

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146791.3 - phase: 0 /pseudo
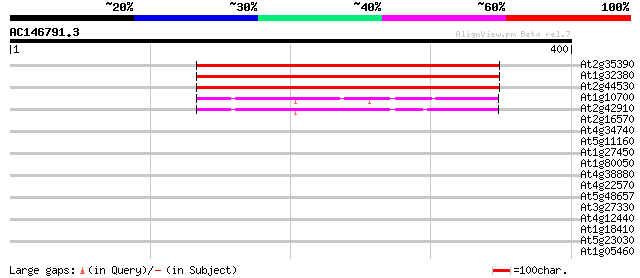
(400 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g35390 phosphoribosyl pyrophosphate synthetase 400 e-112
At1g32380 ribose-phosphate pyrophosphokinase 2 398 e-111
At2g44530 phosphoribosyl pyrophosphate synthetase like protein 395 e-110
At1g10700 putative phosphoribosyl diphosphate synthase 67 1e-11
At2g42910 putative ribose phosphate pyrophosphokinase 57 2e-08
At2g16570 amidophosphoribosyltransferase 37 0.019
At4g34740 amidophosphoribosyltransferase 2 precursor 36 0.033
At5g11160 adenine phosphoribosyltransferase - like protein 33 0.36
At1g27450 adenine phosphoribosyltransferase 1, APRT 32 0.47
At1g80050 adenine phosphoribosyltransferase 31 1.1
At4g38880 amidophosphoribosyltransferase - like protein 31 1.4
At4g22570 adenine phosphoribosyltransferase (EC 2.4.2.7) - like ... 31 1.4
At5g48657 unknown protein 30 2.3
At3g27330 hypothetical protein 30 2.3
At4g12440 putative adenine phosphoribosyltransferase 30 3.1
At1g18410 kinesin-related protein, putative 29 4.0
At5g23030 senescence-associated protein 5-like protein 28 6.8
At1g05460 unknown protein 28 6.8
>At2g35390 phosphoribosyl pyrophosphate synthetase
Length = 403
Score = 400 bits (1027), Expect = e-112
Identities = 196/216 (90%), Positives = 208/216 (95%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGC+VYL+QPTC P NENLMEL +M+DACRRASAK +TAVIPYFGYARADR
Sbjct: 129 IYVQLQESVRGCDVYLVQPTCTPTNENLMELLIMVDACRRASAKKVTAVIPYFGYARADR 188
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLIT+AGADRVLACDLHSGQSMGYFDIPVDHV+CQPVILDYLASK+
Sbjct: 189 KTQGRESIAAKLVANLITEAGADRVLACDLHSGQSMGYFDIPVDHVYCQPVILDYLASKS 248
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
I S DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDV+GKVA+M
Sbjct: 249 IPSEDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVRGKVAIM 308
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTI KGAALLH+EGAREVYACCTH VFS
Sbjct: 309 VDDMIDTAGTIVKGAALLHQEGAREVYACCTHAVFS 344
>At1g32380 ribose-phosphate pyrophosphokinase 2
Length = 400
Score = 398 bits (1022), Expect = e-111
Identities = 196/216 (90%), Positives = 208/216 (95%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQL+ESVRGC+V+L+QPTC P NENLMEL +M+DACRRASAK +TAVIPYFGYARADR
Sbjct: 126 IYVQLKESVRGCDVFLVQPTCTPTNENLMELLIMVDACRRASAKKVTAVIPYFGYARADR 185
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLIT+AGADRVLACDLHSGQSMGYFDIPVDHV+CQPVILDYLASK+
Sbjct: 186 KTQGRESIAAKLVANLITEAGADRVLACDLHSGQSMGYFDIPVDHVYCQPVILDYLASKS 245
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
ISS DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRR GHNVAEVMNLIGDVKGKVAVM
Sbjct: 246 ISSEDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRHGHNVAEVMNLIGDVKGKVAVM 305
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDD+IDTAGTI KGAALLHEEGAREVYACCTH VFS
Sbjct: 306 VDDIIDTAGTIVKGAALLHEEGAREVYACCTHAVFS 341
>At2g44530 phosphoribosyl pyrophosphate synthetase like protein
Length = 394
Score = 395 bits (1016), Expect = e-110
Identities = 198/216 (91%), Positives = 208/216 (95%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGC+V+L+QPTCPPANENLMEL VMIDACRRASAK ITAVIPYFGYARADR
Sbjct: 114 IYVQLQESVRGCDVFLVQPTCPPANENLMELLVMIDACRRASAKTITAVIPYFGYARADR 173
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLIT++GADRVLACDLHSGQSMGYFDIPVDHV+ QPVILDYLASK
Sbjct: 174 KTQGRESIAAKLVANLITQSGADRVLACDLHSGQSMGYFDIPVDHVYGQPVILDYLASKA 233
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
ISS DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRR GHNVAEVMNLIGDVKGKVA+M
Sbjct: 234 ISSEDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRHGHNVAEVMNLIGDVKGKVAIM 293
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTI+KGAALLH+EGAREVYAC TH VFS
Sbjct: 294 VDDMIDTAGTISKGAALLHQEGAREVYACTTHAVFS 329
>At1g10700 putative phosphoribosyl diphosphate synthase
Length = 411
Score = 67.4 bits (163), Expect = 1e-11
Identities = 58/224 (25%), Positives = 111/224 (48%), Gaps = 18/224 (8%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+++Q + +RG +V + PA + E +I A + + T V+P+F ++R
Sbjct: 140 LFIQNAQGIRGQHVAFLASFSSPAV--IFEQLSVIYALPKLFVSSFTLVLPFFPTGTSER 197
Query: 194 -KTQGRESIA---AKLVANLIT-KAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDY 248
+ +G + A A++++N+ T + G ++ D+H+ Q YF + + C +
Sbjct: 198 MEDEGDVATAFTLARILSNIPTSRGGPTSLVTFDIHALQERFYFGDTI--LPCFESGIPL 255
Query: 249 LASKTIS---SNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHN-VAEVMNLIG 304
L S+ S S+++ + PD G R F K+L P + +K R G + + G
Sbjct: 256 LKSRLQSLPDSDNISIAFPDDGAWKR---FHKQLQHYPTIVCNKVRMGDKRIVRIKE--G 310
Query: 305 DVKGKVAVMVDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVF 348
D +G+ V+VDD++ + GT+ + +L GA ++ A THG+F
Sbjct: 311 DAEGRHVVIVDDLVQSGGTLIECQKVLAAHGAAKISAYVTHGIF 354
>At2g42910 putative ribose phosphate pyrophosphokinase
Length = 337
Score = 56.6 bits (135), Expect = 2e-08
Identities = 59/222 (26%), Positives = 101/222 (44%), Gaps = 14/222 (6%)
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+++ +RG +V + PA + E +I R + T V+P+F +R
Sbjct: 67 LFINNAHDIRGQHVAFLASFSSPAV--IFEQISVIYLLPRLFVASFTLVLPFFPTGSFER 124
Query: 194 -KTQGRESIA---AKLVANL-ITKAGADRVLACDLHSGQSMGYF-DIPVDHVHCQPVILD 247
+ +G + A A++V+N+ I++ G V+ D+H+ Q YF D + +L
Sbjct: 125 MEEEGDVATAFTMARIVSNIPISRGGPTSVVIYDIHALQERFYFADQVLPLFETGIPLLT 184
Query: 248 YLASKTISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLI-GDV 306
+ + ++V PD G R F K L P + K R G ++ L G+
Sbjct: 185 KRLQQLPETEKVIVAFPDDGAWKR---FHKLLDHYPTVVCTKVREGDK--RIVRLKEGNP 239
Query: 307 KGKVAVMVDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVF 348
G V+VDD++ + GT+ + +L GA +V A THGVF
Sbjct: 240 AGCHVVIVDDLVQSGGTLIECQKVLAAHGAVKVSAYVTHGVF 281
>At2g16570 amidophosphoribosyltransferase
Length = 566
Score = 37.0 bits (84), Expect = 0.019
Identities = 40/145 (27%), Positives = 57/145 (38%), Gaps = 28/145 (19%)
Query: 231 YFDIPVDHV------HCQPVILDYLASKTISSNDLVVVSPDVGGVARARAFAKKLSDAPL 284
YF +P V + V + LA+++ D+V+ PD G VA AK
Sbjct: 333 YFSLPNSIVFGRSVYESRHVFGEILATESPVECDVVIAVPDSGVVAALGYAAKSGVPFQQ 392
Query: 285 AIVDKRRSGHNVAEVMNLIGD-------------VKGKVAVMVDDMIDTAGTIAKGAALL 331
++ G E I D ++GK V+VDD I T +K LL
Sbjct: 393 GLIRSHYVGRTFIEPSQKIRDFGVKLKLSPVRGVLEGKRVVVVDDSIVRGTTSSKIVRLL 452
Query: 332 HEEGAREVY---------ACCTHGV 347
E GA+EV+ A C +GV
Sbjct: 453 REAGAKEVHMRIASPPIVASCYYGV 477
>At4g34740 amidophosphoribosyltransferase 2 precursor
Length = 561
Score = 36.2 bits (82), Expect = 0.033
Identities = 40/145 (27%), Positives = 57/145 (38%), Gaps = 28/145 (19%)
Query: 231 YFDIPVDHV------HCQPVILDYLASKTISSNDLVVVSPDVGGVARARAFAKKLSDAPL 284
YF +P V + V + LA+++ D+V+ PD G VA AK
Sbjct: 329 YFSLPNSIVFGRSVYESRHVFGEILATESPVDCDVVIAVPDSGVVAALGYAAKAGVAFQQ 388
Query: 285 AIVDKRRSGHNVAEVMNLIGD-------------VKGKVAVMVDDMIDTAGTIAKGAALL 331
++ G E I D ++GK V+VDD I T +K LL
Sbjct: 389 GLIRSHYVGRTFIEPSQKIRDFGVKLKLSPVRGVLEGKRVVVVDDSIVRGTTSSKIVRLL 448
Query: 332 HEEGAREVY---------ACCTHGV 347
E GA+EV+ A C +GV
Sbjct: 449 REAGAKEVHMRIASPPIIASCYYGV 473
>At5g11160 adenine phosphoribosyltransferase - like protein
Length = 191
Score = 32.7 bits (73), Expect = 0.36
Identities = 12/36 (33%), Positives = 22/36 (60%)
Query: 312 VMVDDMIDTAGTIAKGAALLHEEGAREVYACCTHGV 347
+++DD++ T GT++ +LL +GA V C G+
Sbjct: 129 IIIDDLVATGGTLSAAMSLLESQGAEVVECACVIGL 164
>At1g27450 adenine phosphoribosyltransferase 1, APRT
Length = 243
Score = 32.3 bits (72), Expect = 0.47
Identities = 14/36 (38%), Positives = 21/36 (57%)
Query: 308 GKVAVMVDDMIDTAGTIAKGAALLHEEGAREVYACC 343
G+ A+++DD+I T GT+A LL G + V C
Sbjct: 182 GERAIIIDDLIATGGTLAAAIRLLERVGVKIVECAC 217
>At1g80050 adenine phosphoribosyltransferase
Length = 192
Score = 31.2 bits (69), Expect = 1.1
Identities = 13/37 (35%), Positives = 21/37 (56%)
Query: 311 AVMVDDMIDTAGTIAKGAALLHEEGAREVYACCTHGV 347
A+++DD++ T GT++ LL GA V C G+
Sbjct: 128 ALIIDDLVATGGTLSASINLLERAGAEVVECACVVGL 164
>At4g38880 amidophosphoribosyltransferase - like protein
Length = 532
Score = 30.8 bits (68), Expect = 1.4
Identities = 30/112 (26%), Positives = 44/112 (38%), Gaps = 22/112 (19%)
Query: 258 DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGD------------ 305
D+V+ PD G VA AK + ++ + E I D
Sbjct: 352 DVVIAVPDSGTVAALGYAAKAGVPFQIGLLRSHYAKRTFIEPTQEIRDFAVKVKLSPVRA 411
Query: 306 -VKGKVAVMVDDMIDTAGTIAKGAALLHEEGAREVY---------ACCTHGV 347
++GK V+VDD I T K +L + GA+EV+ A C +GV
Sbjct: 412 VLEGKRVVVVDDSIVRGTTSLKIVRMLRDAGAKEVHMRIALPPMIASCYYGV 463
>At4g22570 adenine phosphoribosyltransferase (EC 2.4.2.7) - like
protein
Length = 183
Score = 30.8 bits (68), Expect = 1.4
Identities = 18/42 (42%), Positives = 23/42 (53%), Gaps = 1/42 (2%)
Query: 303 IGDVK-GKVAVMVDDMIDTAGTIAKGAALLHEEGAREVYACC 343
IG V+ G A++VDD+I T GT+ LL GA V C
Sbjct: 117 IGAVEAGDRALVVDDLIATGGTLCAAINLLERVGAEVVECAC 158
>At5g48657 unknown protein
Length = 245
Score = 30.0 bits (66), Expect = 2.3
Identities = 17/61 (27%), Positives = 29/61 (46%), Gaps = 2/61 (3%)
Query: 22 SFFSGVSHGLTFPTLGFVDLSRSRTVAYNTVVITFF--LFFILLCFIFVFFFTFFIFLLE 79
SF + + LT +G + S ++I+ F + F +CF +FF F+ L+E
Sbjct: 185 SFINPLFLQLTLLCVGLLFSSMEPKGKLMKMIISLFYIILFFFICFCLIFFNNIFVILME 244
Query: 80 M 80
M
Sbjct: 245 M 245
>At3g27330 hypothetical protein
Length = 913
Score = 30.0 bits (66), Expect = 2.3
Identities = 14/22 (63%), Positives = 14/22 (63%)
Query: 55 TFFLFFILLCFIFVFFFTFFIF 76
TFF F IL F FV F T FIF
Sbjct: 21 TFFWFVILFVFSFVLFSTMFIF 42
>At4g12440 putative adenine phosphoribosyltransferase
Length = 182
Score = 29.6 bits (65), Expect = 3.1
Identities = 14/36 (38%), Positives = 19/36 (51%)
Query: 308 GKVAVMVDDMIDTAGTIAKGAALLHEEGAREVYACC 343
G A++VDD+I T GT+ LL GA + C
Sbjct: 122 GDRALVVDDLIATGGTLCAAMNLLKRVGAEVIECAC 157
>At1g18410 kinesin-related protein, putative
Length = 1081
Score = 29.3 bits (64), Expect = 4.0
Identities = 15/61 (24%), Positives = 32/61 (51%), Gaps = 3/61 (4%)
Query: 260 VVVSPDVGGVARARA---FAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVMVDD 316
V ++PD+ + + + FA+++S L + G +V E+M +G +K +A D+
Sbjct: 927 VQLNPDITSYSESMSTLKFAERVSGVELGAAKSSKDGRDVRELMEQLGSLKDTIARKDDE 986
Query: 317 M 317
+
Sbjct: 987 I 987
>At5g23030 senescence-associated protein 5-like protein
Length = 264
Score = 28.5 bits (62), Expect = 6.8
Identities = 10/27 (37%), Positives = 18/27 (66%)
Query: 52 VVITFFLFFILLCFIFVFFFTFFIFLL 78
++IT +LFF+ L + + + FIFL+
Sbjct: 70 IIITLYLFFLFLSILLLLVLSVFIFLV 96
>At1g05460 unknown protein
Length = 1002
Score = 28.5 bits (62), Expect = 6.8
Identities = 19/64 (29%), Positives = 26/64 (39%), Gaps = 1/64 (1%)
Query: 212 KAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKTISSNDLVVVSPDVGGVAR 271
K+ +R+ CD + Y V + C P ILD L SK +LV D V
Sbjct: 594 KSYLERLFECDYYCEGDENYVTKLVKNYRCHPEILD-LPSKLFYDGELVASKEDTDSVLA 652
Query: 272 ARAF 275
+ F
Sbjct: 653 SLNF 656
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.337 0.146 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,708,545
Number of Sequences: 26719
Number of extensions: 287034
Number of successful extensions: 1311
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1292
Number of HSP's gapped (non-prelim): 18
length of query: 400
length of database: 11,318,596
effective HSP length: 101
effective length of query: 299
effective length of database: 8,619,977
effective search space: 2577373123
effective search space used: 2577373123
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146791.3


