G + AL+++ S K+
Sbjct: 564 ARNCKYEGAQEQAARALANLAAHGDSNGNNAAVGQEAGALEALVQLTQ---SPHEGVKQE 620
Query: 389 TMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGEVSIQELALKVAFRLSG--TSEEAK 446
A+ +L F + SI ++G V+ L+ + + L +VA L G SE
Sbjct: 621 AAGALWNLAFDDKNRESIA-AFGGVEALVALAKSSSNASTGLQERVAGALWGLSVSEANS 679
Query: 447 KAMGDAGFMVEFVKFLNAKSFEVREMAAEALSGMVTVPRNRKRFVQDDHNIALL 500
A+G G + + + +++ +V E AA AL + P N R V++ +AL+
Sbjct: 680 IAIGHEGGIPPLIALVRSEAEDVHETAAGALWNLSFNPGNALRIVEEGGVVALV 733
Score = 37.4 bits (85), Expect = 0.023
Identities = 30/127 (23%), Positives = 54/127 (41%)
Query: 170 GMKKQALRNLLEVVVEDEKYVKVIVVDVSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGF 229
G+K++A L + +D+ + + + L + +QE A + L+
Sbjct: 616 GVKQEAAGALWNLAFDDKNRESIAAFGGVEALVALAKSSSNASTGLQERVAGALWGLSVS 675
Query: 230 DSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNAWAVSAHGGVTALLNI 289
++ + G I PLI ++ +E AA L L+ N NA + GGV AL+ +
Sbjct: 676 EANSIAIGHEGGIPPLIALVRSEAEDVHETAAGALWNLSFNPGNALRIVEEGGVVALVQL 735
Query: 290 CGNDDCK 296
C + K
Sbjct: 736 CSSSVSK 742
>At5g67340 unknown proteins
Length = 707
Score = 53.5 bits (127), Expect = 3e-07
Identities = 56/206 (27%), Positives = 91/206 (43%), Gaps = 13/206 (6%)
Query: 171 MKKQALRNLLEVVVEDEKYVKVIVVD---VSDVVHVL-VGFLGSNEVEIQEESAKVVCVL 226
++ A+ LL + + D K ++ + + ++HVL G+L E + SA + L
Sbjct: 479 IQADAVTCLLNLSINDNN--KSLIAESGAIVPLIHVLKTGYLE----EAKANSAATLFSL 532
Query: 227 AGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNAWAVSAHGGVTAL 286
+ + YK + AG I PL+ +L GS GK AA L L+ + +N V G V L
Sbjct: 533 SVIEEYKTEIGEAGAIEPLVDLLGSGSLSGKKDAATALFNLSIHHENKTKVIEAGAVRYL 592
Query: 287 LNICGNDDCKGDLVGPACGVLRNLVGVEEVKRFMVEEDAVSTFIRLVKSKEEAIQVNSIG 346
+ + D +V A VL NL V E K + EE + + +V+ + N+
Sbjct: 593 VELM---DPAFGMVEKAVVVLANLATVREGKIAIGEEGGIPVLVEVVELGSARGKENATA 649
Query: 347 FIQNIAFGDELVRQMVIREGGIRALL 372
+ + VIREG I L+
Sbjct: 650 ALLQLCTHSPKFCNNVIREGVIPPLV 675
>At3g46510 arm repeat containing protein homolog
Length = 660
Score = 50.8 bits (120), Expect = 2e-06
Identities = 45/202 (22%), Positives = 80/202 (39%), Gaps = 39/202 (19%)
Query: 193 IVVDVSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCG 252
+ + + + +LVG L + + IQE S + L+ ++ KG ++SAG I +++VL G
Sbjct: 388 VAIAEAGAIPLLVGLLSTPDSRIQEHSVTALLNLSICENNKGAIVSAGAIPGIVQVLKKG 447
Query: 253 SELGKVAAARCLMKLTENSDNAWAVSAHGGV-------------------TALLNICGND 293
S + AA L L+ +N + A G + TAL N+C
Sbjct: 448 SMEARENAAATLFSLSVIDENKVTIGALGAIPPLVVLLNEGTQRGKKDAATALFNLCIYQ 507
Query: 294 DCKG--------------------DLVGPACGVLRNLVGVEEVKRFMVEEDAVSTFIRLV 333
KG +V A +L L E K + DAV + + +
Sbjct: 508 GNKGKAIRAGVIPTLTRLLTEPGSGMVDEALAILAILSSHPEGKAIIGSSDAVPSLVEFI 567
Query: 334 KSKEEAIQVNSIGFIQNIAFGD 355
++ + N+ + ++ GD
Sbjct: 568 RTGSPRNRENAAAVLVHLCSGD 589
>At2g44900 hypothetical protein
Length = 930
Score = 50.4 bits (119), Expect = 3e-06
Identities = 76/348 (21%), Positives = 149/348 (41%), Gaps = 20/348 (5%)
Query: 160 ILTRMKIGELGMKKQALRNLLEVVVEDEKYVKVIVVDVSDV-----VHVLVGFLGSNEVE 214
+L M+ + +++++ L VV D++ + V + +L+ S
Sbjct: 394 LLNLMQSSQEDVQERSATGLATFVVVDDENASIDCGRAEAVMKDGGIRLLLELAKSWREG 453
Query: 215 IQEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNA 274
+Q E+AK + L+ + + G I L + + L AA L L+ ++
Sbjct: 454 LQSEAAKAIANLSVNANIAKSVAEEGGIKILAGLAKSMNRLVAEEAAGGLWNLSVGEEHK 513
Query: 275 WAVSAHGGVTALLNICGN--DDCKGDLVGPACGVLRNLVGVEEVKRFMVEEDAVSTFIRL 332
A++ GGV AL+++ + C G ++ A G L NL ++ + + V + L
Sbjct: 514 NAIAQAGGVKALVDLIFRWPNGCDG-VLERAAGALANLAADDKCSMEVAKAGGVHALVML 572
Query: 333 VKS-KEEAIQVNSIGFIQNIA-FGDELVRQMVIRE--GGIRALLRVLDPKWSYSSKTKEI 388
++ K E +Q + + N+A GD + + G + AL+++ S ++
Sbjct: 573 ARNCKYEGVQEQAARALANLAAHGDSNNNNAAVGQEAGALEALVQLTK---SPHEGVRQE 629
Query: 389 TMRAIESLCFTSSSSVSILMSYGFVDQLLYYVR---HGEVSIQELALKVAFRLSGTSEEA 445
A+ +L F + SI ++ G V+ L+ + + +QE A + LS SE
Sbjct: 630 AAGALWNLSFDDKNRESISVAGG-VEALVALAQSCSNASTGLQERAAGALWGLS-VSEAN 687
Query: 446 KKAMGDAGFMVEFVKFLNAKSFEVREMAAEALSGMVTVPRNRKRFVQD 493
A+G G + + +++ +V E AA AL + P N R V++
Sbjct: 688 SVAIGREGGVPPLIALARSEAEDVHETAAGALWNLAFNPGNALRIVEE 735
Score = 37.4 bits (85), Expect = 0.023
Identities = 29/127 (22%), Positives = 55/127 (42%)
Query: 170 GMKKQALRNLLEVVVEDEKYVKVIVVDVSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGF 229
G++++A L + +D+ + V + + L + +QE +A + L+
Sbjct: 625 GVRQEAAGALWNLSFDDKNRESISVAGGVEALVALAQSCSNASTGLQERAAGALWGLSVS 684
Query: 230 DSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNAWAVSAHGGVTALLNI 289
++ + G + PLI + +E AA L L N NA + GGV AL+++
Sbjct: 685 EANSVAIGREGGVPPLIALARSEAEDVHETAAGALWNLAFNPGNALRIVEEGGVPALVHL 744
Query: 290 CGNDDCK 296
C + K
Sbjct: 745 CSSSVSK 751
>At1g77460 unknown protein
Length = 2110
Score = 50.4 bits (119), Expect = 3e-06
Identities = 78/341 (22%), Positives = 142/341 (40%), Gaps = 29/341 (8%)
Query: 201 VHVLVGFLGSNEVEIQEESAKVVCVL-AGFDSYKGVLISAGVIAPLIRVLDCGSELGKVA 259
+ + + FLG + + QE + +++ +L A D K + +AG I PL+++L+ GS+ K
Sbjct: 447 IQLFISFLGLSSEQHQEYAVEMLKILTAQVDDSKWAVTAAGGIPPLVQLLETGSQKAKED 506
Query: 260 AARCLMKLTENSDNAW-AVSAHGGVTALLNI--CGNDDCKGDLVGPACGVLRNL--VGVE 314
AA L L +S+ V GG+ A L + G + + ++ +
Sbjct: 507 AACILWNLCCHSEEIRDCVERAGGIPAFLWLLKTGGPNSQETSAKTLVKLVHTADPATIN 566
Query: 315 EVKRFMVEEDAVSTFIRLVKSKEEAIQVNSIGFIQNIAFGDELVRQMVIREGGIRALLRV 374
++ ++ +D SK + I+V +G + + A ++LV + G+R+L+
Sbjct: 567 QLLALLLGDDPT--------SKIQVIEV--LGHVLSKASQEDLVHRGCAANKGLRSLVES 616
Query: 375 LDPKWSYSSKTKEITMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGEVSIQELALKV 434
L S +TKE T + L SS I D + +++ + Q +A +V
Sbjct: 617 LT---SSREETKEHTASVLADL---FSSRQDICGHLATDDIINPWIKLLTNNTQNVAKQV 670
Query: 435 AFRLSGTS------EEAKKAMGDAGFMVEFVKFLNAKSFEVREMAAEALSGMVTVPRNRK 488
A L S KK+ G + +K S E E A AL+ +++ P
Sbjct: 671 ARALDALSRPVKNNNNKKKSYIAEGDIKSLIKLAKNSSIESAENAVSALANLLSDPDIAA 730
Query: 489 RFVQDDHNIALLLQLLD-PEEGNSGNKKFLISILMSLTSCN 528
+ +D A L D EG + L +L + C+
Sbjct: 731 EALAEDVVSAFTRILADGSPEGKRNASRALHQLLKNFPVCD 771
Score = 49.3 bits (116), Expect = 6e-06
Identities = 57/270 (21%), Positives = 115/270 (42%), Gaps = 28/270 (10%)
Query: 183 VVEDEKYVKVIVVDVSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGFDSYKGVLISAGVI 242
+ + ++ + ++ + + + L + + A ++CVL + ++ G I
Sbjct: 40 IAKGKREARRLIGSYGQAMPLFISMLRNGTTLAKVNVASILCVLCKDKDLRLKVLLGGCI 99
Query: 243 APLIRVLDCGSELGKVAAARCLMKLT----ENSDNAWAVSAHGGVTAL----LNICGNDD 294
PL+ VL G+ + AAA + +++ N + GV L++ GN D
Sbjct: 100 PPLLSVLKSGTMETRKAAAEAIYEVSSAGISNDHIGMKIFITEGVVPTLWDQLSLKGNQD 159
Query: 295 CKGDLVGPACGVLRNLVGVEE-VKRFMVEEDAVSTFIRLVKSKEEAIQVNSIGFIQN--I 351
+ G G LRNL GV++ R +E V + L+ S Q N+ + +
Sbjct: 160 --KVVEGYVTGALRNLCGVDDGYWRLTLEGSGVDIVVSLLSSDNPNSQANAASLLARLVL 217
Query: 352 AFGDELVRQMVIREGGIRALLRVLDPKWSYSSKTKEITMRAIESLCFTSSSSVSILMSYG 411
+F D + Q ++ G +++L+++L+ K + + A+E+L S + + G
Sbjct: 218 SFCDSI--QKILNSGVVKSLIQLLEQKNDINVRAS--AADALEALSANSDEAKKCVKDAG 273
Query: 412 FVDQLLYYV----------RHGEVSIQELA 431
V L+ + +HG+ S+QE A
Sbjct: 274 GVHALIEAIVAPSKECMQGKHGQ-SLQEHA 302
Score = 43.1 bits (100), Expect = 4e-04
Identities = 111/521 (21%), Positives = 209/521 (39%), Gaps = 53/521 (10%)
Query: 22 LRRALELICSLLSLTLSIRVFAGKWQLIRNKLEELHSGLIAAENSD-------SGENPSL 74
++ + L+ S S + VFA K L +E H L AA N SG+N +
Sbjct: 1363 MKPLITLMQSERSAAVEAAVFAIKILLD----DEQHLELAAAHNIQELLVGLVSGKNYVI 1418
Query: 75 SRLVTSIVATVKECHDLGQRCVDFSYSGKLLMQSDLDVAFSKLDGLAKKLSEIYRTGILT 134
+ S+ A +K D R +D +G +++ L++ L + E++R ILT
Sbjct: 1419 --IEASLSALIKLGKDRVPRKLDMVEAG--IIERCLELLPGASSSLCSAVVELFR--ILT 1472
Query: 135 NGFALVVSKPCLGACKEDMRFYVRDILTRMKIGELGMKK--QALRNLLEVVVEDEKYVKV 192
N ++ +P + E + +L R + G QAL N+LE ++ ++
Sbjct: 1473 NS-GVIARRPDVAKTVEPLFA----VLLRSDLTLWGQHSALQALVNILE----KQQTLEA 1523
Query: 193 IVVDVSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCG 252
S+ + L+ FL S+ IQ+ A+++ + ++ + + + PL+R+
Sbjct: 1524 FSFTPSEAIVPLISFLESSSQAIQQLGAELLSHFLTMEDFQQDITTQSAVVPLVRL---- 1579
Query: 253 SELGKVAAARCLMKLTENSDNAW--AVSAHGGVTALLNICGNDDCKG--DLVGPACGVLR 308
+ +G ++ +K E +W AV G+ L + +D + DL A VL
Sbjct: 1580 AGIGILSLQETAIKALEKISASWPKAVLDAEGIFELSKVILQEDPQPPLDLWESAAFVLS 1639
Query: 309 NLVGVEEVKRFMVEEDAVSTFIRLVKSKEEAIQVNSIGFIQNIAFGDELVRQMVIREGGI 368
N++ + F VE + ++L+ S E+ + ++ + D + G I
Sbjct: 1640 NILQYDAECFFRVE---LPVLVKLLFSTIESTVLLALKALMLHEKNDASSTVQMAELGAI 1696
Query: 369 RALLRVLDPKWSYSSKTKEITMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGEVSIQ 428
ALL +L S E ++ + F + + + + L Y+ +
Sbjct: 1697 DALLDLL------RSHQCEEESGSLLEVIFNNPRVRELKLCKYAIAPLSQYLLDPHTRSE 1750
Query: 429 ELALKVAFRLSGTS--EEAKKAMGDAGFMVEFVKFLNAK-SFEVREMAAEALSGMVTVPR 485
L A L S E ++ G + L + + E++ +A AL V R
Sbjct: 1751 PGRLLAALALGDLSQHEGLSRSSGSVSACRALISVLEEQPTEEMKVVAICALQNFVMNSR 1810
Query: 486 NRKRFVQDDHNIALLLQLL---DPEEGNSGNKKFLISILMS 523
+R V + + L+ +LL +PE SG ++ L S
Sbjct: 1811 TNRRAVAEAGGVLLIQELLLSCNPEV--SGQAALMVKFLFS 1849
Score = 38.1 bits (87), Expect = 0.013
Identities = 77/373 (20%), Positives = 151/373 (39%), Gaps = 56/373 (15%)
Query: 176 LRNLLEVVVEDEKYVKVIVVDV----------SDVVH----------VLVGFLGSNEVEI 215
+ LL +++ D+ K+ V++V D+VH LV L S+ E
Sbjct: 565 INQLLALLLGDDPTSKIQVIEVLGHVLSKASQEDLVHRGCAANKGLRSLVESLTSSREET 624
Query: 216 QEESAKVVCVLAGFDSYK---GVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTE--- 269
+E +A V+ L F S + G L + +I P I++L ++ AR L L+
Sbjct: 625 KEHTASVLADL--FSSRQDICGHLATDDIINPWIKLLTNNTQNVAKQVARALDALSRPVK 682
Query: 270 -NSDNAWAVSAHGGVTALLNICGNDDCKGDLVGPACGVLRNLVGVEEVKRFMVEEDAVST 328
N++ + A G + +L+ + N + A L NL+ ++ + ED VS
Sbjct: 683 NNNNKKKSYIAEGDIKSLIKLAKNSSIES--AENAVSALANLLSDPDIAAEALAEDVVSA 740
Query: 329 FIRLVKSKEEAIQVNSI----GFIQNIAFGDELVRQMVIREGGIRALLRVLDPKWSYSSK 384
F R++ + N+ ++N D L R A+L ++D S
Sbjct: 741 FTRILADGSPEGKRNASRALHQLLKNFPVCDVLKGSAQCR----FAILSLVDSLKSIDVD 796
Query: 385 TKEITMRAIESLCFTSSSSVSILMSY----------GFVDQLLYYVRHGEVSIQELALKV 434
+ + +E + + + + SY ++ L+ + G +Q+ A++V
Sbjct: 797 SAD-AFNILEVVALLAKTKSGVNFSYPPWIALAEVPSSLETLVQCLAEGHTLVQDKAIEV 855
Query: 435 AFRLSGTSEEAKKAM---GDAGFMVEFVKFLNAKSFEVREMAAEALSGMVTVPRNRKRFV 491
RL + + +V + +NA S EVR + + AL ++ + +K+ +
Sbjct: 856 LSRLCSDQQFLLSELIVSRPKSMLVLADRIVNASSLEVR-VGSTAL--LLCAAKEKKQLI 912
Query: 492 QDDHNIALLLQLL 504
+ + + L+LL
Sbjct: 913 TETLDQSGFLKLL 925
>At5g40140 putative protein
Length = 550
Score = 50.1 bits (118), Expect = 3e-06
Identities = 61/278 (21%), Positives = 122/278 (42%), Gaps = 12/278 (4%)
Query: 160 ILTRMKIGELGMKKQALRNLLEVVVEDEKYVKVIVVDVSDVVHVLVGFLGSNEVEIQEES 219
+LT++K + ++AL ++ + DE I + + V+ L + S +Q
Sbjct: 233 LLTKLKSNRISEIEEALISIRRITRIDES--SRISLCTTRVISALKSLIVSRYATVQVNV 290
Query: 220 AKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNAWAVSA 279
V+ L+ S K ++ +G++ PLI VL CGS + +A + L +N A+
Sbjct: 291 TAVLVNLSLEKSNKVKIVRSGIVPPLIDVLKCGSVEAQEHSAGVIFSLALEDENKTAIGV 350
Query: 280 HGGVTALLNI--CGNDDCKGDLVGPACGVLRNLVGVEEVKRFMVEEDAVSTFIRLVKSKE 337
GG+ LL++ G + + D + L +L V+ + +V+ AV + +V +
Sbjct: 351 LGGLEPLLHLIRVGTELTRHD----SALALYHLSLVQSNRGKLVKLGAVQMLLGMVSLGQ 406
Query: 338 EAIQVNSIGFIQNIAFGDELVRQMVIREGGIRALLRVLDPKWSYSSKTKEITMRAIESLC 397
+V I + N+A + R ++ GG+ ++ VL + T+E + + L
Sbjct: 407 MIGRVLLI--LCNMA-SCPVSRPALLDSGGVECMVGVLRRDREVNESTRESCVAVLYGLS 463
Query: 398 FTSSSSV-SILMSYGFVDQLLYYVRHGEVSIQELALKV 434
+ M+ V++L+ R G ++ A +V
Sbjct: 464 HDGGLRFKGLAMAANAVEELVKVERSGRERAKQKARRV 501
>At2g22130 unknown protein
Length = 2048
Score = 49.3 bits (116), Expect = 6e-06
Identities = 88/391 (22%), Positives = 149/391 (37%), Gaps = 78/391 (19%)
Query: 172 KKQALRNLLEVVVEDEKYVKVIVVDVSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGFDS 231
++ L+ LL+++ E V S V VLV L S V ++ ++A V+ L +
Sbjct: 28 REYCLKQLLDLIEMRENAFSA-VGSHSQAVPVLVSLLRSGSVGVKIQAATVLGSLCKENE 86
Query: 232 YKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTEN--SDNAWA--VSAHGGVTALL 287
+ ++ G I PL+ +L S G++AAA+ + ++E D+ + S G V L
Sbjct: 87 LRVKVLLGGCIPPLLGLLKSSSVEGQIAAAKTIYAVSEGGVKDHVGSKIFSTEGVVPVLW 146
Query: 288 NICGNDDCKGDLVGPACGVLRNLVGVEEVKRFMVEEDAVSTFIRLVKSKEEAIQVNSIGF 347
+ + + KG++ G G L+NL E GF
Sbjct: 147 DQLRSGNKKGEVDGLLTGALKNLSSTTE------------------------------GF 176
Query: 348 IQNIAFGDELVRQMVIREGGIRALLRVLDPKWSYSSKTKEITMRAIESLCFTSSSSVSIL 407
IR GG+ L+++L S S T + + +S S +
Sbjct: 177 WSE-----------TIRAGGVDVLVKLLT---SGQSSTLSNVCFLLACMMMEDASVCSSV 222
Query: 408 MSYGFVDQLLYYVRHG-EVSIQELALKVAFRLSGTSEEAKKAMGDAGFMVEFVKFLNAKS 466
++ QLL + G E ++ A LS S+EAK+ + ++ + + A S
Sbjct: 223 LTADITKQLLKLLGSGNEAPVRAEAAAALKSLSAQSKEAKREIANSNGIPVLINATIAPS 282
Query: 467 FEVREMAAEALSGMVTVPRNRKRFVQDDHNIALLLQLLDPEEGNSGNKKFLISIL-MSLT 525
K F+Q ++ AL + SG ++IS L SL
Sbjct: 283 ---------------------KEFMQGEYAQALQENAMCALANISGGLSYVISSLGQSLE 321
Query: 526 SCNSARKKIVSSGYAKNIDKLADAEVSCDAK 556
SC+S + A + LA A + D K
Sbjct: 322 SCSSPAQT------ADTLGALASALMIYDGK 346
Score = 47.8 bits (112), Expect = 2e-05
Identities = 44/141 (31%), Positives = 65/141 (45%), Gaps = 5/141 (3%)
Query: 198 SDVVHVLVGFLGSNEVEIQEESAKVVCVLAGFD-SYKGVLISAGVIAPLIRVLDCGSELG 256
SD +LVG + E+Q+E K + +L + S L I LI +L SE
Sbjct: 398 SDAKRLLVGLITMAVNEVQDELVKALLMLCNHEGSLWQALQGREGIQLLISLLGLSSEQQ 457
Query: 257 KVAAARCLMKLT-ENSDNAWAVSAHGGVTALLNICGNDDCKGDLVGPACGVLRNLVGVEE 315
+ A L L+ EN ++ WA++A GG+ L+ I K + +LRNL E
Sbjct: 458 QECAVALLCLLSNENDESKWAITAAGGIPPLVQILETGSAKAR--EDSATILRNLCNHSE 515
Query: 316 VKRFMVEE-DAVSTFIRLVKS 335
R VE DAV + L+K+
Sbjct: 516 DIRACVESADAVPALLWLLKN 536
Score = 43.1 bits (100), Expect = 4e-04
Identities = 51/214 (23%), Positives = 91/214 (41%), Gaps = 38/214 (17%)
Query: 201 VHVLVGFLGSNEVEIQEESAKVVCVLAGF-DSYKGVLISAGVIAPLIRVLDCGSELGKVA 259
+ +L+ LG + + QE + ++C+L+ D K + +AG I PL+++L+ GS +
Sbjct: 443 IQLLISLLGLSSEQQQECAVALLCLLSNENDESKWAITAAGGIPPLVQILETGSAKARED 502
Query: 260 AARCLMKLTENSDNAWA-VSAHGGVTALLNICGNDDCKG--------------------- 297
+A L L +S++ A V + V ALL + N G
Sbjct: 503 SATILRNLCNHSEDIRACVESADAVPALLWLLKNGSPNGKEIAAKTLNHLIHKSDTATIS 562
Query: 298 --------DLVGPACGVLRNLVGVEEVKRF--MVEE-----DAVSTFIRLVKSKEEAIQV 342
DL VL L + V F M+ E DA+ T I+L+ S +E Q
Sbjct: 563 QLTALLTSDLPESKIYVLDALKSMLSVVPFNDMLREGSASNDAIETMIKLMSSGKEETQA 622
Query: 343 NSIGFIQNIAFGDELVRQMVIREGGIRALLRVLD 376
NS + I + +R+ + + + +++L+
Sbjct: 623 NSASALAAIFQSRKDLRESALALKTLLSAIKLLN 656
Score = 40.4 bits (93), Expect = 0.003
Identities = 47/222 (21%), Positives = 104/222 (46%), Gaps = 14/222 (6%)
Query: 160 ILTRMKIGELGMKKQALRNLLEVVVEDEKYVKVIVVDVSDVVHVLVGFLGSNEVEIQEES 219
+++ ++ G +G+K QA L + E+E VKV+ + + L+G L S+ VE Q +
Sbjct: 59 LVSLLRSGSVGVKIQAATVLGSLCKENELRVKVL---LGGCIPPLLGLLKSSSVEGQIAA 115
Query: 220 AKVVCVLA--GFDSYKG--VLISAGVIAPLIRVLDCGSELGKV--AAARCLMKLTENSDN 273
AK + ++ G + G + + GV+ L L G++ G+V L L+ ++
Sbjct: 116 AKTIYAVSEGGVKDHVGSKIFSTEGVVPVLWDQLRSGNKKGEVDGLLTGALKNLSSTTEG 175
Query: 274 AWAVSAH-GGVTALLNICGNDDCKGDLVGPACGVLRNLVGVE-EVKRFMVEEDAVSTFIR 331
W+ + GGV L+ + + + + C +L ++ + V ++ D ++
Sbjct: 176 FWSETIRAGGVDVLVKLLTSG--QSSTLSNVCFLLACMMMEDASVCSSVLTADITKQLLK 233
Query: 332 LVKSKEEA-IQVNSIGFIQNIAFGDELVRQMVIREGGIRALL 372
L+ S EA ++ + +++++ + ++ + GI L+
Sbjct: 234 LLGSGNEAPVRAEAAAALKSLSAQSKEAKREIANSNGIPVLI 275
Score = 38.9 bits (89), Expect = 0.008
Identities = 31/140 (22%), Positives = 64/140 (45%), Gaps = 1/140 (0%)
Query: 151 EDMRFYVRDILTRMKIGELGMKKQALRNLLEVVVEDEKYVKVIVVDVSDVVHVLVGFLGS 210
E R V+ ++ + G + A+ L+ ++ ++ + + V VL L S
Sbjct: 1267 ESSRQAVQPLVEILNTGSEREQHAAIAALVRLLSDNPSRALAVADVEMNAVDVLCRILSS 1326
Query: 211 N-EVEIQEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTE 269
N +E++ ++A++ VL + + +A + PL+ +L + + R L KL +
Sbjct: 1327 NYTMELKGDAAELCYVLFANTRIRSTVAAARCVEPLVSLLVTEFSPAQHSVVRALDKLVD 1386
Query: 270 NSDNAWAVSAHGGVTALLNI 289
+ A V+AHG V L+ +
Sbjct: 1387 DEQLAELVAAHGAVVPLVGL 1406
Score = 33.5 bits (75), Expect = 0.33
Identities = 55/273 (20%), Positives = 116/273 (42%), Gaps = 12/273 (4%)
Query: 161 LTRMKIGELGMKKQALRNLLEVVVEDEKYVKVIVVDVSDVVHVLVGFL-GSNEVEIQEES 219
L + + E + ++ L+ +V+DE+ +++ V LVG L G N V + S
Sbjct: 1362 LVSLLVTEFSPAQHSVVRALDKLVDDEQLAELVAAH--GAVVPLVGLLYGKNYVLHEAIS 1419
Query: 220 AKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMKLTENSDNAWAVSA 279
+V + + K ++ AGVI ++ +L + A + L LT N+ A SA
Sbjct: 1420 RALVKLGKDRPACKLEMVKAGVIDCVLDILHEAPDFLCAAFSELLRILTNNATIAKGQSA 1479
Query: 280 HGGVTALLNICGNDDCKGDLVGPACGVLRNLVGVEEVKR--FMVEEDAVSTFIRLVKSKE 337
V L ++ + D A VL N++ + + + + I L++S
Sbjct: 1480 AKVVEPLFHLLTRLEFGADGQHSALQVLVNILEHPQCRADYTLTPHQVIEPLIPLLESPS 1539
Query: 338 EAIQVNSIGFIQNIAFGDELVRQMVIREGGIRALLRVLDPKWSYSSKTKEITMRAIESLC 397
A+Q + + ++ + +E +++ + + I L+ VL S + RA+++L
Sbjct: 1540 PAVQQLAAELLSHLLY-EEHLQKDPLTQLAIGPLIHVL------GSGIHLLQQRAVKALV 1592
Query: 398 FTSSSSVSILMSYGFVDQLLYYVRHGEVSIQEL 430
+ + + + G V +L + + S+ +
Sbjct: 1593 SIALTWPNEIAKEGGVSELSKVILQADPSLSNV 1625
Score = 32.7 bits (73), Expect = 0.56
Identities = 51/239 (21%), Positives = 102/239 (42%), Gaps = 21/239 (8%)
Query: 324 DAVSTFIRLVKSKEEAIQVNSIGFIQNIAFGDELVRQMVIREGGIRALLRVLDPKWSYSS 383
+ + I L+ E Q ++ + ++ ++ + + GGI L+++L+ + S+
Sbjct: 441 EGIQLLISLLGLSSEQQQECAVALLCLLSNENDESKWAITAAGGIPPLVQILE---TGSA 497
Query: 384 KTKEITMRAIESLCFTSSSSVSILMSYGFVDQLLYYVRHGEVSIQELALKVAFRLSGTSE 443
K +E + + +LC S + + S V LL+ +++G + +E+A K L S
Sbjct: 498 KAREDSATILRNLCNHSEDIRACVESADAVPALLWLLKNGSPNGKEIAAKTLNHLIHKS- 556
Query: 444 EAKKAMGDAGFMVEFVKFLNAKSFEVREMAAEALSGMVT-VPRN---RKRFVQDDHNIAL 499
D + + L + E + +AL M++ VP N R+ +D I
Sbjct: 557 -------DTATISQLTALLTSDLPESKIYVLDALKSMLSVVPFNDMLREGSASND-AIET 608
Query: 500 LLQLLD--PEEGNSGNKKFLISILMS---LTSCNSARKKIVSSGYAKNIDKLADAEVSC 553
+++L+ EE + + L +I S L A K ++S+ N+D SC
Sbjct: 609 MIKLMSSGKEETQANSASALAAIFQSRKDLRESALALKTLLSAIKLLNVDSERILVESC 667
Score = 30.8 bits (68), Expect = 2.1
Identities = 35/160 (21%), Positives = 69/160 (42%), Gaps = 5/160 (3%)
Query: 219 SAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMK-LTENSDNAWAV 277
+AK + L D + S + PL+ +L+ GSE + AA L++ L++N A AV
Sbjct: 1250 AAKALDSLFTADHIRNAESSRQAVQPLVEILNTGSEREQHAAIAALVRLLSDNPSRALAV 1309
Query: 278 S--AHGGVTALLNICGNDDCKGDLVGPACGVLRNLVGVEEVKRFMVEEDAVSTFIRLVKS 335
+ V L I ++ +L G A + L ++ + V + L+ +
Sbjct: 1310 ADVEMNAVDVLCRILSSNYTM-ELKGDAAELCYVLFANTRIRSTVAAARCVEPLVSLLVT 1368
Query: 336 KEEAIQVNSIGFIQNIAFGDELVRQMVIREGGIRALLRVL 375
+ Q + + + + DE + ++V G + L+ +L
Sbjct: 1369 EFSPAQHSVVRALDKLV-DDEQLAELVAAHGAVVPLVGLL 1407
Score = 29.3 bits (64), Expect = 6.2
Identities = 25/119 (21%), Positives = 50/119 (42%), Gaps = 4/119 (3%)
Query: 193 IVVDVSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCG 252
+ + + + +V S+ +E+ E+ + L +I +I R+L G
Sbjct: 683 VAISAREALPTIVSLANSSVLEVAEQGMCALANLILDSEVSEKVIVEDIILSATRILREG 742
Query: 253 SELGKVAAARCLMKLTENS--DNAW--AVSAHGGVTALLNICGNDDCKGDLVGPACGVL 307
+ GK AA + +L D+A +V+ G V L+++ + D + D + A L
Sbjct: 743 TVSGKTLAAAAIARLLSRRRIDSALTDSVNRAGTVLTLVSLLESADGRSDAISEALDAL 801
>At1g29340 arm repeat-containing protein, putative
Length = 729
Score = 48.9 bits (115), Expect = 8e-06
Identities = 45/181 (24%), Positives = 81/181 (43%), Gaps = 8/181 (4%)
Query: 197 VSDVVHVLVGFLGSNEVEIQEESAKVVCVLAGFDSYKG-VLISAGVIAPLIRVLDCGSEL 255
+ +V VLV L VE QE +A + L+ YK + I + L +L G+
Sbjct: 492 LESIVSVLVSGL---TVEAQENAAATLFSLSAVHEYKKRIAIVDQCVEALALLLQNGTPR 548
Query: 256 GKVAAARCLMKLTENSDNAWAVSAHGGVTALLNICGNDDCKGDLVGPACGVLRNLVGVEE 315
GK A L L+ + DN + GGV++L+ N+ + G ++R +G E
Sbjct: 549 GKKDAVTALYNLSTHPDNCSRMIEGGGVSSLVGALKNEGVAEEAAGALALLVRQSLGAEA 608
Query: 316 VKRFMVEEDAVSTFIRLVKSKEEAIQVNSIGFIQNIA-FGDELVRQMVIREGGIRALLRV 374
+ + E+ AV+ + +++ + N++ + + G V + V+R I LL+
Sbjct: 609 IGK---EDSAVAGLMGMMRCGTPRGKENAVAALLELCRSGGAAVAEKVLRAPAIAGLLQT 665
Query: 375 L 375
L
Sbjct: 666 L 666
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,670,724
Number of Sequences: 26719
Number of extensions: 465215
Number of successful extensions: 1481
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 32
Number of HSP's that attempted gapping in prelim test: 1313
Number of HSP's gapped (non-prelim): 132
length of query: 577
length of database: 11,318,596
effective HSP length: 105
effective length of query: 472
effective length of database: 8,513,101
effective search space: 4018183672
effective search space used: 4018183672
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146792.2


