

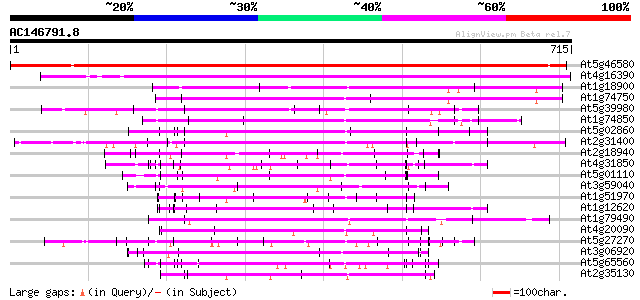
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146791.8 + phase: 0
(715 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g46580 putative protein 943 0.0
At4g16390 salt-inducible protein homolog 452 e-127
At1g18900 unknown protein 173 3e-43
At1g74750 166 3e-41
At5g39980 putative protein 157 2e-38
At1g74850 hypothetical protein 150 2e-36
At5g02860 putative protein 149 4e-36
At2g31400 unknown protein 147 3e-35
At2g18940 putative salt-inducible protein 139 7e-33
At4g31850 putative protein 135 8e-32
At5g01110 unknown protein (At5g01110) 134 2e-31
At3g59040 unknown protein (At3g59040) 130 2e-30
At1g51970 hypothetical protein 130 3e-30
At1g12620 putative protein 130 3e-30
At1g79490 unknown protein 127 2e-29
At4g20090 membrane-associated salt-inducible-like protein 127 3e-29
At5g27270 putative protein 126 4e-29
At3g06920 hypothetical protein 126 4e-29
At5g65560 putative protein 126 5e-29
At2g35130 hypothetical protein 126 5e-29
>At5g46580 putative protein
Length = 711
Score = 943 bits (2437), Expect = 0.0
Identities = 467/710 (65%), Positives = 570/710 (79%), Gaps = 7/710 (0%)
Query: 1 MATSLSSSIHISFLDTKT-TRTRFKFPPTTTTLKSHRRFLISSSKSIPDSETTPPNNNNN 59
MAT L+++I + F + T+ F + +S R L S S+ +T
Sbjct: 1 MATVLTTAIDVCFNPQNSDTKKHSLFLKPSLFRQSRSRKLNISCSSLKQPKTLEEEPITT 60
Query: 60 KKNSSLSDQLASLANTTLSTVPENQPKVLSKPKPTWVNPTKTKRPVLSHQRHKRSSVSYN 119
K SLS+QL L+ TTL + Q ++LSKPK WVNPT+ KR VLS QR KRS+ SYN
Sbjct: 61 K-TPSLSEQLKPLSATTLR---QEQTQILSKPKSVWVNPTRPKRSVLSLQRQKRSAYSYN 116
Query: 120 PQLREFQRFAQRLNNCDVSSSDEEFMVCLEEIPSSLTRGNALLVLNSLRPWQKTHMFFNW 179
PQ+++ + FA +LN+ + + EF+ L+EIP R NALLVLNSLR WQKTH FFNW
Sbjct: 117 PQIKDLRAFALKLNS-SIFTEKSEFLSLLDEIPHPPNRDNALLVLNSLREWQKTHTFFNW 175
Query: 180 IKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCN 239
+K+++L PMETIFYNVTMKSLRFGRQF +IEE+A +M+ GVELDNITYSTII+CAK+CN
Sbjct: 176 VKSKSLFPMETIFYNVTMKSLRFGRQFQLIEEMALEMVKDGVELDNITYSTIITCAKRCN 235
Query: 240 LFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFS 299
L++KA+ WFERMYKTGLMPDEVT+SAILDVY++ GKVEEV++L+ER ATGWKPD I FS
Sbjct: 236 LYNKAIEWFERMYKTGLMPDEVTYSAILDVYSKSGKVEEVLSLYERAVATGWKPDAIAFS 295
Query: 300 VLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDS 359
VLGKMFGEAGDYDGIRYVLQEMKS+ V+PN+VVYNTLLEAMG+AGKPG ARSLF EM+++
Sbjct: 296 VLGKMFGEAGDYDGIRYVLQEMKSMDVKPNVVVYNTLLEAMGRAGKPGLARSLFNEMLEA 355
Query: 360 GIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEA 419
G+ PNEKTLTA++KIYGKARW++DAL+LW+ MK WPMDFILYNTLLNMCAD+GL EEA
Sbjct: 356 GLTPNEKTLTALVKIYGKARWARDALQLWEEMKAKKWPMDFILYNTLLNMCADIGLEEEA 415
Query: 420 ETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQ 479
E LF DMK+S C+PD++SYTAMLNIYGS G +KAM+LFEEM K G+++NVMGCTCL+Q
Sbjct: 416 ERLFNDMKESVQCRPDNFSYTAMLNIYGSGGKAEKAMELFEEMLKAGVQVNVMGCTCLVQ 475
Query: 480 CLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQRANPKL 539
CLGKA IDD+V VFD+S++RGVKPDDRLCGCLLSV++L + S+D EKV+ACL+RAN KL
Sbjct: 476 CLGKAKRIDDVVYVFDLSIKRGVKPDDRLCGCLLSVMALCESSEDAEKVMACLERANKKL 535
Query: 540 VAFIQLIVDEETSFETVKEEFKAIMSNAVVEVRRPFCNCLIDICRNKDLVERAHELLYLG 599
V F+ LIVDE+T +ETVKEEFK +++ VE RRPFCNCLIDICR + ERAHELLYLG
Sbjct: 536 VTFVNLIVDEKTEYETVKEEFKLVINATQVEARRPFCNCLIDICRGNNRHERAHELLYLG 595
Query: 600 TLYGFYPSLHNKTQYEWCLDVRTLSVGAALTALEEWMTTLTKIVKREEALPDLFLAQTGT 659
TL+G YP LHNKT EW LDVR+LSVGAA TALEEWM TL I+KR+E LP+LFLAQTGT
Sbjct: 596 TLFGLYPGLHNKTIKEWSLDVRSLSVGAAETALEEWMRTLANIIKRQEELPELFLAQTGT 655
Query: 660 GAHKFAQGLNISFASHLRKLAAPFRQSEDKVGCFIATKEDLISWVQSNSP 709
G H+F+QGL SFA HL++L+APFRQS D+ G F+ATKEDL+SW++S P
Sbjct: 656 GTHRFSQGLANSFALHLQQLSAPFRQS-DRPGIFVATKEDLVSWLESKFP 704
>At4g16390 salt-inducible protein homolog
Length = 777
Score = 452 bits (1163), Expect = e-127
Identities = 245/680 (36%), Positives = 402/680 (59%), Gaps = 17/680 (2%)
Query: 40 ISSSKSIPDSETTPPNNNNNKKNSSLSDQLASLANTTLSTVPENQPKVLSKPKPTWVNPT 99
+SS +IP N + S+ + + + L V P+ + WVNP
Sbjct: 109 VSSPLTIPILPHFHSRNLLQATHVSVQEAIPQSEKSKLVDVDLPIPEPTASKSYVWVNP- 167
Query: 100 KTKRPVLSHQRHKRSSVSYNPQLREFQRFAQRLNNCDVSSSDEEFMVC--LEEIPSSLTR 157
K P S R K SY+ + + A+ L+ C + +D VC + L
Sbjct: 168 --KSPRASQLRRK----SYDSRYSSLIKLAESLDACKPNEAD----VCDVITGFGGKLFE 217
Query: 158 GNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMI 217
+A++ LN++ + + N + E I YNVTMK R + E+L +M+
Sbjct: 218 QDAVVTLNNMTNPETAPLVLNNLLETMKPSREVILYNVTMKVFRKSKDLEKSEKLFDEML 277
Query: 218 DGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVE 277
+ G++ DN T++TIISCA++ + +AV WFE+M G PD VT +A++D Y R G V+
Sbjct: 278 ERGIKPDNATFTTIISCARQNGVPKRAVEWFEKMSSFGCEPDNVTMAAMIDAYGRAGNVD 337
Query: 278 EVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLL 337
++L++R R W+ D +TFS L +++G +G+YDG + +EMK+LGV+PNLV+YN L+
Sbjct: 338 MALSLYDRARTEKWRIDAVTFSTLIRIYGVSGNYDGCLNIYEEMKALGVKPNLVIYNRLI 397
Query: 338 EAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWP 397
++MG+A +P A+ +++++I +G PN T A+++ YG+AR+ DAL +++ MKE G
Sbjct: 398 DSMGRAKRPWQAKIIYKDLITNGFTPNWSTYAALVRAYGRARYGDDALAIYREMKEKGLS 457
Query: 398 MDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMK 457
+ ILYNTLL+MCAD ++EA +F+DMK E C PDSW++++++ +Y G V KA
Sbjct: 458 LTVILYNTLLSMCADNRHVDEAFEIFQDMKNCETCDPDSWTFSSLITVYACSGRVSKAEA 517
Query: 458 LFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVS 517
++ + G E + T +IQC GKA ++DD+V+ FD +E G+ PDDR CGCLL+V++
Sbjct: 518 ALLQIREAGFEPTLFVLTSVIQCYGKAKQVDDVVRTFDQVLELGITPDDRFCGCLLNVMT 577
Query: 518 LSQGSKDQEKVLACLQRANPKLVAFIQLIVDEETSFETV-KEEFKAIMSNAVVEVRRPFC 576
+ S++ K++ C+++A PKL ++++V+E+ E V K+E ++ + +V++ +
Sbjct: 578 QTP-SEEIGKLIGCVEKAKPKLGQVVKMLVEEQNCEEGVFKKEASELIDSIGSDVKKAYL 636
Query: 577 NCLIDICRNKDLVERAHELLYLGTLYGFYPSLHNKTQYEWCLDVRTLSVGAALTALEEWM 636
NCLID+C N + +ERA E+L LG Y Y L +K+ +W L +++LS+GAALTAL WM
Sbjct: 637 NCLIDLCVNLNKLERACEILQLGLEYDIYTGLQSKSATQWSLHLKSLSLGAALTALHVWM 696
Query: 637 TTLTK-IVKREEALPDLFLAQTGTGAHKFA-QGLNISFASHLRKLAAPFRQSEDKVGCFI 694
L++ ++ E P L TG G HK++ +GL F SHL++L APF ++ DKVG F+
Sbjct: 697 NDLSEAALESGEEFPPLLGINTGHGKHKYSDKGLAAVFESHLKELNAPFHEAPDKVGWFL 756
Query: 695 ATKEDLISWVQSNSPSASIA 714
T +W++S + ++
Sbjct: 757 TTSVAAKAWLESRRSAGGVS 776
>At1g18900 unknown protein
Length = 860
Score = 173 bits (438), Expect = 3e-43
Identities = 128/531 (24%), Positives = 235/531 (44%), Gaps = 13/531 (2%)
Query: 183 QNL-LPMETIFYNVTMKSLR-FGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNL 240
QNL L ++ N +K + +G G L Q G + D TY+T++ +
Sbjct: 323 QNLGLRIDAYQANQVLKQMNDYGNALGFFYWLKRQP---GFKHDGHTYTTMVGNLGRAKQ 379
Query: 241 FDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSV 300
F + M + G P+ VT++ ++ Y R + E +N+F + + G KPD +T+
Sbjct: 380 FGAINKLLDEMVRDGCQPNTVTYNRLIHSYGRANYLNEAMNVFNQMQEAGCKPDRVTYCT 439
Query: 301 LGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSG 360
L + +AG D + Q M++ G+ P+ Y+ ++ +GKAG A LF EM+D G
Sbjct: 440 LIDIHAKAGFLDIAMDMYQRMQAGGLSPDTFTYSVIINCLGKAGHLPAAHKLFCEMVDQG 499
Query: 361 IAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAE 420
PN T ++ ++ KAR ++AL+L++ M+ G+ D + Y+ ++ + G +EEAE
Sbjct: 500 CTPNLVTYNIMMDLHAKARNYQNALKLYRDMQNAGFEPDKVTYSIVMEVLGHCGYLEEAE 559
Query: 421 TLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQC 480
+F +M+Q PD Y +++++G G V+KA + ++ M G+ NV C L+
Sbjct: 560 AVFTEMQQKNWI-PDEPVYGLLVDLWGKAGNVEKAWQWYQAMLHAGLRPNVPTCNSLLST 618
Query: 481 LGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQRANPKLV 540
+ +I + ++ + G++P + LLS + + D +
Sbjct: 619 FLRVNKIAEAYELLQNMLALGLRPSLQTYTLLLSCCTDGRSKLDMGFCGQLMASTGHPAH 678
Query: 541 AFIQLIVDEETSFETVK---EEFKAIMSNAVVEVRRPFCNCLIDICRNKDLVERAHELLY 597
F+ + E V+ F +M + E +R + ++D E A +
Sbjct: 679 MFLLKMPAAGPDGENVRNHANNFLDLMHSEDRESKRGLVDAVVDFLHKSGQKEEAGSVWE 738
Query: 598 LGTLYGFYP-SLHNKTQYEWCLDVRTLSVGAALTALEEWMTTLTKIVKREEALPDLFLAQ 656
+ +P +L K+ W +++ +S G A+TAL + K + P
Sbjct: 739 VAAQKNVFPDALREKSCSYWLINLHVMSEGTAVTALSRTLAWFRKQMLASGTCPSRIDIV 798
Query: 657 TGTGAHKFAQGLNI---SFASHLRKLAAPFRQSEDKVGCFIATKEDLISWV 704
TG G G ++ + L +PF GCF+ + E L W+
Sbjct: 799 TGWGRRSRVTGTSMVRQAVEELLNIFGSPFFTESGNSGCFVGSGEPLNRWL 849
Score = 68.6 bits (166), Expect = 1e-11
Identities = 62/278 (22%), Positives = 124/278 (44%), Gaps = 8/278 (2%)
Query: 319 QEMKSLGVQPNLVVYNTLLEAMGKAGKP-GFARSLFEEMIDSGIAPNEKTLTAVIKIYGK 377
+ +++LG++ + N +L+ M G GF L + G + T T ++ G+
Sbjct: 320 EALQNLGLRIDAYQANQVLKQMNDYGNALGFFYWLKRQ---PGFKHDGHTYTTMVGNLGR 376
Query: 378 ARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSW 437
A+ +L M +G + + YN L++ + EA +F M+++ CKPD
Sbjct: 377 AKQFGAINKLLDEMVRDGCQPNTVTYNRLIHSYGRANYLNEAMNVFNQMQEAG-CKPDRV 435
Query: 438 SYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDIS 497
+Y +++I+ G +D AM +++ M G+ + + +I CLGKA + K+F
Sbjct: 436 TYCTLIDIHAKAGFLDIAMDMYQRMQAGGLSPDTFTYSVIINCLGKAGHLPAAHKLFCEM 495
Query: 498 VERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQRANPKLVAFIQLIVDEETSFETVK 557
V++G P+ ++ + + ++ ++ K+ +Q A + IV E
Sbjct: 496 VDQGCTPNLVTYNIMMDLHAKARNYQNALKLYRDMQNAGFEPDKVTYSIVMEVLGHCGYL 555
Query: 558 EEFKAIMSNAVVE---VRRPFCNCLIDICRNKDLVERA 592
EE +A+ + + P L+D+ VE+A
Sbjct: 556 EEAEAVFTEMQQKNWIPDEPVYGLLVDLWGKAGNVEKA 593
>At1g74750
Length = 855
Score = 166 bits (421), Expect = 3e-41
Identities = 117/492 (23%), Positives = 221/492 (44%), Gaps = 8/492 (1%)
Query: 220 GVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEV 279
G + D TY+T++ + F + + M + G P+ VT++ ++ Y R ++E
Sbjct: 354 GFKHDGHTYTTMVGNLGRAKQFGEINKLLDEMVRDGCKPNTVTYNRLIHSYGRANYLKEA 413
Query: 280 VNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEA 339
+N+F + + G +PD +T+ L + +AG D + Q M+ G+ P+ Y+ ++
Sbjct: 414 MNVFNQMQEAGCEPDRVTYCTLIDIHAKAGFLDIAMDMYQRMQEAGLSPDTFTYSVIINC 473
Query: 340 MGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMD 399
+GKAG A LF EM+ G PN T +I ++ KAR + AL+L++ M+ G+ D
Sbjct: 474 LGKAGHLPAAHRLFCEMVGQGCTPNLVTFNIMIALHAKARNYETALKLYRDMQNAGFQPD 533
Query: 400 FILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLF 459
+ Y+ ++ + G +EEAE +F +M Q ++ PD Y +++++G G VDKA + +
Sbjct: 534 KVTYSIVMEVLGHCGFLEEAEGVFAEM-QRKNWVPDEPVYGLLVDLWGKAGNVDKAWQWY 592
Query: 460 EEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLS 519
+ M + G+ NV C L+ + + + + + G+ P + LLS + +
Sbjct: 593 QAMLQAGLRPNVPTCNSLLSTFLRVHRMSEAYNLLQSMLALGLHPSLQTYTLLLSCCTDA 652
Query: 520 QGSKDQEKVLACLQRANPKLVAFIQLIVDEETSFETVKE---EFKAIMSNAVVEVRRPFC 576
+ + D + + F+ + + V++ F M + E +R
Sbjct: 653 RSNFDMGFCGQLMAVSGHPAHMFLLKMPPAGPDGQKVRDHVSNFLDFMHSEDRESKRGLM 712
Query: 577 NCLIDICRNKDLVERAHELLYLGTLYGFYP-SLHNKTQYEWCLDVRTLSVGAALTALEEW 635
+ ++D L E A + + YP +L K+ W +++ +S G A+ AL
Sbjct: 713 DAVVDFLHKSGLKEEAGSVWEVAAGKNVYPDALREKSYSYWLINLHVMSEGTAVIALSRT 772
Query: 636 MTTLTKIVKREEALPDLFLAQTGTGAHKFAQGLNI---SFASHLRKLAAPFRQSEDKVGC 692
+ K + P TG G G ++ + L PF GC
Sbjct: 773 LAWFRKQMLVSGDCPSRIDIVTGWGRRSRVTGTSMVRQAVEELLNIFNFPFFTENGNSGC 832
Query: 693 FIATKEDLISWV 704
F+ + E L +W+
Sbjct: 833 FVGSGEPLKNWL 844
Score = 85.9 bits (211), Expect = 7e-17
Identities = 63/275 (22%), Positives = 121/275 (43%), Gaps = 1/275 (0%)
Query: 186 LPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAV 245
L +T Y+V + L L +M+ G + +T++ +I+ K ++ A+
Sbjct: 460 LSPDTFTYSVIINCLGKAGHLPAAHRLFCEMVGQGCTPNLVTFNIMIALHAKARNYETAL 519
Query: 246 YWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMF 305
+ M G PD+VT+S +++V G +EE +F + W PD + +L ++
Sbjct: 520 KLYRDMQNAGFQPDKVTYSIVMEVLGHCGFLEEAEGVFAEMQRKNWVPDEPVYGLLVDLW 579
Query: 306 GEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNE 365
G+AG+ D Q M G++PN+ N+LL + + A +L + M+ G+ P+
Sbjct: 580 GKAGNVDKAWQWYQAMLQAGLRPNVPTCNSLLSTFLRVHRMSEAYNLLQSMLALGLHPSL 639
Query: 366 KTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRD 425
+T T ++ AR + D + M +G P L + D + + + F D
Sbjct: 640 QTYTLLLSCCTDARSNFDMGFCGQLMAVSGHPAHMFLLK-MPPAGPDGQKVRDHVSNFLD 698
Query: 426 MKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFE 460
SE + A+++ G ++A ++E
Sbjct: 699 FMHSEDRESKRGLMDAVVDFLHKSGLKEEAGSVWE 733
Score = 41.6 bits (96), Expect = 0.002
Identities = 40/154 (25%), Positives = 63/154 (39%), Gaps = 7/154 (4%)
Query: 371 VIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSE 430
V I + +W A E + G+ MD N +L + A F +K+
Sbjct: 300 VSSILRRFKWGHAAEEA---LHNFGFRMDAYQANQVLKQMDNYA---NALGFFYWLKRQP 353
Query: 431 HCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDL 490
K D +YT M+ G + KL +EM + G + N + LI G+A + +
Sbjct: 354 GFKHDGHTYTTMVGNLGRAKQFGEINKLLDEMVRDGCKPNTVTYNRLIHSYGRANYLKEA 413
Query: 491 VKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKD 524
+ VF+ E G +P DR+ C L + G D
Sbjct: 414 MNVFNQMQEAGCEP-DRVTYCTLIDIHAKAGFLD 446
>At5g39980 putative protein
Length = 678
Score = 157 bits (397), Expect = 2e-38
Identities = 122/478 (25%), Positives = 227/478 (46%), Gaps = 23/478 (4%)
Query: 41 SSSKSIPDSETTPPNNNNNKKNSSLSDQLASLANTTLSTVPENQPKVLSKPKPTW----- 95
SSS S+P T P+ + S++ + + + TV + ++ K W
Sbjct: 8 SSSLSLPLLPLTRPHIYTSIPFSTIPE---ARQRNLIFTVSASSSSESTQNKKVWRKQPE 64
Query: 96 VNPTKTKRPVLSHQRHKRSS-VSYNPQLREFQRFAQRLNN----CDVSSSDEEFMVCLEE 150
N T + + + H+R++RS+ + +N + E + N + S+ ++ + +
Sbjct: 65 KNTTSSFQALRKHRRYQRSAFLDHNVDMDELLASIHQTQNEKELFSLLSTYKDRQLSIRF 124
Query: 151 IPSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIE 210
+ S L+R N WQ++ +W+ + YNV ++++ +QF I
Sbjct: 125 MVSLLSREN---------DWQRSLALLDWVHEEAKYTPSVFAYNVVLRNVLRAKQFDIAH 175
Query: 211 ELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVY 270
L +M + D TYST+I+ K +FD A+ W ++M + + D V +S ++++
Sbjct: 176 GLFDEMRQRALAPDRYTYSTLITSFGKEGMFDSALSWLQKMEQDRVSGDLVLYSNLIELS 235
Query: 271 ARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNL 330
RL + +++F R + +G PD + ++ + ++G+A + R +++EM GV PN
Sbjct: 236 RRLCDYSKAISIFSRLKRSGITPDLVAYNSMINVYGKAKLFREARLLIKEMNEAGVLPNT 295
Query: 331 VVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKR 390
V Y+TLL + K A S+F EM + A + T +I +YG+ K+A L+
Sbjct: 296 VSYSTLLSVYVENHKFLEALSVFAEMKEVNCALDLTTCNIMIDVYGQLDMVKEADRLFWS 355
Query: 391 MKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEG 450
+++ + + YNT+L + + L EA LFR + Q + + + +Y M+ IYG
Sbjct: 356 LRKMDIEPNVVSYNTILRVYGEAELFGEAIHLFR-LMQRKDIEQNVVTYNTMIKIYGKTM 414
Query: 451 AVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRL 508
+KA L +EM GIE N + + +I GKA ++D +F GV+ D L
Sbjct: 415 EHEKATNLVQEMQSRGIEPNAITYSTIISIWGKAGKLDRAATLFQKLRSSGVEIDQVL 472
Score = 128 bits (322), Expect = 1e-29
Identities = 97/412 (23%), Positives = 194/412 (46%), Gaps = 10/412 (2%)
Query: 159 NALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMID 218
N + + L + K F+ +K + P + + YN + + F L +M +
Sbjct: 230 NLIELSRRLCDYSKAISIFSRLKRSGITP-DLVAYNSMINVYGKAKLFREARLLIKEMNE 288
Query: 219 GGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEE 278
GV + ++YST++S + + F +A+ F M + D T + ++DVY +L V+E
Sbjct: 289 AGVLPNTVSYSTLLSVYVENHKFLEALSVFAEMKEVNCALDLTTCNIMIDVYGQLDMVKE 348
Query: 279 VVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLE 338
LF R +P+ ++++ + +++GEA + ++ + M+ ++ N+V YNT+++
Sbjct: 349 ADRLFWSLRKMDIEPNVVSYNTILRVYGEAELFGEAIHLFRLMQRKDIEQNVVTYNTMIK 408
Query: 339 AMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPM 398
GK + A +L +EM GI PN T + +I I+GKA A L+++++ +G +
Sbjct: 409 IYGKTMEHEKATNLVQEMQSRGIEPNAITYSTIISIWGKAGKLDRAATLFQKLRSSGVEI 468
Query: 399 DFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKL 458
D +LY T++ VGL+ A+ L ++K PD+ + I G ++A +
Sbjct: 469 DQVLYQTMIVAYERVGLMGHAKRLLHELK-----LPDNIPRETAITILAKAGRTEEATWV 523
Query: 459 FEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSL 518
F + + G ++ C+I + ++++VF+ G PD + +L+
Sbjct: 524 FRQAFESGEVKDISVFGCMINLYSRNQRYVNVIEVFEKMRTAGYFPDSNVIAMVLNAYGK 583
Query: 519 SQGSKDQEKVLACLQRAN---PKLVAFIQL-IVDEETSFETVKEEFKAIMSN 566
+ + + V +Q P V F L + + FE V+ F+ + S+
Sbjct: 584 QREFEKADTVYREMQEEGCVFPDEVHFQMLSLYSSKKDFEMVESLFQRLESD 635
Score = 60.8 bits (146), Expect = 2e-09
Identities = 43/173 (24%), Positives = 82/173 (46%), Gaps = 2/173 (1%)
Query: 224 DNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLF 283
DNI T I+ K ++A + F + +++G + D F ++++Y+R + V+ +F
Sbjct: 500 DNIPRETAITILAKAGRTEEATWVFRQAFESGEVKDISVFGCMINLYSRNQRYVNVIEVF 559
Query: 284 ERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKA 343
E+ R G+ PD +++ +G+ +++ V +EM+ G V+ +L
Sbjct: 560 EKMRTAGYFPDSNVIAMVLNAYGKQREFEKADTVYREMQEEGCVFPDEVHFQMLSLYSSK 619
Query: 344 GKPGFARSLFEEMIDSGIAPNEKTLTAVIK-IYGKARWSKDALELWKRMKENG 395
SLF+ + +S N K L V+ +Y +A DA + RM+E G
Sbjct: 620 KDFEMVESLFQRL-ESDPNVNSKELHLVVAALYERADKLNDASRVMNRMRERG 671
Score = 50.4 bits (119), Expect = 3e-06
Identities = 50/255 (19%), Positives = 104/255 (40%), Gaps = 25/255 (9%)
Query: 364 NEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFIL----------------YNTLL 407
NEK L +++ Y K R + +EN W L YN +L
Sbjct: 104 NEKELFSLLSTY-KDRQLSIRFMVSLLSRENDWQRSLALLDWVHEEAKYTPSVFAYNVVL 162
Query: 408 NMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGI 467
+ A LF +M+Q PD ++Y+ ++ +G EG D A+ ++M + +
Sbjct: 163 RNVLRAKQFDIAHGLFDEMRQRA-LAPDRYTYSTLITSFGKEGMFDSALSWLQKMEQDRV 221
Query: 468 ELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEK 527
+++ + LI+ + + + +F G+ PD +++V ++ ++
Sbjct: 222 SGDLVLYSNLIELSRRLCDYSKAISIFSRLKRSGITPDLVAYNSMINVYGKAKLFREARL 281
Query: 528 VLACLQRAN--PKLVAFIQLI---VDEETSFETVKEEFKAIMSNAVVEVRRPFCNCLIDI 582
++ + A P V++ L+ V+ E + + N +++ CN +ID+
Sbjct: 282 LIKEMNEAGVLPNTVSYSTLLSVYVENHKFLEALSVFAEMKEVNCALDLTT--CNIMIDV 339
Query: 583 CRNKDLVERAHELLY 597
D+V+ A L +
Sbjct: 340 YGQLDMVKEADRLFW 354
>At1g74850 hypothetical protein
Length = 862
Score = 150 bits (379), Expect = 2e-36
Identities = 118/522 (22%), Positives = 220/522 (41%), Gaps = 42/522 (8%)
Query: 170 WQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYS 229
W+ F ++ + + P + + YN + + E + M DGG+ D TYS
Sbjct: 228 WEGLLGLFAEMRHEGIQP-DIVTYNTLLSACAIRGLGDEAEMVFRTMNDGGIVPDLTTYS 286
Query: 230 TIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRAT 289
++ K +K M G +PD +++ +L+ YA+ G ++E + +F + +A
Sbjct: 287 HLVETFGKLRRLEKVCDLLGEMASGGSLPDITSYNVLLEAYAKSGSIKEAMGVFHQMQAA 346
Query: 290 GWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFA 349
G P+ T+SVL +FG++G YD +R + EMKS P+ YN L+E G+ G
Sbjct: 347 GCTPNANTYSVLLNLFGQSGRYDDVRQLFLEMKSSNTDPDAATYNILIEVFGEGGYFKEV 406
Query: 350 RSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNM 409
+LF +M++ I P+ +T +I GK +DA ++ + M N Y ++
Sbjct: 407 VTLFHDMVEENIEPDMETYEGIIFACGKGGLHEDARKILQYMTANDIVPSSKAYTGVIEA 466
Query: 410 CADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIEL 469
L EEA F M + P ++ ++L + G V ++ + + GI
Sbjct: 467 FGQAALYEEALVAFNTMHEVGS-NPSIETFHSLLYSFARGGLVKESEAILSRLVDSGIPR 525
Query: 470 NVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVL 529
N I+ + + ++ VK + + PD+R +LSV S ++ + +
Sbjct: 526 NRDTFNAQIEAYKQGGKFEEAVKTYVDMEKSRCDPDERTLEAVLSVYSFARLVDECREQF 585
Query: 530 ACLQRAN--PKLVAFIQLIV--DEETSFETVKEEFKAIMSNAVVEVRRP----------- 574
++ ++ P ++ + ++ + ++ V E + ++SN V + +
Sbjct: 586 EEMKASDILPSIMCYCMMLAVYGKTERWDDVNELLEEMLSNRVSNIHQVIGQMIKGDYDD 645
Query: 575 ------------------------FCNCLIDICRNKDLVERAHELLYLGTLYGFYPSLHN 610
F N L+D ERA +L T G +P L
Sbjct: 646 DSNWQIVEYVLDKLNSEGCGLGIRFYNALLDALWWLGQKERAARVLNEATKRGLFPELFR 705
Query: 611 KTQYEWCLDVRTLSVGAALTALEEWMTTLTKIVKREEALPDL 652
K + W +DV +S G TAL W+ + ++ + + LP L
Sbjct: 706 KNKLVWSVDVHRMSEGGMYTALSVWLNDINDMLLKGD-LPQL 746
Score = 135 bits (341), Expect = 6e-32
Identities = 100/373 (26%), Positives = 171/373 (45%), Gaps = 19/373 (5%)
Query: 228 YSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGR 287
Y+ +IS + L DK + F+ M G+ +++A+++ Y R G+ E + L +R +
Sbjct: 144 YTIMISLLGREGLLDKCLEVFDEMPSQGVSRSVFSYTALINAYGRNGRYETSLELLDRMK 203
Query: 288 ATGWKPDPITFSVLGKMFGEAG-DYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKP 346
P +T++ + G D++G+ + EM+ G+QP++V YNTLL A G
Sbjct: 204 NEKISPSILTYNTVINACARGGLDWEGLLGLFAEMRHEGIQPDIVTYNTLLSACAIRGLG 263
Query: 347 GFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTL 406
A +F M D GI P+ T + +++ +GK R + +L M G D YN L
Sbjct: 264 DEAEMVFRTMNDGGIVPDLTTYSHLVETFGKLRRLEKVCDLLGEMASGGSLPDITSYNVL 323
Query: 407 LNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFG 466
L A G I+EA +F M Q+ C P++ +Y+ +LN++G G D +LF EM
Sbjct: 324 LEAYAKSGSIKEAMGVFHQM-QAAGCTPNANTYSVLLNLFGQSGRYDDVRQLFLEMKSSN 382
Query: 467 IELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQE 526
+ + LI+ G+ ++V +F VE ++PD ++ +D
Sbjct: 383 TDPDAATYNILIEVFGEGGYFKEVVTLFHDMVEENIEPDMETYEGIIFACGKGGLHEDAR 442
Query: 527 KVLACLQRANPKLVAFIQLIVDEETSFETVKEEF--KAIMSNAVVEVRRPFCNCLIDICR 584
K+L + + IV ++ V E F A+ A+V N + ++
Sbjct: 443 KILQYMTAND---------IVPSSKAYTGVIEAFGQAALYEEALVAF-----NTMHEVGS 488
Query: 585 NKDLVERAHELLY 597
N +E H LLY
Sbjct: 489 NPS-IETFHSLLY 500
Score = 84.0 bits (206), Expect = 3e-16
Identities = 67/275 (24%), Positives = 124/275 (44%), Gaps = 7/275 (2%)
Query: 298 FSVLGKMFGEAGDYD-GIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEM 356
F+++ K F GD+ +R + + +PN +Y ++ +G+ G +F+EM
Sbjct: 108 FALVFKEFAGRGDWQRSLRLFKYMQRQIWCKPNEHIYTIMISLLGREGLLDKCLEVFDEM 167
Query: 357 IDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLI 416
G++ + + TA+I YG+ + +LEL RMK + YNT++N CA GL
Sbjct: 168 PSQGVSRSVFSYTALINAYGRNGRYETSLELLDRMKNEKISPSILTYNTVINACARGGLD 227
Query: 417 EEAET-LFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCT 475
E LF +M+ E +PD +Y +L+ G D+A +F M+ GI ++ +
Sbjct: 228 WEGLLGLFAEMRH-EGIQPDIVTYNTLLSACAIRGLGDEAEMVFRTMNDGGIVPDLTTYS 286
Query: 476 CLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQRA 535
L++ GK ++ + + G PD LL + S K+ V +Q A
Sbjct: 287 HLVETFGKLRRLEKVCDLLGEMASGGSLPDITSYNVLLEAYAKSGSIKEAMGVFHQMQAA 346
Query: 536 ----NPKLVAFIQLIVDEETSFETVKEEFKAIMSN 566
N + + + + ++ V++ F + S+
Sbjct: 347 GCTPNANTYSVLLNLFGQSGRYDDVRQLFLEMKSS 381
>At5g02860 putative protein
Length = 819
Score = 149 bits (377), Expect = 4e-36
Identities = 95/327 (29%), Positives = 169/327 (51%), Gaps = 4/327 (1%)
Query: 223 LDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNL 282
LDN + IIS K A F + + G D ++++++ +A G+ E VN+
Sbjct: 171 LDNSVVAIIISMLGKEGRVSSAANMFNGLQEDGFSLDVYSYTSLISAFANSGRYREAVNV 230
Query: 283 FERGRATGWKPDPITFSVLGKMFGEAG-DYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMG 341
F++ G KP IT++V+ +FG+ G ++ I ++++MKS G+ P+ YNTL+
Sbjct: 231 FKKMEEDGCKPTLITYNVILNVFGKMGTPWNKITSLVEKMKSDGIAPDAYTYNTLITCCK 290
Query: 342 KAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFI 401
+ A +FEEM +G + ++ T A++ +YGK+ K+A+++ M NG+ +
Sbjct: 291 RGSLHQEAAQVFEEMKAAGFSYDKVTYNALLDVYGKSHRPKEAMKVLNEMVLNGFSPSIV 350
Query: 402 LYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEE 461
YN+L++ A G+++EA L M + + KPD ++YT +L+ + G V+ AM +FEE
Sbjct: 351 TYNSLISAYARDGMLDEAMELKNQMAE-KGTKPDVFTYTTLLSGFERAGKVESAMSIFEE 409
Query: 462 MSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQG 521
M G + N+ I+ G + +++K+FD G+ PD LL+V +
Sbjct: 410 MRNAGCKPNICTFNAFIKMYGNRGKFTEMMKIFDEINVCGLSPDIVTWNTLLAVFGQNGM 469
Query: 522 SKDQEKVLACLQRAN--PKLVAFIQLI 546
+ V ++RA P+ F LI
Sbjct: 470 DSEVSGVFKEMKRAGFVPERETFNTLI 496
Score = 147 bits (371), Expect = 2e-35
Identities = 95/379 (25%), Positives = 176/379 (46%), Gaps = 5/379 (1%)
Query: 211 ELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVY 270
++ ++M+ G +TY+++IS + + D+A+ +M + G PD T++ +L +
Sbjct: 335 KVLNEMVLNGFSPSIVTYNSLISAYARDGMLDEAMELKNQMAEKGTKPDVFTYTTLLSGF 394
Query: 271 ARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNL 330
R GKVE +++FE R G KP+ TF+ KM+G G + + + E+ G+ P++
Sbjct: 395 ERAGKVESAMSIFEEMRNAGCKPNICTFNAFIKMYGNRGKFTEMMKIFDEINVCGLSPDI 454
Query: 331 VVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKR 390
V +NTLL G+ G +F+EM +G P +T +I Y + + A+ +++R
Sbjct: 455 VTWNTLLAVFGQNGMDSEVSGVFKEMKRAGFVPERETFNTLISAYSRCGSFEQAMTVYRR 514
Query: 391 MKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEG 450
M + G D YNT+L A G+ E++E + +M+ CKP+ +Y ++L+ Y +
Sbjct: 515 MLDAGVTPDLSTYNTVLAALARGGMWEQSEKVLAEMEDGR-CKPNELTYCSLLHAYANGK 573
Query: 451 AVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCG 510
+ L EE+ IE + L+ K + + + F ERG PD
Sbjct: 574 EIGLMHSLAEEVYSGVIEPRAVLLKTLVLVCSKCDLLPEAERAFSELKERGFSPDITTLN 633
Query: 511 CLLSVVSLSQGSKDQEKVLACLQRA--NPKLVAFIQLIVDEETSFETVKEE--FKAIMSN 566
++S+ Q VL ++ P + + L+ S + K E + I++
Sbjct: 634 SMVSIYGRRQMVAKANGVLDYMKERGFTPSMATYNSLMYMHSRSADFGKSEEILREILAK 693
Query: 567 AVVEVRRPFCNCLIDICRN 585
+ + + CRN
Sbjct: 694 GIKPDIISYNTVIYAYCRN 712
Score = 140 bits (354), Expect = 2e-33
Identities = 104/422 (24%), Positives = 185/422 (43%), Gaps = 40/422 (9%)
Query: 191 IFYNVTMKSL-RFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFE 249
I YNV + + G + I L +M G+ D TY+T+I+C K+ +L +A FE
Sbjct: 244 ITYNVILNVFGKMGTPWNKITSLVEKMKSDGIAPDAYTYNTLITCCKRGSLHQEAAQVFE 303
Query: 250 RMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAG 309
M G D+VT++A+LDVY + + +E + + G+ P +T++ L + G
Sbjct: 304 EMKAAGFSYDKVTYNALLDVYGKSHRPKEAMKVLNEMVLNGFSPSIVTYNSLISAYARDG 363
Query: 310 DYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLT 369
D + +M G +P++ Y TLL +AGK A S+FEEM ++G PN T
Sbjct: 364 MLDEAMELKNQMAEKGTKPDVFTYTTLLSGFERAGKVESAMSIFEEMRNAGCKPNICTFN 423
Query: 370 AVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQS 429
A IK+YG + ++++ + G D + +NTLL + G+ E +F++MK++
Sbjct: 424 AFIKMYGNRGKFTEMMKIFDEINVCGLSPDIVTWNTLLAVFGQNGMDSEVSGVFKEMKRA 483
Query: 430 EHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDD 489
P+ ++ +++ Y G+ ++AM ++ M
Sbjct: 484 GFV-PERETFNTLISAYSRCGSFEQAMTVYRRM--------------------------- 515
Query: 490 LVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQ--RANPKLVAFIQLIV 547
++ GV PD +L+ ++ + EKVLA ++ R P + + L+
Sbjct: 516 --------LDAGVTPDLSTYNTVLAALARGGMWEQSEKVLAEMEDGRCKPNELTYCSLLH 567
Query: 548 DEETSFET-VKEEFKAIMSNAVVEVRRPFCNCLIDICRNKDLVERAHELLYLGTLYGFYP 606
E + + + V+E R L+ +C DL+ A GF P
Sbjct: 568 AYANGKEIGLMHSLAEEVYSGVIEPRAVLLKTLVLVCSKCDLLPEAERAFSELKERGFSP 627
Query: 607 SL 608
+
Sbjct: 628 DI 629
Score = 101 bits (252), Expect = 1e-21
Identities = 73/294 (24%), Positives = 138/294 (46%), Gaps = 7/294 (2%)
Query: 215 QMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLG 274
+M G + T++T+IS +C F++A+ + RM G+ PD T++ +L AR G
Sbjct: 479 EMKRAGFVPERETFNTLISAYSRCGSFEQAMTVYRRMLDAGVTPDLSTYNTVLAALARGG 538
Query: 275 ---KVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLV 331
+ E+V+ E GR KP+ +T+ L + + + + +E+ S ++P V
Sbjct: 539 MWEQSEKVLAEMEDGRC---KPNELTYCSLLHAYANGKEIGLMHSLAEEVYSGVIEPRAV 595
Query: 332 VYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRM 391
+ TL+ K A F E+ + G +P+ TL +++ IYG+ + A + M
Sbjct: 596 LLKTLVLVCSKCDLLPEAERAFSELKERGFSPDITTLNSMVSIYGRRQMVAKANGVLDYM 655
Query: 392 KENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGA 451
KE G+ YN+L+ M + ++E + R++ ++ KPD SY ++ Y
Sbjct: 656 KERGFTPSMATYNSLMYMHSRSADFGKSEEILREI-LAKGIKPDIISYNTVIYAYCRNTR 714
Query: 452 VDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPD 505
+ A ++F EM GI +V+ I ++ + V ++ G +P+
Sbjct: 715 MRDASRIFSEMRNSGIVPDVITYNTFIGSYAADSMFEEAIGVVRYMIKHGCRPN 768
Score = 95.1 bits (235), Expect = 1e-19
Identities = 60/283 (21%), Positives = 132/283 (46%), Gaps = 1/283 (0%)
Query: 152 PSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEE 211
P T L L W+++ ++ P E + ++ + + G++ G++
Sbjct: 522 PDLSTYNTVLAALARGGMWEQSEKVLAEMEDGRCKPNELTYCSL-LHAYANGKEIGLMHS 580
Query: 212 LAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYA 271
LA ++ G +E + T++ KC+L +A F + + G PD T ++++ +Y
Sbjct: 581 LAEEVYSGVIEPRAVLLKTLVLVCSKCDLLPEAERAFSELKERGFSPDITTLNSMVSIYG 640
Query: 272 RLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLV 331
R V + + + + G+ P T++ L M + D+ +L+E+ + G++P+++
Sbjct: 641 RRQMVAKANGVLDYMKERGFTPSMATYNSLMYMHSRSADFGKSEEILREILAKGIKPDII 700
Query: 332 VYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRM 391
YNT++ A + + A +F EM +SGI P+ T I Y ++A+ + + M
Sbjct: 701 SYNTVIYAYCRNTRMRDASRIFSEMRNSGIVPDVITYNTFIGSYAADSMFEEAIGVVRYM 760
Query: 392 KENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKP 434
++G + YN++++ + +EA+ D++ + P
Sbjct: 761 IKHGCRPNQNTYNSIVDGYCKLNRKDEAKLFVEDLRNLDPHAP 803
>At2g31400 unknown protein
Length = 918
Score = 147 bits (370), Expect = 3e-35
Identities = 127/558 (22%), Positives = 244/558 (42%), Gaps = 31/558 (5%)
Query: 176 FFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCA 235
FF+ ++ + P + I +N + G + L +M + +E D +Y+T++
Sbjct: 326 FFDEMQRNGVQP-DRITFNSLLAVCSRGGLWEAARNLFDEMTNRRIEQDVFSYNTLLDAI 384
Query: 236 KKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDP 295
K D A +M +MP+ V++S ++D +A+ G+ +E +NLF R G D
Sbjct: 385 CKGGQMDLAFEILAQMPVKRIMPNVVSYSTVIDGFAKAGRFDEALNLFGEMRYLGIALDR 444
Query: 296 ITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEE 355
++++ L ++ + G + +L+EM S+G++ ++V YN LL GK GK + +F E
Sbjct: 445 VSYNTLLSIYTKVGRSEEALDILREMASVGIKKDVVTYNALLGGYGKQGKYDEVKKVFTE 504
Query: 356 MIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGL 415
M + PN T + +I Y K K+A+E+++ K G D +LY+ L++ GL
Sbjct: 505 MKREHVLPNLLTYSTLIDGYSKGGLYKEAMEIFREFKSAGLRADVVLYSALIDALCKNGL 564
Query: 416 IEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDK---------------AMKLFE 460
+ A +L +M + E P+ +Y ++++ +G +D+ A+
Sbjct: 565 VGSAVSLIDEMTK-EGISPNVVTYNSIIDAFGRSATMDRSADYSNGGSLPFSSSALSALT 623
Query: 461 E------MSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLS 514
E + FG C E+ +++VF + +KP+ +L+
Sbjct: 624 ETEGNRVIQLFGQLTTESNNRTTKDCEEGMQELSCILEVFRKMHQLEIKPNVVTFSAILN 683
Query: 515 VVSLSQGSKDQEKVLACLQRANPKLVAFIQ-LIVDEETSFETVKEEFKAIMSNAVVEVRR 573
S +D +L L+ + K+ + L++ + + + ++
Sbjct: 684 ACSRCNSFEDASMLLEELRLFDNKVYGVVHGLLMGQRENVWLQAQSLFDKVNEMDGSTAS 743
Query: 574 PFCNCLIDICRNKDLVERAHELLYLGTLYGFYPSLHNKTQYEWCLDVRTLSVGAALTALE 633
F N L D+ + +R EL+ L G + + CLD+ +S GAA +
Sbjct: 744 AFYNALTDMLWHFGQ-KRGAELV---ALEGRSRQVWENVWSDSCLDLHLMSSGAARAMVH 799
Query: 634 EWMTTLTKIVKREEALPDLFLAQTGTGAHKFAQG---LNISFASHLRKLAAPFRQSEDKV 690
W+ + IV LP + TG G H G L + LR + APF S+ +
Sbjct: 800 AWLLNIRSIVYEGHELPKVLSILTGWGKHSKVVGDGALRRAVEVLLRGMDAPFHLSKCNM 859
Query: 691 GCFIATKEDLISWVQSNS 708
G F ++ + +W++ ++
Sbjct: 860 GRFTSSGSVVATWLRESA 877
Score = 134 bits (338), Expect = 1e-31
Identities = 88/308 (28%), Positives = 158/308 (50%), Gaps = 6/308 (1%)
Query: 202 FGRQFGIIEE---LAHQMIDGGVELDNITYSTII-SCAKKCNLFDKAVYWFERMYKTGLM 257
+GR G+ EE + + M + G+ + +TY+ +I +C K F + +F+ M + G+
Sbjct: 278 YGRS-GLHEEAISVFNSMKEYGLRPNLVTYNAVIDACGKGGMEFKQVAKFFDEMQRNGVQ 336
Query: 258 PDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYV 317
PD +TF+++L V +R G E NLF+ + D +++ L + G D +
Sbjct: 337 PDRITFNSLLAVCSRGGLWEAARNLFDEMTNRRIEQDVFSYNTLLDAICKGGQMDLAFEI 396
Query: 318 LQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGK 377
L +M + PN+V Y+T+++ KAG+ A +LF EM GIA + + ++ IY K
Sbjct: 397 LAQMPVKRIMPNVVSYSTVIDGFAKAGRFDEALNLFGEMRYLGIALDRVSYNTLLSIYTK 456
Query: 378 ARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSW 437
S++AL++ + M G D + YN LL G +E + +F +MK+ EH P+
Sbjct: 457 VGRSEEALDILREMASVGIKKDVVTYNALLGGYGKQGKYDEVKKVFTEMKR-EHVLPNLL 515
Query: 438 SYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDIS 497
+Y+ +++ Y G +AM++F E G+ +V+ + LI L K + V + D
Sbjct: 516 TYSTLIDGYSKGGLYKEAMEIFREFKSAGLRADVVLYSALIDALCKNGLVGSAVSLIDEM 575
Query: 498 VERGVKPD 505
+ G+ P+
Sbjct: 576 TKEGISPN 583
Score = 134 bits (336), Expect = 2e-31
Identities = 139/541 (25%), Positives = 247/541 (44%), Gaps = 39/541 (7%)
Query: 7 SSIHISFLDTKTTRTRFKFPPTTTTLKSHRRFLISSSKSIPDSETTPPNNNNNKKNSSLS 66
SS H+S T + + P + + + RF+ ++ + + P N N+ +S S
Sbjct: 41 SSAHLS--QTTPNFSPLQTPKSDFSGRQSTRFVSPATNNHRQTRQNP-NYNHRPYGASSS 97
Query: 67 DQLASLANTTLSTVPENQPKVLSKPKPTWVNPTKTKRPVLSHQRHKRSSVSYNPQL---R 123
+ ++ ++++TV P LS+P P + +P +T + LS R S + ++ R
Sbjct: 98 PRGSAPPPSSVATVA---PAQLSQP-PNF-SPLQTPKSDLSSDFSGRRSTRFVSKMHFGR 152
Query: 124 EFQRFAQR---------LNNCDVSSSDEEFMVCLEEIPSSLTRGN----ALLVLNSLRPW 170
+ A R N D S DE F + S L + + L +
Sbjct: 153 QKTTMATRHSSAAEDALQNAIDFSGDDEMFHSLMLSFESKLCGSDDCTYIIRELGNRNEC 212
Query: 171 QKTHMFFNW-IKTQNLLPMETIFYNVTMKSL-RFGRQFGIIEELAHQMIDGGVELDNITY 228
K F+ + +K + + + + +L R+G+ I + + GG +
Sbjct: 213 DKAVGFYEFAVKRERRKNEQGKLASAMISTLGRYGK-VTIAKRIFETAFAGGYGNTVYAF 271
Query: 229 STIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLG-KVEEVVNLFERGR 287
S +IS + L ++A+ F M + GL P+ VT++A++D + G + ++V F+ +
Sbjct: 272 SALISAYGRSGLHEEAISVFNSMKEYGLRPNLVTYNAVIDACGKGGMEFKQVAKFFDEMQ 331
Query: 288 ATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPG 347
G +PD ITF+ L + G ++ R + EM + ++ ++ YNTLL+A+ K G+
Sbjct: 332 RNGVQPDRITFNSLLAVCSRGGLWEAARNLFDEMTNRRIEQDVFSYNTLLDAICKGGQMD 391
Query: 348 FARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLL 407
A + +M I PN + + VI + KA +AL L+ M+ G +D + YNTLL
Sbjct: 392 LAFEILAQMPVKRIMPNVVSYSTVIDGFAKAGRFDEALNLFGEMRYLGIALDRVSYNTLL 451
Query: 408 NMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGI 467
++ VG EEA + R+M S K D +Y A+L YG +G D+ K+F EM + +
Sbjct: 452 SIYTKVGRSEEALDILREM-ASVGIKKDVVTYNALLGGYGKQGKYDEVKKVFTEMKREHV 510
Query: 468 ELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPD--------DRLC--GCLLSVVS 517
N++ + LI K + +++F G++ D D LC G + S VS
Sbjct: 511 LPNLLTYSTLIDGYSKGGLYKEAMEIFREFKSAGLRADVVLYSALIDALCKNGLVGSAVS 570
Query: 518 L 518
L
Sbjct: 571 L 571
Score = 104 bits (259), Expect = 2e-22
Identities = 104/393 (26%), Positives = 169/393 (42%), Gaps = 13/393 (3%)
Query: 224 DNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEV--TFSAILDVYARLGKVEEVVN 281
D+ TY II N DKAV ++E K +E SA++ R GKV
Sbjct: 197 DDCTY--IIRELGNRNECDKAVGFYEFAVKRERRKNEQGKLASAMISTLGRYGKVTIAKR 254
Query: 282 LFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMG 341
+FE A G+ FS L +G +G ++ V MK G++PNLV YN +++A G
Sbjct: 255 IFETAFAGGYGNTVYAFSALISAYGRSGLHEEAISVFNSMKEYGLRPNLVTYNAVIDACG 314
Query: 342 KAGKP-GFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDF 400
K G F+EM +G+ P+ T +++ + + + A L+ M D
Sbjct: 315 KGGMEFKQVAKFFDEMQRNGVQPDRITFNSLLAVCSRGGLWEAARNLFDEMTNRRIEQDV 374
Query: 401 ILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFE 460
YNTLL+ G ++ A + M + P+ SY+ +++ + G D+A+ LF
Sbjct: 375 FSYNTLLDAICKGGQMDLAFEILAQM-PVKRIMPNVVSYSTVIDGFAKAGRFDEALNLFG 433
Query: 461 EMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQ 520
EM GI L+ + L+ K ++ + + G+K D LL Q
Sbjct: 434 EMRYLGIALDRVSYNTLLSIYTKVGRSEEALDILREMASVGIKKDVVTYNALLGGYG-KQ 492
Query: 521 GSKDQ-EKVLACLQRAN--PKLVAFIQLI--VDEETSFETVKEEFKAIMSNAVVEVRRPF 575
G D+ +KV ++R + P L+ + LI + ++ E F+ S A +
Sbjct: 493 GKYDEVKKVFTEMKREHVLPNLLTYSTLIDGYSKGGLYKEAMEIFREFKS-AGLRADVVL 551
Query: 576 CNCLIDICRNKDLVERAHELLYLGTLYGFYPSL 608
+ LID LV A L+ T G P++
Sbjct: 552 YSALIDALCKNGLVGSAVSLIDEMTKEGISPNV 584
>At2g18940 putative salt-inducible protein
Length = 822
Score = 139 bits (349), Expect = 7e-33
Identities = 88/317 (27%), Positives = 154/317 (47%), Gaps = 2/317 (0%)
Query: 191 IFYNVTMKSL-RFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFE 249
+ YNV + + GR + I + +M G++ D T ST++S + L +A +F
Sbjct: 246 VTYNVILDVFGKMGRSWRKILGVLDEMRSKGLKFDEFTCSTVLSACAREGLLREAKEFFA 305
Query: 250 RMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAG 309
+ G P VT++A+L V+ + G E +++ + D +T++ L + AG
Sbjct: 306 ELKSCGYEPGTVTYNALLQVFGKAGVYTEALSVLKEMEENSCPADSVTYNELVAAYVRAG 365
Query: 310 DYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLT 369
V++ M GV PN + Y T+++A GKAGK A LF M ++G PN T
Sbjct: 366 FSKEAAGVIEMMTKKGVMPNAITYTTVIDAYGKAGKEDEALKLFYSMKEAGCVPNTCTYN 425
Query: 370 AVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQS 429
AV+ + GK S + +++ MK NG + +NT+L +C + G+ + +FR+MK S
Sbjct: 426 AVLSLLGKKSRSNEMIKMLCDMKSNGCSPNRATWNTMLALCGNKGMDKFVNRVFREMK-S 484
Query: 430 EHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDD 489
+PD ++ +++ YG G+ A K++ EM++ G V L+ L + +
Sbjct: 485 CGFEPDRDTFNTLISAYGRCGSEVDASKMYGEMTRAGFNACVTTYNALLNALARKGDWRS 544
Query: 490 LVKVFDISVERGVKPDD 506
V +G KP +
Sbjct: 545 GENVISDMKSKGFKPTE 561
Score = 130 bits (327), Expect = 3e-30
Identities = 94/370 (25%), Positives = 176/370 (47%), Gaps = 8/370 (2%)
Query: 161 LLVLNSL--RPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMID 218
++++NS+ +P FF+ +K++ L S + R + E L
Sbjct: 109 VVLVNSIVEQPLTGLSRFFDSVKSELLRTDLVSLVKGLDDSGHWERAVFLFEWLVLSSNS 168
Query: 219 GGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEE 278
G ++LD+ + + + + A +++ + D ++ IL Y+R GK E+
Sbjct: 169 GALKLDHQVIEIFVRILGRESQYSVAAKLLDKIPLQEYLLDVRAYTTILHAYSRTGKYEK 228
Query: 279 VVNLFERGRATGWKPDPITFSVLGKMFGEAG-DYDGIRYVLQEMKSLGVQPNLVVYNTLL 337
++LFER + G P +T++V+ +FG+ G + I VL EM+S G++ + +T+L
Sbjct: 229 AIDLFERMKEMGPSPTLVTYNVILDVFGKMGRSWRKILGVLDEMRSKGLKFDEFTCSTVL 288
Query: 338 EAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWP 397
A + G A+ F E+ G P T A+++++GKA +AL + K M+EN P
Sbjct: 289 SACAREGLLREAKEFFAELKSCGYEPGTVTYNALLQVFGKAGVYTEALSVLKEMEENSCP 348
Query: 398 MDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMK 457
D + YN L+ G +EA + +M + P++ +YT +++ YG G D+A+K
Sbjct: 349 ADSVTYNELVAAYVRAGFSKEAAGVI-EMMTKKGVMPNAITYTTVIDAYGKAGKEDEALK 407
Query: 458 LFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVS 517
LF M + G N ++ LGK ++++K+ G P+ +L++
Sbjct: 408 LFYSMKEAGCVPNTCTYNAVLSLLGKKSRSNEMIKMLCDMKSNGCSPNRATWNTMLALC- 466
Query: 518 LSQGSKDQEK 527
G+K +K
Sbjct: 467 ---GNKGMDK 473
Score = 95.5 bits (236), Expect = 9e-20
Identities = 98/503 (19%), Positives = 206/503 (40%), Gaps = 96/503 (19%)
Query: 122 LREFQRFAQRLNNCDVSSSDEEFMVCLEEIPSSLTRGNALLVLNSLRPWQKTHMFFNWIK 181
LRE + F L +C + L+ + AL VL +
Sbjct: 297 LREAKEFFAELKSCGYEPGTVTYNALLQVFGKAGVYTEALSVLKEME------------- 343
Query: 182 TQNLLPMETIFYNVTMKS-LRFG---RQFGIIEELAHQMIDGGVELDNITYSTIISCAKK 237
+N P +++ YN + + +R G G+IE M GV + ITY+T+I K
Sbjct: 344 -ENSCPADSVTYNELVAAYVRAGFSKEAAGVIE----MMTKKGVMPNAITYTTVIDAYGK 398
Query: 238 CNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPIT 297
D+A+ F M + G +P+ T++A+L + + + E++ + ++ G P+ T
Sbjct: 399 AGKEDEALKLFYSMKEAGCVPNTCTYNAVLSLLGKKSRSNEMIKMLCDMKSNGCSPNRAT 458
Query: 298 FSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAG------------- 344
++ + + G G + V +EMKS G +P+ +NTL+ A G+ G
Sbjct: 459 WNTMLALCGNKGMDKFVNRVFREMKSCGFEPDRDTFNTLISAYGRCGSEVDASKMYGEMT 518
Query: 345 KPGF----------------------ARSLFEEMIDSGIAPNEKTLTAVIKIYGK----- 377
+ GF ++ +M G P E + + +++ Y K
Sbjct: 519 RAGFNACVTTYNALLNALARKGDWRSGENVISDMKSKGFKPTETSYSLMLQCYAKGGNYL 578
Query: 378 --------------------------ARWSKDALELWKR----MKENGWPMDFILYNTLL 407
A + AL +R K++G+ D +++N++L
Sbjct: 579 GIERIENRIKEGQIFPSWMLLRTLLLANFKCRALAGSERAFTLFKKHGYKPDMVIFNSML 638
Query: 408 NMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGI 467
++ + ++AE + +++ + PD +Y +++++Y G KA ++ + + K +
Sbjct: 639 SIFTRNNMYDQAEGILESIRE-DGLSPDLVTYNSLMDMYVRRGECWKAEEILKTLEKSQL 697
Query: 468 ELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEK 527
+ +++ +I+ + + + V++ ERG++P +S + + E
Sbjct: 698 KPDLVSYNTVIKGFCRRGLMQEAVRMLSEMTERGIRPCIFTYNTFVSGYTAMGMFAEIED 757
Query: 528 VLACLQR--ANPKLVAFIQLIVD 548
V+ C+ + P + F +++VD
Sbjct: 758 VIECMAKNDCRPNELTF-KMVVD 779
Score = 76.6 bits (187), Expect = 4e-14
Identities = 47/176 (26%), Positives = 89/176 (49%), Gaps = 6/176 (3%)
Query: 220 GVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLG---KV 276
G + D + +++++S + N++D+A E + + GL PD VT+++++D+Y R G K
Sbjct: 626 GYKPDMVIFNSMLSIFTRNNMYDQAEGILESIREDGLSPDLVTYNSLMDMYVRRGECWKA 685
Query: 277 EEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTL 336
EE++ E+ + KPD ++++ + K F G +L EM G++P + YNT
Sbjct: 686 EEILKTLEKSQL---KPDLVSYNTVIKGFCRRGLMQEAVRMLSEMTERGIRPCIFTYNTF 742
Query: 337 LEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMK 392
+ G + E M + PNE T V+ Y +A +A++ ++K
Sbjct: 743 VSGYTAMGMFAEIEDVIECMAKNDCRPNELTFKMVVDGYCRAGKYSEAMDFVSKIK 798
Score = 75.5 bits (184), Expect = 1e-13
Identities = 65/312 (20%), Positives = 130/312 (40%), Gaps = 38/312 (12%)
Query: 155 LTRGNALL-VLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELA 213
+T NALL L W+ + +K++ P ET Y++ ++ G + IE +
Sbjct: 526 VTTYNALLNALARKGDWRSGENVISDMKSKGFKPTETS-YSLMLQCYAKGGNYLGIERIE 584
Query: 214 HQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARL 273
+++ +G + + T++ KC + F K G PD V F+++L ++ R
Sbjct: 585 NRIKEGQIFPSWMLLRTLLLANFKCRALAGSERAFTLFKKHGYKPDMVIFNSMLSIFTRN 644
Query: 274 GKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVY 333
++ + E R G PD +T++ L M+ G+ +L+ ++ ++P+LV Y
Sbjct: 645 NMYDQAEGILESIREDGLSPDLVTYNSLMDMYVRRGECWKAEEILKTLEKSQLKPDLVSY 704
Query: 334 NTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKE 393
NT+++ + G A + EM + GI P
Sbjct: 705 NTVIKGFCRRGLMQEAVRMLSEMTERGIRP------------------------------ 734
Query: 394 NGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVD 453
YNT ++ +G+ E E + M +++ C+P+ ++ +++ Y G
Sbjct: 735 -----CIFTYNTFVSGYTAMGMFAEIEDVIECMAKND-CRPNELTFKMVVDGYCRAGKYS 788
Query: 454 KAMKLFEEMSKF 465
+AM ++ F
Sbjct: 789 EAMDFVSKIKTF 800
Score = 52.4 bits (124), Expect = 9e-07
Identities = 43/217 (19%), Positives = 90/217 (40%), Gaps = 2/217 (0%)
Query: 332 VYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRM 391
V + +G+ + A L +++ + + T ++ Y + + A++L++RM
Sbjct: 177 VIEIFVRILGRESQYSVAAKLLDKIPLQEYLLDVRAYTTILHAYSRTGKYEKAIDLFERM 236
Query: 392 KENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGA 451
KE G + YN +L++ +G D +S+ K D ++ + +L+ EG
Sbjct: 237 KEMGPSPTLVTYNVILDVFGKMGRSWRKILGVLDEMRSKGLKFDEFTCSTVLSACAREGL 296
Query: 452 VDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGC 511
+ +A + F E+ G E + L+Q GKA + + V E D
Sbjct: 297 LREAKEFFAELKSCGYEPGTVTYNALLQVFGKAGVYTEALSVLKEMEENSCPADSVTYNE 356
Query: 512 LLSVVSLSQGSKDQEKVLACLQRAN--PKLVAFIQLI 546
L++ + SK+ V+ + + P + + +I
Sbjct: 357 LVAAYVRAGFSKEAAGVIEMMTKKGVMPNAITYTTVI 393
>At4g31850 putative protein
Length = 1112
Score = 135 bits (340), Expect = 8e-32
Identities = 105/351 (29%), Positives = 159/351 (44%), Gaps = 5/351 (1%)
Query: 180 IKTQNLLPMETIFYNVTMKSLRFGRQFGIIE--ELAHQMIDGGVELDNITYSTIISCAKK 237
+K L ++ Y T+ GR I E E+ +M D G D +TY+ +I
Sbjct: 246 LKEMETLGLKPNVYTFTICIRVLGRAGKINEAYEILKRMDDEGCGPDVVTYTVLIDALCT 305
Query: 238 CNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPIT 297
D A FE+M PD VT+ +LD ++ ++ V + G PD +T
Sbjct: 306 ARKLDCAKEVFEKMKTGRHKPDRVTYITLLDRFSDNRDLDSVKQFWSEMEKDGHVPDVVT 365
Query: 298 FSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMI 357
F++L +AG++ L M+ G+ PNL YNTL+ + + + A LF M
Sbjct: 366 FTILVDALCKAGNFGEAFDTLDVMRDQGILPNLHTYNTLICGLLRVHRLDDALELFGNME 425
Query: 358 DSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIE 417
G+ P T I YGK+ S ALE +++MK G + + N L A G
Sbjct: 426 SLGVKPTAYTYIVFIDYYGKSGDSVSALETFEKMKTKGIAPNIVACNASLYSLAKAGRDR 485
Query: 418 EAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCL 477
EA+ +F +K PDS +Y M+ Y G +D+A+KL EM + G E +V+ L
Sbjct: 486 EAKQIFYGLKDI-GLVPDSVTYNMMMKCYSKVGEIDEAIKLLSEMMENGCEPDVIVVNSL 544
Query: 478 IQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKV 528
I L KA +D+ K+F E +KP LL+ L + K QE +
Sbjct: 545 INTLYKADRVDEAWKMFMRMKEMKLKPTVVTYNTLLA--GLGKNGKIQEAI 593
Score = 133 bits (334), Expect = 4e-31
Identities = 86/329 (26%), Positives = 158/329 (47%), Gaps = 2/329 (0%)
Query: 177 FNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAK 236
F +KT P + + Y + R +++ +M G D +T++ ++
Sbjct: 316 FEKMKTGRHKP-DRVTYITLLDRFSDNRDLDSVKQFWSEMEKDGHVPDVVTFTILVDALC 374
Query: 237 KCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPI 296
K F +A + M G++P+ T++ ++ R+ ++++ + LF + G KP
Sbjct: 375 KAGNFGEAFDTLDVMRDQGILPNLHTYNTLICGLLRVHRLDDALELFGNMESLGVKPTAY 434
Query: 297 TFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEM 356
T+ V +G++GD ++MK+ G+ PN+V N L ++ KAG+ A+ +F +
Sbjct: 435 TYIVFIDYYGKSGDSVSALETFEKMKTKGIAPNIVACNASLYSLAKAGRDREAKQIFYGL 494
Query: 357 IDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLI 416
D G+ P+ T ++K Y K +A++L M ENG D I+ N+L+N +
Sbjct: 495 KDIGLVPDSVTYNMMMKCYSKVGEIDEAIKLLSEMMENGCEPDVIVVNSLINTLYKADRV 554
Query: 417 EEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTC 476
+EA +F MK+ + KP +Y +L G G + +A++LFE M + G N +
Sbjct: 555 DEAWKMFMRMKEMK-LKPTVVTYNTLLAGLGKNGKIQEAIELFEGMVQKGCPPNTITFNT 613
Query: 477 LIQCLGKAMEIDDLVKVFDISVERGVKPD 505
L CL K E+ +K+ ++ G PD
Sbjct: 614 LFDCLCKNDEVTLALKMLFKMMDMGCVPD 642
Score = 123 bits (308), Expect = 4e-28
Identities = 80/324 (24%), Positives = 148/324 (44%), Gaps = 36/324 (11%)
Query: 218 DGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVE 277
D GV+ TY+ +I + ++ + A F ++ TG +PD T++ +LD Y + GK++
Sbjct: 778 DLGVQPKLPTYNLLIGGLLEADMIEIAQDVFLQVKSTGCIPDVATYNFLLDAYGKSGKID 837
Query: 278 EVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDG------------------------ 313
E+ L++ + + IT +++ +AG+ D
Sbjct: 838 ELFELYKEMSTHECEANTITHNIVISGLVKAGNVDDALDLYYDLMSDRDFSPTACTYGPL 897
Query: 314 ------------IRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGI 361
+ + + M G +PN +YN L+ GKAG+ A +LF+ M+ G+
Sbjct: 898 IDGLSKSGRLYEAKQLFEGMLDYGCRPNCAIYNILINGFGKAGEADAACALFKRMVKEGV 957
Query: 362 APNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAET 421
P+ KT + ++ + L +K +KE+G D + YN ++N +EEA
Sbjct: 958 RPDLKTYSVLVDCLCMVGRVDEGLHYFKELKESGLNPDVVCYNLIINGLGKSHRLEEALV 1017
Query: 422 LFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCL 481
LF +MK S PD ++Y +++ G G V++A K++ E+ + G+E NV LI+
Sbjct: 1018 LFNEMKTSRGITPDLYTYNSLILNLGIAGMVEEAGKIYNEIQRAGLEPNVFTFNALIRGY 1077
Query: 482 GKAMEIDDLVKVFDISVERGVKPD 505
+ + + V+ V G P+
Sbjct: 1078 SLSGKPEHAYAVYQTMVTGGFSPN 1101
Score = 105 bits (262), Expect = 9e-23
Identities = 94/388 (24%), Positives = 178/388 (45%), Gaps = 12/388 (3%)
Query: 123 REFQRFAQRLNNCDVSSSDEEFMVCLEEIPSSLTRGNALLVLNSLRPWQKTHMFFNWIKT 182
+ ++R + R + + SSD + + L+ V L+ + T F++ K+
Sbjct: 50 KHWRRKSMRCSVVSMKSSDFSGSMIRKSSKPDLSSSEE--VTRGLKSFPDTDSSFSYFKS 107
Query: 183 Q--NL-LPMETIFYNVTMKSLRFGRQFGIIEELAHQ---MIDGGVELDNITYSTIISCAK 236
NL L T N +++LR G +EE+A+ M ++ D TY TI
Sbjct: 108 VAGNLNLVHTTETCNYMLEALRVD---GKLEEMAYVFDLMQKRIIKRDTNTYLTIFKSLS 164
Query: 237 KCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPI 296
+A Y +M + G + + +++ ++ + + E + ++ R G++P
Sbjct: 165 VKGGLKQAPYALRKMREFGFVLNAYSYNGLIHLLLKSRFCTEAMEVYRRMILEGFRPSLQ 224
Query: 297 TFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEM 356
T+S L G+ D D + +L+EM++LG++PN+ + + +G+AGK A + + M
Sbjct: 225 TYSSLMVGLGKRRDIDSVMGLLKEMETLGLKPNVYTFTICIRVLGRAGKINEAYEILKRM 284
Query: 357 IDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLI 416
D G P+ T T +I AR A E++++MK D + Y TLL+ +D +
Sbjct: 285 DDEGCGPDVVTYTVLIDALCTARKLDCAKEVFEKMKTGRHKPDRVTYITLLDRFSDNRDL 344
Query: 417 EEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTC 476
+ + + +M++ H PD ++T +++ G +A + M GI N+
Sbjct: 345 DSVKQFWSEMEKDGHV-PDVVTFTILVDALCKAGNFGEAFDTLDVMRDQGILPNLHTYNT 403
Query: 477 LIQCLGKAMEIDDLVKVFDISVERGVKP 504
LI L + +DD +++F GVKP
Sbjct: 404 LICGLLRVHRLDDALELFGNMESLGVKP 431
Score = 98.2 bits (243), Expect = 1e-20
Identities = 90/383 (23%), Positives = 158/383 (40%), Gaps = 55/383 (14%)
Query: 187 PMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVY 246
P TI +N L + + ++ +M+D G D TY+TII K +A+
Sbjct: 605 PPNTITFNTLFDCLCKNDEVTLALKMLFKMMDMGCVPDVFTYNTIIFGLVKNGQVKEAMC 664
Query: 247 WFERMYKTGLMPDEVTFSAILDVYARLGKVEE--------VVNLFERGRATGWKPDPITF 298
+F +M K + PD VT +L + +E+ + N ++ W+
Sbjct: 665 FFHQMKKL-VYPDFVTLCTLLPGVVKASLIEDAYKIITNFLYNCADQPANLFWE------ 717
Query: 299 SVLGKMFGEAG-----------------------------------DYDGIRYVLQEM-K 322
++G + EAG + G R + ++ K
Sbjct: 718 DLIGSILAEAGIDNAVSFSERLVANGICRDGDSILVPIIRYSCKHNNVSGARTLFEKFTK 777
Query: 323 SLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSK 382
LGVQP L YN L+ + +A A+ +F ++ +G P+ T ++ YGK+
Sbjct: 778 DLGVQPKLPTYNLLIGGLLEADMIEIAQDVFLQVKSTGCIPDVATYNFLLDAYGKSGKID 837
Query: 383 DALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAM 442
+ EL+K M + + I +N +++ G +++A L+ D+ P + +Y +
Sbjct: 838 ELFELYKEMSTHECEANTITHNIVISGLVKAGNVDDALDLYYDLMSDRDFSPTACTYGPL 897
Query: 443 LNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGV 502
++ G + +A +LFE M +G N LI GKA E D +F V+ GV
Sbjct: 898 IDGLSKSGRLYEAKQLFEGMLDYGCRPNCAIYNILINGFGKAGEADAACALFKRMVKEGV 957
Query: 503 KPDDR----LCGCLLSVVSLSQG 521
+PD + L CL V + +G
Sbjct: 958 RPDLKTYSVLVDCLCMVGRVDEG 980
Score = 95.1 bits (235), Expect = 1e-19
Identities = 67/291 (23%), Positives = 135/291 (46%), Gaps = 2/291 (0%)
Query: 177 FNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAK 236
F +KT+ + P + N ++ SL + +++ + + D G+ D++TY+ ++ C
Sbjct: 456 FEKMKTKGIAP-NIVACNASLYSLAKAGRDREAKQIFYGLKDIGLVPDSVTYNMMMKCYS 514
Query: 237 KCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPI 296
K D+A+ M + G PD + +++++ + +V+E +F R + KP +
Sbjct: 515 KVGEIDEAIKLLSEMMENGCEPDVIVVNSLINTLYKADRVDEAWKMFMRMKEMKLKPTVV 574
Query: 297 TFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEM 356
T++ L G+ G + + M G PN + +NTL + + K + A + +M
Sbjct: 575 TYNTLLAGLGKNGKIQEAIELFEGMVQKGCPPNTITFNTLFDCLCKNDEVTLALKMLFKM 634
Query: 357 IDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLI 416
+D G P+ T +I K K+A+ + +MK+ +P DF+ TLL LI
Sbjct: 635 MDMGCVPDVFTYNTIIFGLVKNGQVKEAMCFFHQMKKLVYP-DFVTLCTLLPGVVKASLI 693
Query: 417 EEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGI 467
E+A + + + +P + + ++ +E +D A+ E + GI
Sbjct: 694 EDAYKIITNFLYNCADQPANLFWEDLIGSILAEAGIDNAVSFSERLVANGI 744
Score = 87.0 bits (214), Expect = 3e-17
Identities = 55/200 (27%), Positives = 103/200 (51%), Gaps = 1/200 (0%)
Query: 210 EELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDV 269
++L M+D G + Y+ +I+ K D A F+RM K G+ PD T+S ++D
Sbjct: 911 KQLFEGMLDYGCRPNCAIYNILINGFGKAGEADAACALFKRMVKEGVRPDLKTYSVLVDC 970
Query: 270 YARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMK-SLGVQP 328
+G+V+E ++ F+ + +G PD + ++++ G++ + + EMK S G+ P
Sbjct: 971 LCMVGRVDEGLHYFKELKESGLNPDVVCYNLIINGLGKSHRLEEALVLFNEMKTSRGITP 1030
Query: 329 NLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELW 388
+L YN+L+ +G AG A ++ E+ +G+ PN T A+I+ Y + + A ++
Sbjct: 1031 DLYTYNSLILNLGIAGMVEEAGKIYNEIQRAGLEPNVFTFNALIRGYSLSGKPEHAYAVY 1090
Query: 389 KRMKENGWPMDFILYNTLLN 408
+ M G+ + Y L N
Sbjct: 1091 QTMVTGGFSPNTGTYEQLPN 1110
Score = 83.6 bits (205), Expect = 4e-16
Identities = 47/176 (26%), Positives = 89/176 (49%), Gaps = 1/176 (0%)
Query: 193 YNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMY 252
YN+ + + L +M+ GV D TYS ++ C D+ +++F+ +
Sbjct: 929 YNILINGFGKAGEADAACALFKRMVKEGVRPDLKTYSVLVDCLCMVGRVDEGLHYFKELK 988
Query: 253 KTGLMPDEVTFSAILDVYARLGKVEEVVNLF-ERGRATGWKPDPITFSVLGKMFGEAGDY 311
++GL PD V ++ I++ + ++EE + LF E + G PD T++ L G AG
Sbjct: 989 ESGLNPDVVCYNLIINGLGKSHRLEEALVLFNEMKTSRGITPDLYTYNSLILNLGIAGMV 1048
Query: 312 DGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKT 367
+ + E++ G++PN+ +N L+ +GKP A ++++ M+ G +PN T
Sbjct: 1049 EEAGKIYNEIQRAGLEPNVFTFNALIRGYSLSGKPEHAYAVYQTMVTGGFSPNTGT 1104
Score = 60.5 bits (145), Expect = 3e-09
Identities = 68/296 (22%), Positives = 128/296 (42%), Gaps = 10/296 (3%)
Query: 322 KSLGVQPNLV----VYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGK 377
KS+ NLV N +LEA+ GK +F+ M I + T + K
Sbjct: 106 KSVAGNLNLVHTTETCNYMLEALRVDGKLEEMAYVFDLMQKRIIKRDTNTYLTIFKSLSV 165
Query: 378 ARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSW 437
K A ++M+E G+ ++ YN L+++ EA ++R M E +P
Sbjct: 166 KGGLKQAPYALRKMREFGFVLNAYSYNGLIHLLLKSRFCTEAMEVYRRMIL-EGFRPSLQ 224
Query: 438 SYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDIS 497
+Y++++ G +D M L +EM G++ NV T I+ LG+A +I++ ++
Sbjct: 225 TYSSLMVGLGKRRDIDSVMGLLKEMETLGLKPNVYTFTICIRVLGRAGKINEAYEILKRM 284
Query: 498 VERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQ--RANPKLVAFIQLI--VDEETSF 553
+ G PD L+ + ++ ++V ++ R P V +I L+ +
Sbjct: 285 DDEGCGPDVVTYTVLIDALCTARKLDCAKEVFEKMKTGRHKPDRVTYITLLDRFSDNRDL 344
Query: 554 ETVKEEFKAIMSNAVVEVRRPFCNCLIDICRNKDLVERAHELLYLGTLYGFYPSLH 609
++VK+ + + + V F + +C+ + E A + L + G P+LH
Sbjct: 345 DSVKQFWSEMEKDGHVPDVVTFTILVDALCKAGNFGE-AFDTLDVMRDQGILPNLH 399
>At5g01110 unknown protein (At5g01110)
Length = 729
Score = 134 bits (336), Expect = 2e-31
Identities = 89/370 (24%), Positives = 173/370 (46%), Gaps = 23/370 (6%)
Query: 145 MVCLEEIPSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGR 204
+VC + S TR L K M+FN +K L+P + + Y + ++
Sbjct: 375 LVCFSSMMSLFTRSGNL---------DKALMYFNSVKEAGLIP-DNVIYTILIQGYCRKG 424
Query: 205 QFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFS 264
+ L ++M+ G +D +TY+TI+ K + +A F M + L PD T +
Sbjct: 425 MISVAMNLRNEMLQQGCAMDVVTYNTILHGLCKRKMLGEADKLFNEMTERALFPDSYTLT 484
Query: 265 AILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSL 324
++D + +LG ++ + LF++ + + D +T++ L FG+ GD D + + +M S
Sbjct: 485 ILIDGHCKLGNLQNAMELFQKMKEKRIRLDVVTYNTLLDGFGKVGDIDTAKEIWADMVSK 544
Query: 325 GVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDA 384
+ P + Y+ L+ A+ G A +++EMI I P ++IK Y ++ + D
Sbjct: 545 EILPTPISYSILVNALCSKGHLAEAFRVWDEMISKNIKPTVMICNSMIKGYCRSGNASDG 604
Query: 385 LELWKRMKENGWPMDFILYNTLL-------NMCADVGLIEEAETLFRDMKQSEHCKPDSW 437
++M G+ D I YNTL+ NM GL+++ E ++ PD +
Sbjct: 605 ESFLEKMISEGFVPDCISYNTLIYGFVREENMSKAFGLVKKME------EEQGGLVPDVF 658
Query: 438 SYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDIS 497
+Y ++L+ + + + +A + +M + G+ + TC+I + + ++ D
Sbjct: 659 TYNSILHGFCRQNQMKEAEVVLRKMIERGVNPDRSTYTCMINGFVSQDNLTEAFRIHDEM 718
Query: 498 VERGVKPDDR 507
++RG PDD+
Sbjct: 719 LQRGFSPDDK 728
Score = 124 bits (310), Expect = 2e-28
Identities = 84/313 (26%), Positives = 156/313 (49%), Gaps = 1/313 (0%)
Query: 193 YNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMY 252
YN + L ++ +E+ +M+ G+ D+ TY +++ A K + F M
Sbjct: 308 YNTVINGLCKHGKYERAKEVFAEMLRSGLSPDSTTYRSLLMEACKKGDVVETEKVFSDMR 367
Query: 253 KTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYD 312
++PD V FS+++ ++ R G +++ + F + G PD + +++L + + G
Sbjct: 368 SRDVVPDLVCFSSMMSLFTRSGNLDKALMYFNSVKEAGLIPDNVIYTILIQGYCRKGMIS 427
Query: 313 GIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVI 372
+ EM G ++V YNT+L + K G A LF EM + + P+ TLT +I
Sbjct: 428 VAMNLRNEMLQQGCAMDVVTYNTILHGLCKRKMLGEADKLFNEMTERALFPDSYTLTILI 487
Query: 373 KIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHC 432
+ K ++A+EL+++MKE +D + YNTLL+ VG I+ A+ ++ DM S+
Sbjct: 488 DGHCKLGNLQNAMELFQKMKEKRIRLDVVTYNTLLDGFGKVGDIDTAKEIWADM-VSKEI 546
Query: 433 KPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVK 492
P SY+ ++N S+G + +A ++++EM I+ VM C +I+ ++ D
Sbjct: 547 LPTPISYSILVNALCSKGHLAEAFRVWDEMISKNIKPTVMICNSMIKGYCRSGNASDGES 606
Query: 493 VFDISVERGVKPD 505
+ + G PD
Sbjct: 607 FLEKMISEGFVPD 619
Score = 116 bits (290), Expect = 5e-26
Identities = 76/290 (26%), Positives = 141/290 (48%), Gaps = 1/290 (0%)
Query: 215 QMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLG 274
Q+ + GV D +TY+T+IS L ++A M G P T++ +++ + G
Sbjct: 260 QVQEKGVYPDIVTYNTLISAYSSKGLMEEAFELMNAMPGKGFSPGVYTYNTVINGLCKHG 319
Query: 275 KVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYN 334
K E +F +G PD T+ L + GD V +M+S V P+LV ++
Sbjct: 320 KYERAKEVFAEMLRSGLSPDSTTYRSLLMEACKKGDVVETEKVFSDMRSRDVVPDLVCFS 379
Query: 335 TLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKEN 394
+++ ++G A F + ++G+ P+ T +I+ Y + A+ L M +
Sbjct: 380 SMMSLFTRSGNLDKALMYFNSVKEAGLIPDNVIYTILIQGYCRKGMISVAMNLRNEMLQQ 439
Query: 395 GWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDK 454
G MD + YNT+L+ ++ EA+ LF +M + PDS++ T +++ + G +
Sbjct: 440 GCAMDVVTYNTILHGLCKRKMLGEADKLFNEMTE-RALFPDSYTLTILIDGHCKLGNLQN 498
Query: 455 AMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKP 504
AM+LF++M + I L+V+ L+ GK +ID +++ V + + P
Sbjct: 499 AMELFQKMKEKRIRLDVVTYNTLLDGFGKVGDIDTAKEIWADMVSKEILP 548
Score = 79.3 bits (194), Expect = 7e-15
Identities = 65/272 (23%), Positives = 118/272 (42%), Gaps = 14/272 (5%)
Query: 221 VELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTF-------------SAIL 267
+E+ N ST +C ++FD + + + K + T +A++
Sbjct: 148 LEIVNSLDSTFSNCGSNDSVFDLLIRTYVQARKLREAHEAFTLLRSKGFTVSIDACNALI 207
Query: 268 DVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQ 327
R+G VE +++ +G + T +++ + G + + L +++ GV
Sbjct: 208 GSLVRIGWVELAWGVYQEISRSGVGINVYTLNIMVNALCKDGKMEKVGTFLSQVQEKGVY 267
Query: 328 PNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALEL 387
P++V YNTL+ A G A L M G +P T VI K + A E+
Sbjct: 268 PDIVTYNTLISAYSSKGLMEEAFELMNAMPGKGFSPGVYTYNTVINGLCKHGKYERAKEV 327
Query: 388 WKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYG 447
+ M +G D Y +LL G + E E +F DM+ S PD +++M++++
Sbjct: 328 FAEMLRSGLSPDSTTYRSLLMEACKKGDVVETEKVFSDMR-SRDVVPDLVCFSSMMSLFT 386
Query: 448 SEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQ 479
G +DKA+ F + + G+ + + T LIQ
Sbjct: 387 RSGNLDKALMYFNSVKEAGLIPDNVIYTILIQ 418
Score = 51.6 bits (122), Expect = 2e-06
Identities = 47/217 (21%), Positives = 90/217 (40%), Gaps = 3/217 (1%)
Query: 332 VYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRM 391
V++ L+ +A K A F + G + A+I + W + A +++ +
Sbjct: 167 VFDLLIRTYVQARKLREAHEAFTLLRSKGFTVSIDACNALIGSLVRIGWVELAWGVYQEI 226
Query: 392 KENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGA 451
+G ++ N ++N G +E+ T F Q + PD +Y +++ Y S+G
Sbjct: 227 SRSGVGINVYTLNIMVNALCKDGKMEKVGT-FLSQVQEKGVYPDIVTYNTLISAYSSKGL 285
Query: 452 VDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGC 511
+++A +L M G V +I L K + + +VF + G+ PD
Sbjct: 286 MEEAFELMNAMPGKGFSPGVYTYNTVINGLCKHGKYERAKEVFAEMLRSGLSPDSTTYRS 345
Query: 512 LLSVVSLSQGSKDQEKVLACLQRAN--PKLVAFIQLI 546
LL + EKV + ++ + P LV F ++
Sbjct: 346 LLMEACKKGDVVETEKVFSDMRSRDVVPDLVCFSSMM 382
Score = 38.5 bits (88), Expect = 0.013
Identities = 41/236 (17%), Positives = 94/236 (39%), Gaps = 40/236 (16%)
Query: 359 SGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEE 418
S N+ +I+ Y +AR ++A E + ++ G+ + N L+ +G +E
Sbjct: 159 SNCGSNDSVFDLLIRTYVQARKLREAHEAFTLLRSKGFTVSIDACNALIGSLVRIGWVEL 218
Query: 419 AETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLI 478
A W +++E+S+ G+ +NV ++
Sbjct: 219 A-----------------WG-------------------VYQEISRSGVGINVYTLNIMV 242
Query: 479 QCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACL--QRAN 536
L K +++ + E+GV PD L+S S ++ +++ + + +
Sbjct: 243 NALCKDGKMEKVGTFLSQVQEKGVYPDIVTYNTLISAYSSKGLMEEAFELMNAMPGKGFS 302
Query: 537 PKLVAFIQLI--VDEETSFETVKEEFKAIMSNAVVEVRRPFCNCLIDICRNKDLVE 590
P + + +I + + +E KE F ++ + + + + L++ C+ D+VE
Sbjct: 303 PGVYTYNTVINGLCKHGKYERAKEVFAEMLRSGLSPDSTTYRSLLMEACKKGDVVE 358
>At3g59040 unknown protein (At3g59040)
Length = 526
Score = 130 bits (328), Expect = 2e-30
Identities = 91/361 (25%), Positives = 172/361 (47%), Gaps = 8/361 (2%)
Query: 151 IPSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIE 210
+P L G L+ L+ W W++ QN I + + + + F E
Sbjct: 44 LPRDLVLGT-LVRFKQLKKWNLVSEILEWLRYQNWWNFSEIDFLMLITAYGKLGNFNGAE 102
Query: 211 ELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVY 270
+ + G + I+Y+ ++ + + A F RM +G P +T+ IL +
Sbjct: 103 RVLSVLSKMGSTPNVISYTALMESYGRGGKCNNAEAIFRRMQSSGPEPSAITYQIILKTF 162
Query: 271 ARLGKVEEVVNLFER---GRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQ 327
K +E +FE + + KPD + ++ M+ +AG+Y+ R V M GV
Sbjct: 163 VEGDKFKEAEEVFETLLDEKKSPLKPDQKMYHMMIYMYKKAGNYEKARKVFSSMVGKGVP 222
Query: 328 PNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALEL 387
+ V YN+L+ + ++++M S I P+ + +IK YG+AR ++AL +
Sbjct: 223 QSTVTYNSLMSFETSYKE---VSKIYDQMQRSDIQPDVVSYALLIKAYGRARREEEALSV 279
Query: 388 WKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYG 447
++ M + G YN LL+ A G++E+A+T+F+ M++ + PD WSYT ML+ Y
Sbjct: 280 FEEMLDAGVRPTHKAYNILLDAFAISGMVEQAKTVFKSMRR-DRIFPDLWSYTTMLSAYV 338
Query: 448 SEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDR 507
+ ++ A K F+ + G E N++ LI+ KA +++ +++V++ G+K +
Sbjct: 339 NASDMEGAEKFFKRIKVDGFEPNIVTYGTLIKGYAKANDVEKMMEVYEKMRLSGIKANQT 398
Query: 508 L 508
+
Sbjct: 399 I 399
Score = 100 bits (248), Expect = 4e-21
Identities = 63/237 (26%), Positives = 114/237 (47%), Gaps = 3/237 (1%)
Query: 186 LPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAV 245
+P T+ YN M F + + ++ QM ++ D ++Y+ +I + ++A+
Sbjct: 221 VPQSTVTYNSLMS---FETSYKEVSKIYDQMQRSDIQPDVVSYALLIKAYGRARREEEAL 277
Query: 246 YWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMF 305
FE M G+ P ++ +LD +A G VE+ +F+ R PD +++ + +
Sbjct: 278 SVFEEMLDAGVRPTHKAYNILLDAFAISGMVEQAKTVFKSMRRDRIFPDLWSYTTMLSAY 337
Query: 306 GEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNE 365
A D +G + +K G +PN+V Y TL++ KA ++E+M SGI N+
Sbjct: 338 VNASDMEGAEKFFKRIKVDGFEPNIVTYGTLIKGYAKANDVEKMMEVYEKMRLSGIKANQ 397
Query: 366 KTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETL 422
LT ++ G+ + AL +K M+ G P D N LL++ + +EEA+ L
Sbjct: 398 TILTTIMDASGRCKNFGSALGWYKEMESCGVPPDQKAKNVLLSLASTQDELEEAKEL 454
Score = 99.0 bits (245), Expect = 8e-21
Identities = 83/372 (22%), Positives = 160/372 (42%), Gaps = 43/372 (11%)
Query: 191 IFYNVTMKSLRFGRQFGIIEELAHQMID---GGVELDNITYSTIISCAKKCNLFDKAVYW 247
I Y + +K+ G +F EE+ ++D ++ D Y +I KK ++KA
Sbjct: 153 ITYQIILKTFVEGDKFKEAEEVFETLLDEKKSPLKPDQKMYHMMIYMYKKAGNYEKARKV 212
Query: 248 FERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGE 307
F M G+ VT+++++ +EV ++++ + + +PD +++++L K +G
Sbjct: 213 FSSMVGKGVPQSTVTYNSLMSFET---SYKEVSKIYDQMQRSDIQPDVVSYALLIKAYGR 269
Query: 308 AGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKT 367
A + V +EM GV+P YN LL+A +G A+++F+ M I P+ +
Sbjct: 270 ARREEEALSVFEEMLDAGVRPTHKAYNILLDAFAISGMVEQAKTVFKSMRRDRIFPDLWS 329
Query: 368 LTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMK 427
T ++ Y A + A + +KR+K +G+ + + Y TL+
Sbjct: 330 YTTMLSAYVNASDMEGAEKFFKRIKVDGFEPNIVTYGTLIKG------------------ 371
Query: 428 QSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEI 487
Y V+K M+++E+M GI+ N T ++ G+
Sbjct: 372 ------------------YAKANDVEKMMEVYEKMRLSGIKANQTILTTIMDASGRCKNF 413
Query: 488 DDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQRANPKLVAFIQLIV 547
+ + GV PD + LLS+ S +Q ++ K L ++ ++A +
Sbjct: 414 GSALGWYKEMESCGVPPDQKAKNVLLSLAS-TQDELEEAKELTGIRNETATIIARVYGSD 472
Query: 548 DEETSFETVKEE 559
D+E E + E
Sbjct: 473 DDEEGVEDISSE 484
Score = 92.8 bits (229), Expect = 6e-19
Identities = 59/242 (24%), Positives = 123/242 (50%), Gaps = 8/242 (3%)
Query: 291 WKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFAR 350
W I F +L +G+ G+++G VL + +G PN++ Y L+E+ G+ GK A
Sbjct: 78 WNFSEIDFLMLITAYGKLGNFNGAERVLSVLSKMGSTPNVISYTALMESYGRGGKCNNAE 137
Query: 351 SLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRM---KENGWPMDFILYNTLL 407
++F M SG P+ T ++K + + K+A E+++ + K++ D +Y+ ++
Sbjct: 138 AIFRRMQSSGPEPSAITYQIILKTFVEGDKFKEAEEVFETLLDEKKSPLKPDQKMYHMMI 197
Query: 408 NMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGI 467
M G E+A +F M + + +Y ++++ E + + K++++M + I
Sbjct: 198 YMYKKAGNYEKARKVFSSMV-GKGVPQSTVTYNSLMSF---ETSYKEVSKIYDQMQRSDI 253
Query: 468 ELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEK 527
+ +V+ LI+ G+A ++ + VF+ ++ GV+P + LL ++S G +Q K
Sbjct: 254 QPDVVSYALLIKAYGRARREEEALSVFEEMLDAGVRPTHKAYNILLDAFAIS-GMVEQAK 312
Query: 528 VL 529
+
Sbjct: 313 TV 314
Score = 31.2 bits (69), Expect = 2.1
Identities = 25/137 (18%), Positives = 58/137 (42%), Gaps = 7/137 (5%)
Query: 439 YTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISV 498
+ ++ YG G + A ++ +SK G NV+ T L++ G+ + ++ +F
Sbjct: 85 FLMLITAYGKLGNFNGAERVLSVLSKMGSTPNVISYTALMESYGRGGKCNNAEAIFRRMQ 144
Query: 499 ERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACL--QRANP-----KLVAFIQLIVDEET 551
G +P +L K+ E+V L ++ +P K+ + + +
Sbjct: 145 SSGPEPSAITYQIILKTFVEGDKFKEAEEVFETLLDEKKSPLKPDQKMYHMMIYMYKKAG 204
Query: 552 SFETVKEEFKAIMSNAV 568
++E ++ F +++ V
Sbjct: 205 NYEKARKVFSSMVGKGV 221
>At1g51970 hypothetical protein
Length = 782
Score = 130 bits (327), Expect = 3e-30
Identities = 81/297 (27%), Positives = 154/297 (51%), Gaps = 10/297 (3%)
Query: 212 LAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYA 271
L ++MI G+ L+ + Y+T++ K + DKA+ F RM +TG P+E T+S +L++
Sbjct: 424 LFNEMITEGLTLNVVGYNTLMQVLAKGKMVDKAIQVFSRMVETGCRPNEYTYSLLLNLLV 483
Query: 272 RLG---KVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQP 328
G +++ VV + +R G +S L + + G + +M S V+
Sbjct: 484 AEGQLVRLDGVVEISKRYMTQG------IYSYLVRTLSKLGHVSEAHRLFCDMWSFPVKG 537
Query: 329 NLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELW 388
Y ++LE++ AGK A + ++ + G+ + V GK + +L+
Sbjct: 538 ERDSYMSMLESLCGAGKTIEAIEMLSKIHEKGVVTDTMMYNTVFSALGKLKQISHIHDLF 597
Query: 389 KRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGS 448
++MK++G D YN L+ VG ++EA +F ++++S+ CKPD SY +++N G
Sbjct: 598 EKMKKDGPSPDIFTYNILIASFGRVGEVDEAINIFEELERSD-CKPDIISYNSLINCLGK 656
Query: 449 EGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPD 505
G VD+A F+EM + G+ +V+ + L++C GK ++ +F+ + +G +P+
Sbjct: 657 NGDVDEAHVRFKEMQEKGLNPDVVTYSTLMECFGKTERVEMAYSLFEEMLVKGCQPN 713
Score = 117 bits (293), Expect = 2e-26
Identities = 64/196 (32%), Positives = 105/196 (52%)
Query: 211 ELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVY 270
E+ ++ + GV D + Y+T+ S K FE+M K G PD T++ ++ +
Sbjct: 560 EMLSKIHEKGVVTDTMMYNTVFSALGKLKQISHIHDLFEKMKKDGPSPDIFTYNILIASF 619
Query: 271 ARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNL 330
R+G+V+E +N+FE + KPD I+++ L G+ GD D +EM+ G+ P++
Sbjct: 620 GRVGEVDEAINIFEELERSDCKPDIISYNSLINCLGKNGDVDEAHVRFKEMQEKGLNPDV 679
Query: 331 VVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKR 390
V Y+TL+E GK + A SLFEEM+ G PN T ++ K + +A++L+ +
Sbjct: 680 VTYSTLMECFGKTERVEMAYSLFEEMLVKGCQPNIVTYNILLDCLEKNGRTAEAVDLYSK 739
Query: 391 MKENGWPMDFILYNTL 406
MK+ G D I Y L
Sbjct: 740 MKQQGLTPDSITYTVL 755
Score = 101 bits (251), Expect = 2e-21
Identities = 53/200 (26%), Positives = 106/200 (52%), Gaps = 1/200 (0%)
Query: 243 KAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLG 302
+A+ ++++ G++ D + ++ + +L ++ + +LFE+ + G PD T+++L
Sbjct: 557 EAIEMLSKIHEKGVVTDTMMYNTVFSALGKLKQISHIHDLFEKMKKDGPSPDIFTYNILI 616
Query: 303 KMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIA 362
FG G+ D + +E++ +P+++ YN+L+ +GK G A F+EM + G+
Sbjct: 617 ASFGRVGEVDEAINIFEELERSDCKPDIISYNSLINCLGKNGDVDEAHVRFKEMQEKGLN 676
Query: 363 PNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETL 422
P+ T + +++ +GK + A L++ M G + + YN LL+ G EA L
Sbjct: 677 PDVVTYSTLMECFGKTERVEMAYSLFEEMLVKGCQPNIVTYNILLDCLEKNGRTAEAVDL 736
Query: 423 FRDMKQSEHCKPDSWSYTAM 442
+ MKQ + PDS +YT +
Sbjct: 737 YSKMKQ-QGLTPDSITYTVL 755
Score = 98.2 bits (243), Expect = 1e-20
Identities = 60/206 (29%), Positives = 102/206 (49%), Gaps = 4/206 (1%)
Query: 311 YDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTA 370
+D +R +L M V N+ N L+ G L ++ + N T
Sbjct: 281 FDRVRSILDSMVKSNVHGNISTVNILIGFFGNTEDLQMCLRLVKKW---DLKMNSFTYKC 337
Query: 371 VIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSE 430
+++ Y ++R A +++ ++ G +D YN LL+ A I++A +F DMK+
Sbjct: 338 LLQAYLRSRDYSKAFDVYCEIRRGGHKLDIFAYNMLLDALAKDEKIDQACQVFEDMKK-R 396
Query: 431 HCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDL 490
HC+ D ++YT M+ G G D+A+ LF EM G+ LNV+G L+Q L K +D
Sbjct: 397 HCRRDEYTYTIMIRTMGRIGKCDEAVGLFNEMITEGLTLNVVGYNTLMQVLAKGKMVDKA 456
Query: 491 VKVFDISVERGVKPDDRLCGCLLSVV 516
++VF VE G +P++ LL+++
Sbjct: 457 IQVFSRMVETGCRPNEYTYSLLLNLL 482
Score = 94.7 bits (234), Expect = 2e-19
Identities = 54/195 (27%), Positives = 101/195 (51%), Gaps = 4/195 (2%)
Query: 189 ETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWF 248
+T+ YN +L +Q I +L +M G D TY+ +I+ + D+A+ F
Sbjct: 573 DTMMYNTVFSALGKLKQISHIHDLFEKMKKDGPSPDIFTYNILIASFGRVGEVDEAINIF 632
Query: 249 ERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEA 308
E + ++ PD ++++++++ + G V+E F+ + G PD +T+S L + FG+
Sbjct: 633 EELERSDCKPDIISYNSLINCLGKNGDVDEAHVRFKEMQEKGLNPDVVTYSTLMECFGKT 692
Query: 309 GDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTL 368
+ + +EM G QPN+V YN LL+ + K G+ A L+ +M G+ P+ T
Sbjct: 693 ERVEMAYSLFEEMLVKGCQPNIVTYNILLDCLEKNGRTAEAVDLYSKMKQQGLTPDSITY 752
Query: 369 TAVIKI----YGKAR 379
T + ++ +GK+R
Sbjct: 753 TVLERLQSVSHGKSR 767
Score = 93.2 bits (230), Expect = 5e-19
Identities = 67/288 (23%), Positives = 135/288 (46%), Gaps = 10/288 (3%)
Query: 221 VELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVV 280
+++++ TY ++ + + KA + + + G D ++ +LD A+ K+++
Sbjct: 328 LKMNSFTYKCLLQAYLRSRDYSKAFDVYCEIRRGGHKLDIFAYNMLLDALAKDEKIDQAC 387
Query: 281 NLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAM 340
+FE + + D T++++ + G G D + EM + G+ N+V YNTL++ +
Sbjct: 388 QVFEDMKKRHCRRDEYTYTIMIRTMGRIGKCDEAVGLFNEMITEGLTLNVVGYNTLMQVL 447
Query: 341 GKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIY---GKARWSKDALELWKRMKENGWP 397
K A +F M+++G PNE T + ++ + G+ +E+ KR G
Sbjct: 448 AKGKMVDKAIQVFSRMVETGCRPNEYTYSLLLNLLVAEGQLVRLDGVVEISKRYMTQG-- 505
Query: 398 MDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMK 457
+Y+ L+ + +G + EA LF DM S K + SY +ML G +A++
Sbjct: 506 ----IYSYLVRTLSKLGHVSEAHRLFCDM-WSFPVKGERDSYMSMLESLCGAGKTIEAIE 560
Query: 458 LFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPD 505
+ ++ + G+ + M + LGK +I + +F+ + G PD
Sbjct: 561 MLSKIHEKGVVTDTMMYNTVFSALGKLKQISHIHDLFEKMKKDGPSPD 608
Score = 84.0 bits (206), Expect = 3e-16
Identities = 61/278 (21%), Positives = 124/278 (43%), Gaps = 8/278 (2%)
Query: 190 TIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFE 249
++ N+ + + R G++E M G YS ++ K +A F
Sbjct: 476 SLLLNLLVAEGQLVRLDGVVEISKRYMTQG-------IYSYLVRTLSKLGHVSEAHRLFC 528
Query: 250 RMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAG 309
M+ + + ++ ++L+ GK E + + + G D + ++ + G+
Sbjct: 529 DMWSFPVKGERDSYMSMLESLCGAGKTIEAIEMLSKIHEKGVVTDTMMYNTVFSALGKLK 588
Query: 310 DYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLT 369
I + ++MK G P++ YN L+ + G+ G+ A ++FEE+ S P+ +
Sbjct: 589 QISHIHDLFEKMKKDGPSPDIFTYNILIASFGRVGEVDEAINIFEELERSDCKPDIISYN 648
Query: 370 AVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQS 429
++I GK +A +K M+E G D + Y+TL+ +E A +LF +M
Sbjct: 649 SLINCLGKNGDVDEAHVRFKEMQEKGLNPDVVTYSTLMECFGKTERVEMAYSLFEEM-LV 707
Query: 430 EHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGI 467
+ C+P+ +Y +L+ G +A+ L+ +M + G+
Sbjct: 708 KGCQPNIVTYNILLDCLEKNGRTAEAVDLYSKMKQQGL 745
>At1g12620 putative protein
Length = 621
Score = 130 bits (326), Expect = 3e-30
Identities = 87/315 (27%), Positives = 149/315 (46%), Gaps = 1/315 (0%)
Query: 191 IFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFER 250
+ Y +K + Q + EL +M + ++LD + YS II K D A F
Sbjct: 213 VTYGPVLKVMCKSGQTALAMELLRKMEERKIKLDAVKYSIIIDGLCKDGSLDNAFNLFNE 272
Query: 251 MYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGD 310
M G D + ++ ++ + G+ ++ L PD + FS L F + G
Sbjct: 273 MEIKGFKADIIIYTTLIRGFCYAGRWDDGAKLLRDMIKRKITPDVVAFSALIDCFVKEGK 332
Query: 311 YDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTA 370
+ +EM G+ P+ V Y +L++ K + A + + M+ G PN +T
Sbjct: 333 LREAEELHKEMIQRGISPDTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNI 392
Query: 371 VIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSE 430
+I Y KA D LEL+++M G D + YNTL+ ++G +E A+ LF++M S
Sbjct: 393 LINGYCKANLIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMV-SR 451
Query: 431 HCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDL 490
+PD SY +L+ G +KA+++FE++ K +EL++ +I + A ++DD
Sbjct: 452 RVRPDIVSYKILLDGLCDNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDA 511
Query: 491 VKVFDISVERGVKPD 505
+F +GVKPD
Sbjct: 512 WDLFCSLPLKGVKPD 526
Score = 119 bits (298), Expect = 6e-27
Identities = 84/311 (27%), Positives = 146/311 (46%), Gaps = 1/311 (0%)
Query: 204 RQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTF 263
+Q+ ++ +L QM G+ + T S +I+C +C A ++ K G PD VTF
Sbjct: 86 KQYDLVLDLCKQMELKGIAHNLYTLSIMINCCCRCRKLSLAFSAMGKIIKLGYEPDTVTF 145
Query: 264 SAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKS 323
S +++ G+V E + L +R G KP IT + L G ++ M
Sbjct: 146 STLINGLCLEGRVSEALELVDRMVEMGHKPTLITLNALVNGLCLNGKVSDAVLLIDRMVE 205
Query: 324 LGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKD 383
G QPN V Y +L+ M K+G+ A L +M + I + + +I K +
Sbjct: 206 TGFQPNEVTYGPVLKVMCKSGQTALAMELLRKMEERKIKLDAVKYSIIIDGLCKDGSLDN 265
Query: 384 ALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAML 443
A L+ M+ G+ D I+Y TL+ G ++ L RDM + + PD +++A++
Sbjct: 266 AFNLFNEMEIKGFKADIIIYTTLIRGFCYAGRWDDGAKLLRDMIKRK-ITPDVVAFSALI 324
Query: 444 NIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVK 503
+ + EG + +A +L +EM + GI + + T LI K ++D + D+ V +G
Sbjct: 325 DCFVKEGKLREAEELHKEMIQRGISPDTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCG 384
Query: 504 PDDRLCGCLLS 514
P+ R L++
Sbjct: 385 PNIRTFNILIN 395
Score = 114 bits (285), Expect = 2e-25
Identities = 74/272 (27%), Positives = 133/272 (48%), Gaps = 1/272 (0%)
Query: 210 EELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDV 269
EEL +MI G+ D +TY+++I K N DKA + + M G P+ TF+ +++
Sbjct: 337 EELHKEMIQRGISPDTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILING 396
Query: 270 YARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPN 329
Y + +++ + LF + G D +T++ L + F E G + + + QEM S V+P+
Sbjct: 397 YCKANLIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRPD 456
Query: 330 LVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWK 389
+V Y LL+ + G+P A +FE++ S + + +I A DA +L+
Sbjct: 457 IVSYKILLDGLCDNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFC 516
Query: 390 RMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSE 449
+ G D YN ++ G + EA+ LFR M++ H P+ +Y ++ + E
Sbjct: 517 SLPLKGVKPDVKTYNIMIGGLCKKGSLSEADLLFRKMEEDGH-SPNGCTYNILIRAHLGE 575
Query: 450 GAVDKAMKLFEEMSKFGIELNVMGCTCLIQCL 481
G K+ KL EE+ + G ++ ++ L
Sbjct: 576 GDATKSAKLIEEIKRCGFSVDASTVKMVVDML 607
Score = 101 bits (252), Expect = 1e-21
Identities = 69/280 (24%), Positives = 132/280 (46%), Gaps = 1/280 (0%)
Query: 226 ITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFER 285
I +S + S + +D + ++M G+ + T S +++ R K+ + +
Sbjct: 73 IDFSRLFSVVARTKQYDLVLDLCKQMELKGIAHNLYTLSIMINCCCRCRKLSLAFSAMGK 132
Query: 286 GRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGK 345
G++PD +TFS L G ++ M +G +P L+ N L+ + GK
Sbjct: 133 IIKLGYEPDTVTFSTLINGLCLEGRVSEALELVDRMVEMGHKPTLITLNALVNGLCLNGK 192
Query: 346 PGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNT 405
A L + M+++G PNE T V+K+ K+ + A+EL ++M+E +D + Y+
Sbjct: 193 VSDAVLLIDRMVETGFQPNEVTYGPVLKVMCKSGQTALAMELLRKMEERKIKLDAVKYSI 252
Query: 406 LLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKF 465
+++ G ++ A LF +M + + K D YT ++ + G D KL +M K
Sbjct: 253 IIDGLCKDGSLDNAFNLFNEM-EIKGFKADIIIYTTLIRGFCYAGRWDDGAKLLRDMIKR 311
Query: 466 GIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPD 505
I +V+ + LI C K ++ + ++ ++RG+ PD
Sbjct: 312 KITPDVVAFSALIDCFVKEGKLREAEELHKEMIQRGISPD 351
Score = 75.1 bits (183), Expect = 1e-13
Identities = 80/349 (22%), Positives = 151/349 (42%), Gaps = 11/349 (3%)
Query: 264 SAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKS 323
S I+D+ K ++ V+LF+ + +P I FS L + YD + + ++M+
Sbjct: 46 SGIVDI-----KEDDAVDLFQEMTRSRPRPRLIDFSRLFSVVARTKQYDLVLDLCKQMEL 100
Query: 324 LGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKD 383
G+ NL + ++ + K A S ++I G P+ T + +I +
Sbjct: 101 KGIAHNLYTLSIMINCCCRCRKLSLAFSAMGKIIKLGYEPDTVTFSTLINGLCLEGRVSE 160
Query: 384 ALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAML 443
ALEL RM E G I N L+N G + +A L M ++ +P+ +Y +L
Sbjct: 161 ALELVDRMVEMGHKPTLITLNALVNGLCLNGKVSDAVLLIDRMVETGF-QPNEVTYGPVL 219
Query: 444 NIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVK 503
+ G AM+L +M + I+L+ + + +I L K +D+ +F+ +G K
Sbjct: 220 KVMCKSGQTALAMELLRKMEERKIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEIKGFK 279
Query: 504 PDDRLCGCLLSVVSLSQGSKDQEKVLACL--QRANPKLVAFIQLI--VDEETSFETVKEE 559
D + L+ + D K+L + ++ P +VAF LI +E +E
Sbjct: 280 ADIIIYTTLIRGFCYAGRWDDGAKLLRDMIKRKITPDVVAFSALIDCFVKEGKLREAEEL 339
Query: 560 FKAIMSNAVVEVRRPFCNCLIDICRNKDLVERAHELLYLGTLYGFYPSL 608
K ++ + + + + C+ L ++A+ +L L G P++
Sbjct: 340 HKEMIQRGISPDTVTYTSLIDGFCKENQL-DKANHMLDLMVSKGCGPNI 387
Score = 65.5 bits (158), Expect = 1e-10
Identities = 50/203 (24%), Positives = 97/203 (47%), Gaps = 2/203 (0%)
Query: 211 ELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVY 270
EL +M GV D +TY+T+I + + A F+ M + PD V++ +LD
Sbjct: 408 ELFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLDGL 467
Query: 271 ARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNL 330
G+ E+ + +FE+ + + D ++++ A D + + GV+P++
Sbjct: 468 CDNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPDV 527
Query: 331 VVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIY-GKARWSKDALELWK 389
YN ++ + K G A LF +M + G +PN T +I+ + G+ +K A +L +
Sbjct: 528 KTYNIMIGGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSA-KLIE 586
Query: 390 RMKENGWPMDFILYNTLLNMCAD 412
+K G+ +D +++M +D
Sbjct: 587 EIKRCGFSVDASTVKMVVDMLSD 609
Score = 57.4 bits (137), Expect = 3e-08
Identities = 44/199 (22%), Positives = 88/199 (44%)
Query: 189 ETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWF 248
+T+ YN ++ + + +EL +M+ V D ++Y ++ +KA+ F
Sbjct: 421 DTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLDGLCDNGEPEKALEIF 480
Query: 249 ERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEA 308
E++ K+ + D ++ I+ KV++ +LF G KPD T++++ +
Sbjct: 481 EKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPDVKTYNIMIGGLCKK 540
Query: 309 GDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTL 368
G + ++M+ G PN YN L+ A G + L EE+ G + + T+
Sbjct: 541 GSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLIEEIKRCGFSVDASTV 600
Query: 369 TAVIKIYGKARWSKDALEL 387
V+ + R K L++
Sbjct: 601 KMVVDMLSDGRLKKSFLDM 619
>At1g79490 unknown protein
Length = 836
Score = 127 bits (320), Expect = 2e-29
Identities = 99/417 (23%), Positives = 184/417 (43%), Gaps = 19/417 (4%)
Query: 177 FNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNIT---YSTIIS 233
F W K Q Y V L GR F I+ L +M+ +++ Y+ +I
Sbjct: 192 FRWAKKQPWYLPSDECYVVLFDGLNQGRDFVGIQSLFEEMVQDSSSHGDLSFNAYNQVIQ 251
Query: 234 CAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKP 293
K + A F++ ++G D T++ ++ ++ G + ++E T
Sbjct: 252 YLAKAEKLEVAFCCFKKAQESGCKIDTQTYNNLMMLFLNKGLPYKAFEIYESMEKTDSLL 311
Query: 294 DPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLF 353
D T+ ++ ++G D + Q+MK ++P+ V+++L+++MGKAG+ + ++
Sbjct: 312 DGSTYELIIPSLAKSGRLDAAFKLFQQMKERKLRPSFSVFSSLVDSMGKAGRLDTSMKVY 371
Query: 354 EEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADV 413
EM G P+ ++I Y KA AL LW MK++G+ +F LY ++ A
Sbjct: 372 MEMQGFGHRPSATMFVSLIDSYAKAGKLDTALRLWDEMKKSGFRPNFGLYTMIIESHAKS 431
Query: 414 GLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMG 473
G +E A T+F+DM+++ P +Y+ +L ++ G VD AMK++ M+ G+ +
Sbjct: 432 GKLEVAMTVFKDMEKAGFL-PTPSTYSCLLEMHAGSGQVDSAMKIYNSMTNAGLRPGLSS 490
Query: 474 CTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQE-KVLACL 532
L+ L +D K+ G D +C + ++ + S D K L +
Sbjct: 491 YISLLTLLANKRLVDVAGKILLEMKAMGYSVD--VCASDVLMIYIKDASVDLALKWLRFM 548
Query: 533 QRANPKLVAFIQLIVDEETSFETVKEEFKAIMSNAVVEVRRPFCNCLIDICRNKDLV 589
+ K FI +++ F++ M N + + RP L+ DLV
Sbjct: 549 GSSGIKTNNFI------------IRQLFESCMKNGLYDSARPLLETLVHSAGKVDLV 593
Score = 117 bits (293), Expect = 2e-26
Identities = 113/475 (23%), Positives = 198/475 (40%), Gaps = 23/475 (4%)
Query: 223 LDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNL 282
LD TY II K D A F++M + L P FS+++D + G+++ + +
Sbjct: 311 LDGSTYELIIPSLAKSGRLDAAFKLFQQMKERKLRPSFSVFSSLVDSMGKAGRLDTSMKV 370
Query: 283 FERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGK 342
+ + G +P F L + +AG D + EMK G +PN +Y ++E+ K
Sbjct: 371 YMEMQGFGHRPSATMFVSLIDSYAKAGKLDTALRLWDEMKKSGFRPNFGLYTMIIESHAK 430
Query: 343 AGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFIL 402
+GK A ++F++M +G P T + +++++ + A++++ M G
Sbjct: 431 SGKLEVAMTVFKDMEKAGFLPTPSTYSCLLEMHAGSGQVDSAMKIYNSMTNAGLRPGLSS 490
Query: 403 YNTLLNMCADVGLIEEAETLFRDMKQSEH----CKPDSWSYTAMLNIYGSEGAVDKAMKL 458
Y +LL + A+ L++ A + +MK + C D +L IY + +VD A+K
Sbjct: 491 YISLLTLLANKRLVDVAGKILLEMKAMGYSVDVCASD------VLMIYIKDASVDLALKW 544
Query: 459 FEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSL 518
M GI+ N L + K D + + V K D L +L+ +
Sbjct: 545 LRFMGSSGIKTNNFIIRQLFESCMKNGLYDSARPLLETLVHSAGKVDLVLYTSILAHLVR 604
Query: 519 SQGSKDQEKVLACLQRANPKLVAFIQLIVD-----EETSFETVKEEFKAIMSNAVVEVRR 573
Q + ++++ L K AF+ + ++ V+E ++ I R
Sbjct: 605 CQDEDKERQLMSILSATKHKAHAFMCGLFTGPEQRKQPVLTFVREFYQGIDYELEEGAAR 664
Query: 574 PFCNCLIDICRNKDLVERAHELLYLGTLYGFYPSLHNKTQY-EWCLDVRTLSVGAALTAL 632
F N L++ + RA + + +P Q+ W LDVR LSVGAAL A+
Sbjct: 665 YFVNVLLNYLVLMGQINRARCVWKVAYENKLFPKAIVFDQHIAWSLDVRNLSVGAALIAV 724
Query: 633 EEWMTTLTKIVKREEALPDLFLAQTGTGAHKFAQGLNISFASHLRKLAAPFRQSE 687
+ K + +P TG L I A L + +PF S+
Sbjct: 725 VHTLHRFRKRMLYYGVVPRRIKLVTG-------PTLKIVIAQMLSSVESPFEVSK 772
>At4g20090 membrane-associated salt-inducible-like protein
Length = 660
Score = 127 bits (318), Expect = 3e-29
Identities = 86/338 (25%), Positives = 165/338 (48%), Gaps = 4/338 (1%)
Query: 191 IFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFER 250
+ YNV + L + +L M G + +TY+T+I DKAV ER
Sbjct: 258 VIYNVLIDGLCKKGDLTRVTKLVDNMFLKGCVPNEVTYNTLIHGLCLKGKLDKAVSLLER 317
Query: 251 MYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGD 310
M + +P++VT+ +++ + + + V L G+ + +SVL + G
Sbjct: 318 MVSSKCIPNDVTYGTLINGLVKQRRATDAVRLLSSMEERGYHLNQHIYSVLISGLFKEGK 377
Query: 311 YDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTA 370
+ + ++M G +PN+VVY+ L++ + + GKP A+ + MI SG PN T ++
Sbjct: 378 AEEAMSLWRKMAEKGCKPNIVVYSVLVDGLCREGKPNEAKEILNRMIASGCLPNAYTYSS 437
Query: 371 VIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSE 430
++K + K ++A+++WK M + G + Y+ L++ VG ++EA ++ M +
Sbjct: 438 LMKGFFKTGLCEEAVQVWKEMDKTGCSRNKFCYSVLIDGLCGVGRVKEAMMVWSKM-LTI 496
Query: 431 HCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEM---SKFGIELNVMGCTCLIQCLGKAMEI 487
KPD+ +Y++++ G++D A+KL+ EM + + +V+ L+ L +I
Sbjct: 497 GIKPDTVAYSSIIKGLCGIGSMDAALKLYHEMLCQEEPKSQPDVVTYNILLDGLCMQKDI 556
Query: 488 DDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQ 525
V + + ++RG PD C L+ +S S D+
Sbjct: 557 SRAVDLLNSMLDRGCDPDVITCNTFLNTLSEKSNSCDK 594
Score = 114 bits (285), Expect = 2e-25
Identities = 79/346 (22%), Positives = 156/346 (44%), Gaps = 34/346 (9%)
Query: 194 NVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYK 253
NV + + R + + + ++ + + ++++ +I K D+A+ F M +
Sbjct: 156 NVIINEGLYHRGLEFYDYVVNSNMNMNISPNGLSFNLVIKALCKLRFVDRAIEVFRGMPE 215
Query: 254 TGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDG 313
+PD T+ ++D + +++E V L + ++ G P P+ ++VL + GD
Sbjct: 216 RKCLPDGYTYCTLMDGLCKEERIDEAVLLLDEMQSEGCSPSPVIYNVLIDGLCKKGDLTR 275
Query: 314 IRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIK 373
+ ++ M G PN V YNTL+ + GK A SL E M+ S PN+ T +I
Sbjct: 276 VTKLVDNMFLKGCVPNEVTYNTLIHGLCLKGKLDKAVSLLERMVSSKCIPNDVTYGTLIN 335
Query: 374 IYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQ----- 428
K R + DA+ L M+E G+ ++ +Y+ L++ G EEA +L+R M +
Sbjct: 336 GLVKQRRATDAVRLLSSMEERGYHLNQHIYSVLISGLFKEGKAEEAMSLWRKMAEKGCKP 395
Query: 429 -----------------------------SEHCKPDSWSYTAMLNIYGSEGAVDKAMKLF 459
+ C P++++Y++++ + G ++A++++
Sbjct: 396 NIVVYSVLVDGLCREGKPNEAKEILNRMIASGCLPNAYTYSSLMKGFFKTGLCEEAVQVW 455
Query: 460 EEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPD 505
+EM K G N + LI L + + + V+ + G+KPD
Sbjct: 456 KEMDKTGCSRNKFCYSVLIDGLCGVGRVKEAMMVWSKMLTIGIKPD 501
Score = 62.8 bits (151), Expect = 7e-10
Identities = 64/277 (23%), Positives = 121/277 (43%), Gaps = 6/277 (2%)
Query: 262 TFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEM 321
T S++++ YA G + V L R R +F V+ + +G+A D + M
Sbjct: 79 TLSSMIESYANSGDFDSVEKLLSRIRLENRVIIERSFIVVFRAYGKAHLPDKAVDLFHRM 138
Query: 322 -KSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSG----IAPNEKTLTAVIKIYG 376
+ ++ +N++L + G ++ +++S I+PN + VIK
Sbjct: 139 VDEFRCKRSVKSFNSVLNVIINEGLYHRGLEFYDYVVNSNMNMNISPNGLSFNLVIKALC 198
Query: 377 KARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDS 436
K R+ A+E+++ M E D Y TL++ I+EA L +M QSE C P
Sbjct: 199 KLRFVDRAIEVFRGMPERKCLPDGYTYCTLMDGLCKEERIDEAVLLLDEM-QSEGCSPSP 257
Query: 437 WSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDI 496
Y +++ +G + + KL + M G N + LI L ++D V + +
Sbjct: 258 VIYNVLIDGLCKKGDLTRVTKLVDNMFLKGCVPNEVTYNTLIHGLCLKGKLDKAVSLLER 317
Query: 497 SVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQ 533
V P+D G L++ + + + D ++L+ ++
Sbjct: 318 MVSSKCIPNDVTYGTLINGLVKQRRATDAVRLLSSME 354
Score = 29.3 bits (64), Expect = 8.0
Identities = 23/88 (26%), Positives = 40/88 (45%), Gaps = 2/88 (2%)
Query: 189 ETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTII-SCAKKCNLFDKAVYW 247
+ + YN+ + L + +L + M+D G + D IT +T + + ++K N DK +
Sbjct: 539 DVVTYNILLDGLCMQKDISRAVDLLNSMLDRGCDPDVITCNTFLNTLSEKSNSCDKGRSF 598
Query: 248 FERMYKTGLMPDEVTFSAILDVYARLGK 275
E + L V+ A V LGK
Sbjct: 599 LEELVVRLLKRQRVS-GACTIVEVMLGK 625
>At5g27270 putative protein
Length = 1038
Score = 126 bits (317), Expect = 4e-29
Identities = 104/484 (21%), Positives = 210/484 (42%), Gaps = 13/484 (2%)
Query: 45 SIPDSETTPPNNNNNKKNSSLSD----QLASLANTTLSTVPENQPKVLSKPKPTWVNPTK 100
S+ D P + LSD ++ LST+ NQ PK W+ +
Sbjct: 45 SLSDGNPEKPKPRYERPKHPLSDDDARRIIKKKAQYLSTLRRNQGSQAMTPK--WIK--R 100
Query: 101 TKRPVLSHQRHKRSSVSYNPQLREFQRFAQRLNNCDVSSSDEEFMVCLEEIPSSLTRGNA 160
T ++ + R+ Y + + + L+ S D F+ + + L+ +
Sbjct: 101 TPEQMVQYLEDDRNGQMYGKHVVAAIKTVRGLSQRRQGSDDMRFV--MSSFVAKLSFRDM 158
Query: 161 LLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGG 220
+VL R W++ FF+W+K Q + Y + ++ + + EE +M++ G
Sbjct: 159 CVVLKEQRGWRQVRDFFSWMKLQLSYRPSVVVYTIVLRLYGQVGKIKMAEETFLEMLEVG 218
Query: 221 VELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVV 280
E D + T++ + + +++ + + ++ ++ +L + +V+
Sbjct: 219 CEPDAVACGTMLCTYARWGRHSAMLTFYKAVQERRILLSTSVYNFMLSSLQKKSFHGKVI 278
Query: 281 NLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAM 340
+L+ G P+ T++++ + + G + EMKSLG P V Y++++
Sbjct: 279 DLWLEMVEEGVPPNEFTYTLVVSSYAKQGFKEEALKAFGEMKSLGFVPEEVTYSSVISLS 338
Query: 341 GKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDF 400
KAG A L+E+M GI P+ T ++ +Y K AL L+ M+ N P D
Sbjct: 339 VKAGDWEKAIGLYEDMRSQGIVPSNYTCATMLSLYYKTENYPKALSLFADMERNKIPADE 398
Query: 401 ILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFE 460
++ ++ + +GL +A+++F + ++ + D +Y AM ++ + G V KA+ + E
Sbjct: 399 VIRGLIIRIYGKLGLFHDAQSMFEETERL-NLLADEKTYLAMSQVHLNSGNVVKALDVIE 457
Query: 461 EMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVS-LS 519
M I L+ ++QC K +D + F + G+ PD C +L++ + L+
Sbjct: 458 MMKTRDIPLSRFAYIVMLQCYAKIQNVDCAEEAFRALSKTGL-PDASSCNDMLNLYTRLN 516
Query: 520 QGSK 523
G K
Sbjct: 517 LGEK 520
Score = 100 bits (249), Expect = 3e-21
Identities = 81/339 (23%), Positives = 158/339 (45%), Gaps = 9/339 (2%)
Query: 137 VSSSDEEFMVCLEE--IPSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYN 194
+ ++E F+ LE P ++ G L F+ ++ + +L + T YN
Sbjct: 204 IKMAEETFLEMLEVGCEPDAVACGTMLCTYARWGRHSAMLTFYKAVQERRIL-LSTSVYN 262
Query: 195 VTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKT 254
+ SL+ G + +L +M++ GV + TY+ ++S K ++A+ F M
Sbjct: 263 FMLSSLQKKSFHGKVIDLWLEMVEEGVPPNEFTYTLVVSSYAKQGFKEEALKAFGEMKSL 322
Query: 255 GLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGI 314
G +P+EVT+S+++ + + G E+ + L+E R+ G P T + + ++ + +Y
Sbjct: 323 GFVPEEVTYSSVISLSVKAGDWEKAIGLYEDMRSQGIVPSNYTCATMLSLYYKTENYPKA 382
Query: 315 RYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKI 374
+ +M+ + + V+ ++ GK G A+S+FEE + +EKT A+ ++
Sbjct: 383 LSLFADMERNKIPADEVIRGLIIRIYGKLGLFHDAQSMFEETERLNLLADEKTYLAMSQV 442
Query: 375 YGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKP 434
+ + AL++ + MK P+ Y +L A + ++ AE FR + S+ P
Sbjct: 443 HLNSGNVVKALDVIEMMKTRDIPLSRFAYIVMLQCYAKIQNVDCAEEAFRAL--SKTGLP 500
Query: 435 DSWSYTAMLNIYGSEGAVDKA----MKLFEEMSKFGIEL 469
D+ S MLN+Y +KA ++ + F IEL
Sbjct: 501 DASSCNDMLNLYTRLNLGEKAKGFIKQIMVDQVHFDIEL 539
Score = 98.6 bits (244), Expect = 1e-20
Identities = 75/329 (22%), Positives = 149/329 (44%), Gaps = 8/329 (2%)
Query: 210 EELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTG---LMPDEVTFSAI 266
E +A +I G+ ++ T +T+I+ + + +A +R+Y P + ++
Sbjct: 655 EMIADIIIRLGLRMEEETIATLIAVYGRQHKLKEA----KRLYLAAGESKTPGKSVIRSM 710
Query: 267 LDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGV 326
+D Y R G +E+ LF G P +T S+L G + ++ + +
Sbjct: 711 IDAYVRCGWLEDAYGLFMESAEKGCDPGAVTISILVNALTNRGKHREAEHISRTCLEKNI 770
Query: 327 QPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALE 386
+ + V YNTL++AM +AGK A ++E M SG+ + +T +I +YG+ A+E
Sbjct: 771 ELDTVGYNTLIKAMLEAGKLQCASEIYERMHTSGVPCSIQTYNTMISVYGRGLQLDKAIE 830
Query: 387 LWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIY 446
++ + +G +D +Y ++ G + EA +LF +M Q + KP + SY M+ I
Sbjct: 831 IFSNARRSGLYLDEKIYTNMIMHYGKGGKMSEALSLFSEM-QKKGIKPGTPSYNMMVKIC 889
Query: 447 GSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDD 506
+ + +L + M + G ++ LIQ ++ + + K + E+G+
Sbjct: 890 ATSRLHHEVDELLQAMERNGRCTDLSTYLTLIQVYAESSQFAEAEKTITLVKEKGIPLSH 949
Query: 507 RLCGCLLSVVSLSQGSKDQEKVLACLQRA 535
LLS + + ++ E+ + A
Sbjct: 950 SHFSSLLSALVKAGMMEEAERTYCKMSEA 978
Score = 95.9 bits (237), Expect = 7e-20
Identities = 73/340 (21%), Positives = 161/340 (46%), Gaps = 9/340 (2%)
Query: 258 PDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYV 317
P V ++ +L +Y ++GK++ F G +PD + + + G + +
Sbjct: 186 PSVVVYTIVLRLYGQVGKIKMAEETFLEMLEVGCEPDAVACGTMLCTYARWGRHSAMLTF 245
Query: 318 LQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGK 377
+ ++ + + VYN +L ++ K G L+ EM++ G+ PNE T T V+ Y K
Sbjct: 246 YKAVQERRILLSTSVYNFMLSSLQKKSFHGKVIDLWLEMVEEGVPPNEFTYTLVVSSYAK 305
Query: 378 ARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSW 437
+ ++AL+ + MK G+ + + Y++++++ G E+A L+ DM+ S+ P ++
Sbjct: 306 QGFKEEALKAFGEMKSLGFVPEEVTYSSVISLSVKAGDWEKAIGLYEDMR-SQGIVPSNY 364
Query: 438 SYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDIS 497
+ ML++Y KA+ LF +M + I + + +I+ GK D +F+ +
Sbjct: 365 TCATMLSLYYKTENYPKALSLFADMERNKIPADEVIRGLIIRIYGKLGLFHDAQSMFEET 424
Query: 498 VERGVKPDDRLCGCLLSVVSLSQGSKDQE-KVLACLQRANPKLVAFIQLIV----DEETS 552
+ D++ +S V L+ G+ + V+ ++ + L F +++ + +
Sbjct: 425 ERLNLLADEKTY-LAMSQVHLNSGNVVKALDVIEMMKTRDIPLSRFAYIVMLQCYAKIQN 483
Query: 553 FETVKEEFKAIMSNAVVEVRRPFCNCLIDICRNKDLVERA 592
+ +E F+A+ + + CN ++++ +L E+A
Sbjct: 484 VDCAEEAFRALSKTGLPDASS--CNDMLNLYTRLNLGEKA 521
Score = 91.7 bits (226), Expect = 1e-18
Identities = 57/259 (22%), Positives = 123/259 (47%), Gaps = 1/259 (0%)
Query: 210 EELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDV 269
E ++ ++ +ELD + Y+T+I + A +ERM+ +G+ T++ ++ V
Sbjct: 759 EHISRTCLEKNIELDTVGYNTLIKAMLEAGKLQCASEIYERMHTSGVPCSIQTYNTMISV 818
Query: 270 YARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPN 329
Y R ++++ + +F R +G D ++ + +G+ G + EM+ G++P
Sbjct: 819 YGRGLQLDKAIEIFSNARRSGLYLDEKIYTNMIMHYGKGGKMSEALSLFSEMQKKGIKPG 878
Query: 330 LVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWK 389
YN +++ + L + M +G + T +I++Y ++ +A +
Sbjct: 879 TPSYNMMVKICATSRLHHEVDELLQAMERNGRCTDLSTYLTLIQVYAESSQFAEAEKTIT 938
Query: 390 RMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSE 449
+KE G P+ +++LL+ G++EEAE + M ++ PDS +L Y +
Sbjct: 939 LVKEKGIPLSHSHFSSLLSALVKAGMMEEAERTYCKMSEA-GISPDSACKRTILKGYMTC 997
Query: 450 GAVDKAMKLFEEMSKFGIE 468
G +K + +E+M + +E
Sbjct: 998 GDAEKGILFYEKMIRSSVE 1016
Score = 74.7 bits (182), Expect = 2e-13
Identities = 75/423 (17%), Positives = 166/423 (38%), Gaps = 39/423 (9%)
Query: 177 FNWIKTQNLLPMETIFYNVTMKSLRFG---RQFGIIEELAHQMIDGGVELDNITYSTIIS 233
F +K+ +P E + +V S++ G + G+ E++ Q G+ N T +T++S
Sbjct: 316 FGEMKSLGFVPEEVTYSSVISLSVKAGDWEKAIGLYEDMRSQ----GIVPSNYTCATMLS 371
Query: 234 CAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKP 293
K + KA+ F M + + DEV I+ +Y +LG + ++FE
Sbjct: 372 LYYKTENYPKALSLFADMERNKIPADEVIRGLIIRIYGKLGLFHDAQSMFEETERLNLLA 431
Query: 294 DPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLF 353
D T+ + ++ +G+ V++ MK+ + + Y +L+ K A F
Sbjct: 432 DEKTYLAMSQVHLNSGNVVKALDVIEMMKTRDIPLSRFAYIVMLQCYAKIQNVDCAEEAF 491
Query: 354 EEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADV 413
+ +G+ P+ + ++ +Y + + A K++ + D LY T + +
Sbjct: 492 RALSKTGL-PDASSCNDMLNLYTRLNLGEKAKGFIKQIMVDQVHFDIELYKTAMRVYCKE 550
Query: 414 GLIEEAETLFRDMKQSEHCKPDSWSYT-----------------------------AMLN 444
G++ EA+ L M + K + + T MLN
Sbjct: 551 GMVAEAQDLIVKMGREARVKDNRFVQTLAESMHIVNKHDKHEAVLNVSQLDVMALGLMLN 610
Query: 445 IYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKP 504
+ EG +++ + M F +L +I + ++ + DI + G++
Sbjct: 611 LRLKEGNLNETKAILNLM--FKTDLGSSAVNRVISSFVREGDVSKAEMIADIIIRLGLRM 668
Query: 505 DDRLCGCLLSVVSLSQGSKDQEKVLACLQRANPKLVAFIQLIVDEETSFETVKEEFKAIM 564
++ L++V K+ +++ + + I+ ++D +++ + M
Sbjct: 669 EEETIATLIAVYGRQHKLKEAKRLYLAAGESKTPGKSVIRSMIDAYVRCGWLEDAYGLFM 728
Query: 565 SNA 567
+A
Sbjct: 729 ESA 731
Score = 70.9 bits (172), Expect = 2e-12
Identities = 49/213 (23%), Positives = 101/213 (47%), Gaps = 5/213 (2%)
Query: 324 LGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKD 383
L +P++VVY +L G+ GK A F EM++ G P+ ++ Y ARW +
Sbjct: 182 LSYRPSVVVYTIVLRLYGQVGKIKMAEETFLEMLEVGCEPDAVACGTMLCTY--ARWGRH 239
Query: 384 A--LELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTA 441
+ L +K ++E + +YN +L+ + L+ +M + E P+ ++YT
Sbjct: 240 SAMLTFYKAVQERRILLSTSVYNFMLSSLQKKSFHGKVIDLWLEMVE-EGVPPNEFTYTL 298
Query: 442 MLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERG 501
+++ Y +G ++A+K F EM G + + +I KA + + + +++ +G
Sbjct: 299 VVSSYAKQGFKEEALKAFGEMKSLGFVPEEVTYSSVISLSVKAGDWEKAIGLYEDMRSQG 358
Query: 502 VKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQR 534
+ P + C +LS+ ++ + A ++R
Sbjct: 359 IVPSNYTCATMLSLYYKTENYPKALSLFADMER 391
Score = 68.9 bits (167), Expect = 9e-12
Identities = 51/261 (19%), Positives = 112/261 (42%), Gaps = 9/261 (3%)
Query: 291 WKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFAR 350
++P + ++++ +++G+ G EM +G +P+ V T+L + G+
Sbjct: 184 YRPSVVVYTIVLRLYGQVGKIKMAEETFLEMLEVGCEPDAVACGTMLCTYARWGRHSAML 243
Query: 351 SLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMC 410
+ ++ + + I + ++ K + ++LW M E G P + Y +++
Sbjct: 244 TFYKAVQERRILLSTSVYNFMLSSLQKKSFHGKVIDLWLEMVEEGVPPNEFTYTLVVSSY 303
Query: 411 ADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELN 470
A G EEA F +MK S P+ +Y++++++ G +KA+ L+E+M GI +
Sbjct: 304 AKQGFKEEALKAFGEMK-SLGFVPEEVTYSSVISLSVKAGDWEKAIGLYEDMRSQGIVPS 362
Query: 471 VMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLA 530
C ++ K + +F + D+ + G ++ + D + +
Sbjct: 363 NYTCATMLSLYYKTENYPKALSLFADMERNKIPADEVIRGLIIRIYGKLGLFHDAQSMFE 422
Query: 531 CLQRANPKLVAFIQLIVDEET 551
+R N L+ DE+T
Sbjct: 423 ETERLN--------LLADEKT 435
Score = 50.1 bits (118), Expect = 4e-06
Identities = 80/456 (17%), Positives = 161/456 (34%), Gaps = 99/456 (21%)
Query: 150 EIPSS-LTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGI 208
+IP+ + RG + + L + F + NLL E + ++ L G
Sbjct: 393 KIPADEVIRGLIIRIYGKLGLFHDAQSMFEETERLNLLADEKTYLAMSQVHLNSGNVVKA 452
Query: 209 IEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGL------------ 256
++ + M + L Y ++ C K D A F + KTGL
Sbjct: 453 LDVI-EMMKTRDIPLSRFAYIVMLQCYAKIQNVDCAEEAFRALSKTGLPDASSCNDMLNL 511
Query: 257 -----------------MPDEVTFS-----AILDVYARLGKVEEVVNLFER-GRATGWKP 293
M D+V F + VY + G V E +L + GR K
Sbjct: 512 YTRLNLGEKAKGFIKQIMVDQVHFDIELYKTAMRVYCKEGMVAEAQDLIVKMGREARVKD 571
Query: 294 DPITFSVLGKMF--GEAGDYDGIRYVLQ-EMKSLGVQPNLVV------------------ 332
+ ++ M + ++ + V Q ++ +LG+ NL +
Sbjct: 572 NRFVQTLAESMHIVNKHDKHEAVLNVSQLDVMALGLMLNLRLKEGNLNETKAILNLMFKT 631
Query: 333 ------YNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKAR------- 379
N ++ + + G A + + +I G+ E+T+ +I +YG+
Sbjct: 632 DLGSSAVNRVISSFVREGDVSKAEMIADIIIRLGLRMEEETIATLIAVYGRQHKLKEAKR 691
Query: 380 ---------------------------WSKDALELWKRMKENGWPMDFILYNTLLNMCAD 412
W +DA L+ E G + + L+N +
Sbjct: 692 LYLAAGESKTPGKSVIRSMIDAYVRCGWLEDAYGLFMESAEKGCDPGAVTISILVNALTN 751
Query: 413 VGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVM 472
G EAE + R + ++ + D+ Y ++ G + A +++E M G+ ++
Sbjct: 752 RGKHREAEHISRTCLE-KNIELDTVGYNTLIKAMLEAGKLQCASEIYERMHTSGVPCSIQ 810
Query: 473 GCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRL 508
+I G+ +++D +++F + G+ D+++
Sbjct: 811 TYNTMISVYGRGLQLDKAIEIFSNARRSGLYLDEKI 846
Score = 38.1 bits (87), Expect = 0.017
Identities = 37/153 (24%), Positives = 63/153 (40%), Gaps = 3/153 (1%)
Query: 193 YNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMY 252
YN+ +K R ++EL M G D TY T+I + + F +A +
Sbjct: 882 YNMMVKICATSRLHHEVDELLQAMERNGRCTDLSTYLTLIQVYAESSQFAEAEKTITLVK 941
Query: 253 KTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYD 312
+ G+ FS++L + G +EE + + G PD + K + GD +
Sbjct: 942 EKGIPLSHSHFSSLLSALVKAGMMEEAERTYCKMSEAGISPDSACKRTILKGYMTCGDAE 1001
Query: 313 -GIRYVLQEMKSLGVQPNLV--VYNTLLEAMGK 342
GI + + ++S V V L +A+GK
Sbjct: 1002 KGILFYEKMIRSSVEDDRFVSSVVEDLYKAVGK 1034
Score = 37.4 bits (85), Expect = 0.029
Identities = 19/76 (25%), Positives = 39/76 (51%), Gaps = 3/76 (3%)
Query: 207 GIIEELAH---QMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTF 263
G++EE +M + G+ D+ TI+ C +K + ++E+M ++ + D
Sbjct: 963 GMMEEAERTYCKMSEAGISPDSACKRTILKGYMTCGDAEKGILFYEKMIRSSVEDDRFVS 1022
Query: 264 SAILDVYARLGKVEEV 279
S + D+Y +GK ++V
Sbjct: 1023 SVVEDLYKAVGKEQDV 1038
>At3g06920 hypothetical protein
Length = 871
Score = 126 bits (317), Expect = 4e-29
Identities = 89/313 (28%), Positives = 145/313 (45%), Gaps = 2/313 (0%)
Query: 171 QKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYST 230
+K F IK + +P + Y++ + L EL + M + G LD Y+
Sbjct: 534 EKGRAMFEEIKARRFVP-DARSYSILIHGLIKAGFANETYELFYSMKEQGCVLDTRAYNI 592
Query: 231 IISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATG 290
+I KC +KA E M G P VT+ +++D A++ +++E LFE ++
Sbjct: 593 VIDGFCKCGKVNKAYQLLEEMKTKGFEPTVVTYGSVIDGLAKIDRLDEAYMLFEEAKSKR 652
Query: 291 WKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFAR 350
+ + + +S L FG+ G D +L+E+ G+ PNL +N+LL+A+ KA + A
Sbjct: 653 IELNVVIYSSLIDGFGKVGRIDEAYLILEELMQKGLTPNLYTWNSLLDALVKAEEINEAL 712
Query: 351 SLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMC 410
F+ M + PN+ T +I K R A W+ M++ G I Y T+++
Sbjct: 713 VCFQSMKELKCTPNQVTYGILINGLCKVRKFNKAFVFWQEMQKQGMKPSTISYTTMISGL 772
Query: 411 ADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELN 470
A G I EA LF D ++ PDS Y AM+ + A LFEE + G+ ++
Sbjct: 773 AKAGNIAEAGALF-DRFKANGGVPDSACYNAMIEGLSNGNRAMDAFSLFEETRRRGLPIH 831
Query: 471 VMGCTCLIQCLGK 483
C L+ L K
Sbjct: 832 NKTCVVLLDTLHK 844
Score = 121 bits (304), Expect = 1e-27
Identities = 94/383 (24%), Positives = 179/383 (46%), Gaps = 6/383 (1%)
Query: 151 IPSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIE 210
IPS + L L + + F +K + T YN+ + L +
Sbjct: 340 IPSVIAYNCILTCLRKMGKVDEALKVFEEMKKDAAPNLST--YNILIDMLCRAGKLDTAF 397
Query: 211 ELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVY 270
EL M G+ + T + ++ K D+A FE M PDE+TF +++D
Sbjct: 398 ELRDSMQKAGLFPNVRTVNIMVDRLCKSQKLDEACAMFEEMDYKVCTPDEITFCSLIDGL 457
Query: 271 ARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNL 330
++G+V++ ++E+ + + + I ++ L K F G + + ++M + P+L
Sbjct: 458 GKVGRVDDAYKVYEKMLDSDCRTNSIVYTSLIKNFFNHGRKEDGHKIYKDMINQNCSPDL 517
Query: 331 VVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKR 390
+ NT ++ M KAG+P R++FEE+ P+ ++ + +I KA ++ + EL+
Sbjct: 518 QLLNTYMDCMFKAGEPEKGRAMFEEIKARRFVPDARSYSILIHGLIKAGFANETYELFYS 577
Query: 391 MKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEG 450
MKE G +D YN +++ G + +A L +MK ++ +P +Y ++++
Sbjct: 578 MKEQGCVLDTRAYNIVIDGFCKCGKVNKAYQLLEEMK-TKGFEPTVVTYGSVIDGLAKID 636
Query: 451 AVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCG 510
+D+A LFEE IELNV+ + LI GK ID+ + + +++G+ P+
Sbjct: 637 RLDEAYMLFEEAKSKRIELNVVIYSSLIDGFGKVGRIDEAYLILEELMQKGLTPNLYTWN 696
Query: 511 CLLSVVSLSQGSKDQEKVLACLQ 533
LL + ++ + + L C Q
Sbjct: 697 SLLDALVKAE---EINEALVCFQ 716
Score = 120 bits (301), Expect = 3e-27
Identities = 87/359 (24%), Positives = 170/359 (47%), Gaps = 4/359 (1%)
Query: 163 VLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVE 222
VL L+ + +F W + + LP YN + + R F ++++ +M G
Sbjct: 71 VLRRLKDVNRAIEYFRWYERRTELPHCPESYNSLLLVMARCRNFDALDQILGEMSVAGFG 130
Query: 223 LD-NITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVN 281
N ++ C K N + + M K P ++ ++ ++ + + ++
Sbjct: 131 PSVNTCIEMVLGCVK-ANKLREGYDVVQMMRKFKFRPAFSAYTTLIGAFSAVNHSDMMLT 189
Query: 282 LFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMG 341
LF++ + G++P F+ L + F + G D +L EMKS + ++V+YN +++ G
Sbjct: 190 LFQQMQELGYEPTVHLFTTLIRGFAKEGRVDSALSLLDEMKSSSLDADIVLYNVCIDSFG 249
Query: 342 KAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFI 401
K GK A F E+ +G+ P+E T T++I + KA +A+E+++ +++N
Sbjct: 250 KVGKVDMAWKFFHEIEANGLKPDEVTYTSMIGVLCKANRLDEAVEMFEHLEKNRRVPCTY 309
Query: 402 LYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEE 461
YNT++ G +EA +L + ++++ P +Y +L G VD+A+K+FEE
Sbjct: 310 AYNTMIMGYGSAGKFDEAYSLL-ERQRAKGSIPSVIAYNCILTCLRKMGKVDEALKVFEE 368
Query: 462 MSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQ 520
M K N+ LI L +A ++D ++ D + G+ P+ R ++ + SQ
Sbjct: 369 MKKDAAP-NLSTYNILIDMLCRAGKLDTAFELRDSMQKAGLFPNVRTVNIMVDRLCKSQ 426
Score = 95.9 bits (237), Expect = 7e-20
Identities = 74/308 (24%), Positives = 137/308 (44%), Gaps = 3/308 (0%)
Query: 216 MIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGK 275
MI+ D +T + C K +K FE + +PD ++S ++ + G
Sbjct: 508 MINQNCSPDLQLLNTYMDCMFKAGEPEKGRAMFEEIKARRFVPDARSYSILIHGLIKAGF 567
Query: 276 VEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNT 335
E LF + G D ++++ F + G + +L+EMK+ G +P +V Y +
Sbjct: 568 ANETYELFYSMKEQGCVLDTRAYNIVIDGFCKCGKVNKAYQLLEEMKTKGFEPTVVTYGS 627
Query: 336 LLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENG 395
+++ + K + A LFEE I N +++I +GK +A + + + + G
Sbjct: 628 VIDGLAKIDRLDEAYMLFEEAKSKRIELNVVIYSSLIDGFGKVGRIDEAYLILEELMQKG 687
Query: 396 WPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKA 455
+ +N+LL+ I EA F+ MK+ + C P+ +Y ++N +KA
Sbjct: 688 LTPNLYTWNSLLDALVKAEEINEALVCFQSMKELK-CTPNQVTYGILINGLCKVRKFNKA 746
Query: 456 MKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSV 515
++EM K G++ + + T +I L KA I + +FD G PD ++
Sbjct: 747 FVFWQEMQKQGMKPSTISYTTMISGLAKAGNIAEAGALFDRFKANGGVPDSACYNAMIE- 805
Query: 516 VSLSQGSK 523
LS G++
Sbjct: 806 -GLSNGNR 812
Score = 63.2 bits (152), Expect = 5e-10
Identities = 48/248 (19%), Positives = 110/248 (44%), Gaps = 9/248 (3%)
Query: 152 PSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSL----RFGRQFG 207
P+ +T G+ + L + + +M F K++ + + + Y+ + R +
Sbjct: 620 PTVVTYGSVIDGLAKIDRLDEAYMLFEEAKSKRI-ELNVVIYSSLIDGFGKVGRIDEAYL 678
Query: 208 IIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAIL 267
I+EEL + G+ + T+++++ K ++A+ F+ M + P++VT+ ++
Sbjct: 679 ILEELMQK----GLTPNLYTWNSLLDALVKAEEINEALVCFQSMKELKCTPNQVTYGILI 734
Query: 268 DVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQ 327
+ ++ K + ++ + G KP I+++ + +AG+ + K+ G
Sbjct: 735 NGLCKVRKFNKAFVFWQEMQKQGMKPSTISYTTMISGLAKAGNIAEAGALFDRFKANGGV 794
Query: 328 PNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALEL 387
P+ YN ++E + + A SLFEE G+ + KT ++ K + A +
Sbjct: 795 PDSACYNAMIEGLSNGNRAMDAFSLFEETRRRGLPIHNKTCVVLLDTLHKNDCLEQAAIV 854
Query: 388 WKRMKENG 395
++E G
Sbjct: 855 GAVLRETG 862
>At5g65560 putative protein
Length = 915
Score = 126 bits (316), Expect = 5e-29
Identities = 92/357 (25%), Positives = 180/357 (49%), Gaps = 6/357 (1%)
Query: 193 YNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTII-SCAKKCNLFDKAVYWFERM 251
Y V +KSL + L +M + G++ + TY+ +I S +C F+KA +M
Sbjct: 326 YTVLIKSLCGSERKSEALNLVKEMEETGIKPNIHTYTVLIDSLCSQCK-FEKARELLGQM 384
Query: 252 YKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDY 311
+ GLMP+ +T++A+++ Y + G +E+ V++ E + P+ T++ L K + ++ +
Sbjct: 385 LEKGLMPNVITYNALINGYCKRGMIEDAVDVVELMESRKLSPNTRTYNELIKGYCKSNVH 444
Query: 312 DGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAV 371
+ VL +M V P++V YN+L++ ++G A L M D G+ P++ T T++
Sbjct: 445 KAMG-VLNKMLERKVLPDVVTYNSLIDGQCRSGNFDSAYRLLSLMNDRGLVPDQWTYTSM 503
Query: 372 IKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEH 431
I K++ ++A +L+ +++ G + ++Y L++ G ++EA + M S++
Sbjct: 504 IDSLCKSKRVEEACDLFDSLEQKGVNPNVVMYTALIDGYCKAGKVDEAHLMLEKM-LSKN 562
Query: 432 CKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLV 491
C P+S ++ A+++ ++G + +A L E+M K G++ V T LI L K + D
Sbjct: 563 CLPNSLTFNALIHGLCADGKLKEATLLEEKMVKIGLQPTVSTDTILIHRLLKDGDFDHAY 622
Query: 492 KVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQR--ANPKLVAFIQLI 546
F + G KPD + D E ++A ++ +P L + LI
Sbjct: 623 SRFQQMLSSGTKPDAHTYTTFIQTYCREGRLLDAEDMMAKMRENGVSPDLFTYSSLI 679
Score = 102 bits (255), Expect = 6e-22
Identities = 82/339 (24%), Positives = 152/339 (44%), Gaps = 19/339 (5%)
Query: 214 HQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARL 273
++M++ V D +TY+++I + FD A M GL+PD+ T+++++D +
Sbjct: 451 NKMLERKVLPDVVTYNSLIDGQCRSGNFDSAYRLLSLMNDRGLVPDQWTYTSMIDSLCKS 510
Query: 274 GKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVY 333
+VEE +LF+ G P+ + ++ L + +AG D +L++M S PN + +
Sbjct: 511 KRVEEACDLFDSLEQKGVNPNVVMYTALIDGYCKAGKVDEAHLMLEKMLSKNCLPNSLTF 570
Query: 334 NTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKE 393
N L+ + GK A L E+M+ G+ P T T +I K A +++M
Sbjct: 571 NALIHGLCADGKLKEATLLEEKMVKIGLQPTVSTDTILIHRLLKDGDFDHAYSRFQQMLS 630
Query: 394 NGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVD 453
+G D Y T + G + +AE + M+++ PD ++Y++++ YG G +
Sbjct: 631 SGTKPDAHTYTTFIQTYCREGRLLDAEDMMAKMREN-GVSPDLFTYSSLIKGYGDLGQTN 689
Query: 454 KAMKLFEEMSKFGIELNVMGCTCLIQ------------------CLGKAMEIDDLVKVFD 495
A + + M G E + LI+ + ME D +V++ +
Sbjct: 690 FAFDVLKRMRDTGCEPSQHTFLSLIKHLLEMKYGKQKGSEPELCAMSNMMEFDTVVELLE 749
Query: 496 ISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQR 534
VE V P+ + L+ + + EKV +QR
Sbjct: 750 KMVEHSVTPNAKSYEKLILGICEVGNLRVAEKVFDHMQR 788
Score = 102 bits (254), Expect = 7e-22
Identities = 86/371 (23%), Positives = 163/371 (43%), Gaps = 21/371 (5%)
Query: 178 NWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKK 237
N + + +LP + + YN + F L M D G+ D TY+++I K
Sbjct: 451 NKMLERKVLP-DVVTYNSLIDGQCRSGNFDSAYRLLSLMNDRGLVPDQWTYTSMIDSLCK 509
Query: 238 CNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPIT 297
++A F+ + + G+ P+ V ++A++D Y + GKV+E + E+ + P+ +T
Sbjct: 510 SKRVEEACDLFDSLEQKGVNPNVVMYTALIDGYCKAGKVDEAHLMLEKMLSKNCLPNSLT 569
Query: 298 FSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMI 357
F+ L G + ++M +G+QP + L+ + K G A S F++M+
Sbjct: 570 FNALIHGLCADGKLKEATLLEEKMVKIGLQPTVSTDTILIHRLLKDGDFDHAYSRFQQML 629
Query: 358 DSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIE 417
SG P+ T T I+ Y + DA ++ +M+ENG D Y++L+ D+G
Sbjct: 630 SSGTKPDAHTYTTFIQTYCREGRLLDAEDMMAKMRENGVSPDLFTYSSLIKGYGDLGQTN 689
Query: 418 EAETLFRDMKQSEHCKPDSWSYTAMLN-----IYGSEGA-------------VDKAMKLF 459
A + + M+ + C+P ++ +++ YG + D ++L
Sbjct: 690 FAFDVLKRMRDT-GCEPSQHTFLSLIKHLLEMKYGKQKGSEPELCAMSNMMEFDTVVELL 748
Query: 460 EEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFD-ISVERGVKPDDRLCGCLLSVVSL 518
E+M + + N LI + + + KVFD + G+ P + + LLS
Sbjct: 749 EKMVEHSVTPNAKSYEKLILGICEVGNLRVAEKVFDHMQRNEGISPSELVFNALLSCCCK 808
Query: 519 SQGSKDQEKVL 529
+ + KV+
Sbjct: 809 LKKHNEAAKVV 819
Score = 88.2 bits (217), Expect = 1e-17
Identities = 75/295 (25%), Positives = 130/295 (43%), Gaps = 9/295 (3%)
Query: 211 ELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVY 270
EL +++I G Y+T+++ + L D+ + M + + P+ T++ +++ Y
Sbjct: 176 ELKYKLIIG-------CYNTLLNSLARFGLVDEMKQVYMEMLEDKVCPNIYTYNKMVNGY 228
Query: 271 ARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNL 330
+LG VEE + G PD T++ L + + D D V EM G + N
Sbjct: 229 CKLGNVEEANQYVSKIVEAGLDPDFFTYTSLIMGYCQRKDLDSAFKVFNEMPLKGCRRNE 288
Query: 331 VVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKR 390
V Y L+ + A + A LF +M D P +T T +IK + +AL L K
Sbjct: 289 VAYTHLIHGLCVARRIDEAMDLFVKMKDDECFPTVRTYTVLIKSLCGSERKSEALNLVKE 348
Query: 391 MKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEG 450
M+E G + Y L++ E+A L M + + P+ +Y A++N Y G
Sbjct: 349 MEETGIKPNIHTYTVLIDSLCSQCKFEKARELLGQMLE-KGLMPNVITYNALINGYCKRG 407
Query: 451 AVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPD 505
++ A+ + E M + N LI+ K+ + + V + +ER V PD
Sbjct: 408 MIEDAVDVVELMESRKLSPNTRTYNELIKGYCKS-NVHKAMGVLNKMLERKVLPD 461
Score = 85.9 bits (211), Expect = 7e-17
Identities = 74/312 (23%), Positives = 139/312 (43%), Gaps = 18/312 (5%)
Query: 220 GVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEV 279
GV + + Y+ +I K D+A E+M +P+ +TF+A++ GK++E
Sbjct: 527 GVNPNVVMYTALIDGYCKAGKVDEAHLMLEKMLSKNCLPNSLTFNALIHGLCADGKLKEA 586
Query: 280 VNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEA 339
L E+ G +P T ++L + GD+D Q+M S G +P+ Y T ++
Sbjct: 587 TLLEEKMVKIGLQPTVSTDTILIHRLLKDGDFDHAYSRFQQMLSSGTKPDAHTYTTFIQT 646
Query: 340 MGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMD 399
+ G+ A + +M ++G++P+ T +++IK YG + A ++ KRM++ G
Sbjct: 647 YCREGRLLDAEDMMAKMRENGVSPDLFTYSSLIKGYGDLGQTNFAFDVLKRMRDTGCEPS 706
Query: 400 FILYNTLL----------------NMCADVGLIEEAETLFRDMKQSEH-CKPDSWSYTAM 442
+ +L+ +CA ++E + K EH P++ SY +
Sbjct: 707 QHTFLSLIKHLLEMKYGKQKGSEPELCAMSNMMEFDTVVELLEKMVEHSVTPNAKSYEKL 766
Query: 443 LNIYGSEGAVDKAMKLFEEMSK-FGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERG 501
+ G + A K+F+ M + GI + + L+ C K + ++ KV D + G
Sbjct: 767 ILGICEVGNLRVAEKVFDHMQRNEGISPSELVFNALLSCCCKLKKHNEAAKVVDDMICVG 826
Query: 502 VKPDDRLCGCLL 513
P C L+
Sbjct: 827 HLPQLESCKVLI 838
Score = 81.3 bits (199), Expect = 2e-15
Identities = 75/357 (21%), Positives = 155/357 (43%), Gaps = 33/357 (9%)
Query: 172 KTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTI 231
+ H+ + ++N LP ++ +N + L + L +M+ G++ T + +
Sbjct: 550 EAHLMLEKMLSKNCLP-NSLTFNALIHGLCADGKLKEATLLEEKMVKIGLQPTVSTDTIL 608
Query: 232 ISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGW 291
I K FD A F++M +G PD T++ + Y R G++ + ++ + R G
Sbjct: 609 IHRLLKDGDFDHAYSRFQQMLSSGTKPDAHTYTTFIQTYCREGRLLDAEDMMAKMRENGV 668
Query: 292 KPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAM-------GKAG 344
PD T+S L K +G+ G + VL+ M+ G +P+ + +L++ + K
Sbjct: 669 SPDLFTYSSLIKGYGDLGQTNFAFDVLKRMRDTGCEPSQHTFLSLIKHLLEMKYGKQKGS 728
Query: 345 KPGFAR-----------SLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKE 393
+P L E+M++ + PN K+ +I + + A +++ M+
Sbjct: 729 EPELCAMSNMMEFDTVVELLEKMVEHSVTPNAKSYEKLILGICEVGNLRVAEKVFDHMQR 788
Query: 394 N-GWPMDFILYNTLLNMCADVGLIEEAETLFRDM------KQSEHCKPDSWSYTAMLNIY 446
N G +++N LL+ C + EA + DM Q E CK + +Y
Sbjct: 789 NEGISPSELVFNALLSCCCKLKKHNEAAKVVDDMICVGHLPQLESCK------VLICGLY 842
Query: 447 GSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVK 503
+G ++ +F+ + + G + + +I +GK ++ ++F++ + G K
Sbjct: 843 -KKGEKERGTSVFQNLLQCGYYEDELAWKIIIDGVGKQGLVEAFYELFNVMEKNGCK 898
Score = 43.5 bits (101), Expect = 4e-04
Identities = 35/168 (20%), Positives = 70/168 (40%), Gaps = 1/168 (0%)
Query: 241 FDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFER-GRATGWKPDPITFS 299
FD V E+M + + P+ ++ ++ +G + +F+ R G P + F+
Sbjct: 741 FDTVVELLEKMVEHSVTPNAKSYEKLILGICEVGNLRVAEKVFDHMQRNEGISPSELVFN 800
Query: 300 VLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDS 359
L + ++ V+ +M +G P L L+ + K G+ S+F+ ++
Sbjct: 801 ALLSCCCKLKKHNEAAKVVDDMICVGHLPQLESCKVLICGLYKKGEKERGTSVFQNLLQC 860
Query: 360 GIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLL 407
G +E +I GK + EL+ M++NG Y+ L+
Sbjct: 861 GYYEDELAWKIIIDGVGKQGLVEAFYELFNVMEKNGCKFSSQTYSLLI 908
Score = 41.2 bits (95), Expect = 0.002
Identities = 36/170 (21%), Positives = 72/170 (42%), Gaps = 1/170 (0%)
Query: 205 QFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKT-GLMPDEVTF 263
+F + EL +M++ V + +Y +I + A F+ M + G+ P E+ F
Sbjct: 740 EFDTVVELLEKMVEHSVTPNAKSYEKLILGICEVGNLRVAEKVFDHMQRNEGISPSELVF 799
Query: 264 SAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKS 323
+A+L +L K E + + G P + VL + G+ + V Q +
Sbjct: 800 NALLSCCCKLKKHNEAAKVVDDMICVGHLPQLESCKVLICGLYKKGEKERGTSVFQNLLQ 859
Query: 324 LGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIK 373
G + + + +++ +GK G LF M +G + +T + +I+
Sbjct: 860 CGYYEDELAWKIIIDGVGKQGLVEAFYELFNVMEKNGCKFSSQTYSLLIE 909
>At2g35130 hypothetical protein
Length = 591
Score = 126 bits (316), Expect = 5e-29
Identities = 85/321 (26%), Positives = 153/321 (47%), Gaps = 7/321 (2%)
Query: 227 TYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERG 286
TY+ +I+ K + + + M P+ T++A+++ +AR G E+ +FE+
Sbjct: 266 TYNLMINLYGKASKSYMSWKLYCEMRSHQCKPNICTYTALVNAFAREGLCEKAEEIFEQL 325
Query: 287 RATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKP 346
+ G +PD ++ L + + AG G + M+ +G +P+ YN +++A G+AG
Sbjct: 326 QEDGLEPDVYVYNALMESYSRAGYPYGAAEIFSLMQHMGCEPDRASYNIMVDAYGRAGLH 385
Query: 347 GFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTL 406
A ++FEEM GIAP K+ ++ Y KAR + K M ENG D + N++
Sbjct: 386 SDAEAVFEEMKRLGIAPTMKSHMLLLSAYSKARDVTKCEAIVKEMSENGVEPDTFVLNSM 445
Query: 407 LNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFG 466
LN+ +G + E + +M+ C D +Y ++NIYG G +++ +LF E+ +
Sbjct: 446 LNLYGRLGQFTKMEKILAEMENGP-CTADISTYNILINIYGKAGFLERIEELFVELKEKN 504
Query: 467 IELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQE 526
+V+ T I + ++VF+ ++ G PD LLS S + +
Sbjct: 505 FRPDVVTWTSRIGAYSRKKLYVKCLEVFEEMIDSGCAPDGGTAKVLLSACSSEEQVEQVT 564
Query: 527 KVLACLQRA------NPKLVA 541
VL + + PKL+A
Sbjct: 565 SVLRTMHKGVTVSSLVPKLMA 585
Score = 103 bits (257), Expect = 3e-22
Identities = 86/345 (24%), Positives = 145/345 (41%), Gaps = 40/345 (11%)
Query: 224 DNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLF 283
D I S + KK + W R K+ PD + F+ ++D Y + + +E +L+
Sbjct: 121 DLINVSVQLRLNKKWDSIILVCEWILR--KSSFQPDVICFNLLIDAYGQKFQYKEAESLY 178
Query: 284 ERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLV---VYNTLLEA- 339
+ + + P T+++L K + AG + VL EM++ V P + VYN +E
Sbjct: 179 VQLLESRYVPTEDTYALLIKAYCMAGLIERAEVVLVEMQNHHVSPKTIGVTVYNAYIEGL 238
Query: 340 MGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMD 399
M + G A +F+ M P +T +I +YGKA S + +L+ M+ + +
Sbjct: 239 MKRKGNTEEAIDVFQRMKRDRCKPTTETYNLMINLYGKASKSYMSWKLYCEMRSHQCKPN 298
Query: 400 FILYNTLLNMCADVGLIEEAETLFRDMKQS------------------------------ 429
Y L+N A GL E+AE +F +++
Sbjct: 299 ICTYTALVNAFAREGLCEKAEEIFEQLQEDGLEPDVYVYNALMESYSRAGYPYGAAEIFS 358
Query: 430 --EH--CKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAM 485
+H C+PD SY M++ YG G A +FEEM + GI + L+ KA
Sbjct: 359 LMQHMGCEPDRASYNIMVDAYGRAGLHSDAEAVFEEMKRLGIAPTMKSHMLLLSAYSKAR 418
Query: 486 EIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLA 530
++ + E GV+PD + +L++ EK+LA
Sbjct: 419 DVTKCEAIVKEMSENGVEPDTFVLNSMLNLYGRLGQFTKMEKILA 463
Score = 63.2 bits (152), Expect = 5e-10
Identities = 45/183 (24%), Positives = 87/183 (46%), Gaps = 6/183 (3%)
Query: 193 YNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMY 252
YN+ + + E + +M G+ ++ ++S K K + M
Sbjct: 372 YNIMVDAYGRAGLHSDAEAVFEEMKRLGIAPTMKSHMLLLSAYSKARDVTKCEAIVKEMS 431
Query: 253 KTGLMPDEVTFSAILDVYARLG---KVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAG 309
+ G+ PD +++L++Y RLG K+E+++ E G T D T+++L ++G+AG
Sbjct: 432 ENGVEPDTFVLNSMLNLYGRLGQFTKMEKILAEMENGPCTA---DISTYNILINIYGKAG 488
Query: 310 DYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLT 369
+ I + E+K +P++V + + + A + +FEEMIDSG AP+ T
Sbjct: 489 FLERIEELFVELKEKNFRPDVVTWTSRIGAYSRKKLYVKCLEVFEEMIDSGCAPDGGTAK 548
Query: 370 AVI 372
++
Sbjct: 549 VLL 551
Score = 40.4 bits (93), Expect = 0.003
Identities = 30/122 (24%), Positives = 55/122 (44%), Gaps = 2/122 (1%)
Query: 202 FGR--QFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPD 259
+GR QF +E++ +M +G D TY+ +I+ K ++ F + + PD
Sbjct: 449 YGRLGQFTKMEKILAEMENGPCTADISTYNILINIYGKAGFLERIEELFVELKEKNFRPD 508
Query: 260 EVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQ 319
VT+++ + Y+R + + +FE +G PD T VL + + VL+
Sbjct: 509 VVTWTSRIGAYSRKKLYVKCLEVFEEMIDSGCAPDGGTAKVLLSACSSEEQVEQVTSVLR 568
Query: 320 EM 321
M
Sbjct: 569 TM 570
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,132,525
Number of Sequences: 26719
Number of extensions: 713325
Number of successful extensions: 11179
Number of sequences better than 10.0: 497
Number of HSP's better than 10.0 without gapping: 457
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 2343
Number of HSP's gapped (non-prelim): 2867
length of query: 715
length of database: 11,318,596
effective HSP length: 106
effective length of query: 609
effective length of database: 8,486,382
effective search space: 5168206638
effective search space used: 5168206638
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146791.8


