

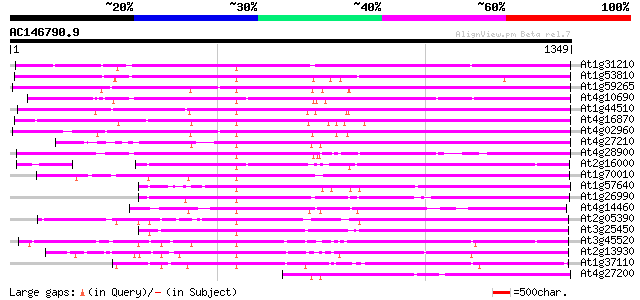
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146790.9 + phase: 0
(1349 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g31210 putative reverse transcriptase 978 0.0
At1g53810 959 0.0
At1g59265 polyprotein, putative 911 0.0
At4g10690 retrotransposon like protein 901 0.0
At1g44510 polyprotein, putative 896 0.0
At4g16870 retrotransposon like protein 893 0.0
At4g02960 putative polyprotein of LTR transposon 884 0.0
At4g27210 putative protein 848 0.0
At4g28900 putative protein 845 0.0
At2g16000 putative retroelement pol polyprotein 660 0.0
At1g70010 hypothetical protein 659 0.0
At1g57640 615 e-176
At1g26990 polyprotein, putative 596 e-170
At4g14460 retrovirus-related like polyprotein 581 e-165
At2g05390 putative retroelement pol polyprotein 566 e-161
At3g25450 hypothetical protein 563 e-160
At3g45520 copia-like polyprotein 538 e-153
At2g13930 putative retroelement pol polyprotein 533 e-151
At1g37110 523 e-148
At4g27200 putative protein 523 e-148
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 978 bits (2528), Expect = 0.0
Identities = 557/1378 (40%), Positives = 798/1378 (57%), Gaps = 71/1378 (5%)
Query: 14 SSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEE---FITSSDSSKSNNPAFE 70
SSV++KL +NY LWK+ ++ KL G++ G P + + +S+ NP +E
Sbjct: 15 SSVTLKLTDSNYLLWKTQFESLLSSQKLIGFVNGAVNAPSQSRLVVNGEVTSEEPNPLYE 74
Query: 71 EWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQI---IYLKFQF 127
W DQ + W+ +++ E+ + + TS+Q+W SLA +S + L+
Sbjct: 75 SWFCTDQLVRSWLFGTLSEEVLGHVHNLSTSRQIW---VSLAENFNKSSVAREFSLRQNL 131
Query: 128 HSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSNLIIQTLNGLDSEYNPIVV----KLS 183
+ K E Y + K + D L G P+ S I LNGL +Y+PI LS
Sbjct: 132 QLLSKKEKPFSVYCREFKTICDALSSIGKPVDESMKIFGFLNGLGRDYDPITTVIQSSLS 191
Query: 184 DHTTLSWVDLQAQLLTFESRIEQLNTLTNLNLNATANVANKFDHRDNRFNSNNNWRG--- 240
T ++ D+ +++ F+S+++ ++ + N+ + + ++N N RG
Sbjct: 192 KLPTPTFNDVVSEVQGFDSKLQSYEEAASVTPHLAFNI-ERSESGSPQYNPNQKGRGRSG 250
Query: 241 -SNFRGWRGGRGRGRSS----------KAPCQVYGKTNHTAINCFHRFDKNYSRSNYSAD 289
+ RG RGRG S + CQ+ G+T HTA+ C++RFD NY
Sbjct: 251 QNKGRGGYSTRGRGFSQHQSSPQVSGPRPVCQICGRTGHTALKCYNRFDNNY-------- 302
Query: 290 SDKQGSHNAFIASQNSVED-YDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKL 348
Q AF + S + +W+ DS A+ HVT T+ Q TE+ G ++ +VG+G L
Sbjct: 303 ---QAEIQAFSTLRVSDDTGKEWHPDSAATAHVTSSTNGLQSATEYEGDDAVLVGDGTYL 359
Query: 349 EIVATGSSKLKSLN----LDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLT 404
I TGS+ +KS N L++VL VPNI K+LLSVSKL D V FD N + + T
Sbjct: 360 PITHTGSTTIKSSNGKIPLNEVLVVPNIQKSLLSVSKLCDDYPCGVYFDANKVCIIDLQT 419
Query: 405 GKVILKGLLKNGLYQLSDTKG----NPYAFVSVKESWHRRLGHPNNKVLDKVLKSCNVKV 460
KV+ G +NGLY L + + + + +E WH RLGH N+K L + S +++
Sbjct: 420 QKVVTTGPRRNGLYVLENQEFVALYSNRQCAATEEVWHHRLGHANSKALQHLQNSKAIQI 479
Query: 461 PPSDNFSFCEACQYGKMHLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFID 520
S CE CQ GK LPF S S PL+ +H D+WGP+P++S+ G KYY F+D
Sbjct: 480 NKSRTSPVCEPCQMGKSSRLPFLISDSRVLHPLDRIHCDLWGPSPVVSNQGLKYYAIFVD 539
Query: 521 DFSRFTWIYPLKQKSETVQAFTT-----ENQFNKRIKVIQCDGGGEY--KPVQKLAIDVG 573
D+SR++W YPL KSE + F + ENQ N +IKV Q DGGGE+ ++ + G
Sbjct: 540 DYSRYSWFYPLHNKSEFLSVFISFQKLVENQLNTKIKVFQSDGGGEFVSNKLKTHLSEHG 599
Query: 574 IQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQV 633
I R+SCPYT QQNG AERKHRH+ E GL++L + P +W E+F TA Y+INRLPS V
Sbjct: 600 IHHRISCPYTPQQNGLAERKHRHLVELGLSMLFHSHTPQKFWVESFFTANYIINRLPSSV 659
Query: 634 TQNESPYSLIFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKC 693
+N SPY +F ++P+Y L+ FG ACYPCL+P Q+K + +CVFLGY++ +KGY+C
Sbjct: 660 LKNLSPYEALFGEKPDYSSLRVFGSACYPCLRPLAQNKFDPRSLQCVFLGYNSQYKGYRC 719
Query: 694 LNSH-GRTFISRHVIFNEDLFPFHEGFLNTRSPLKTTINNPSTSFPLCSAG--NSINDAS 750
G+ +ISR+VIFNE PF E + + P S PL A N I++ S
Sbjct: 720 FYPPTGKVYISRNVIFNESELPFKEKYQSLV---------PQYSTPLLQAWQHNKISEIS 770
Query: 751 IPIIEEENQDETNEEDSQGVTSDTEQ-TDNGSSEGDTTHEETLDIVQQQNVGESSLDTNT 809
+P + + + ++ + TEQ TD + + +E ++ V ++ N
Sbjct: 771 VPAAPVQLFSKPIDLNTYAGSQVTEQLTDPEPTSNNEGSDEEVNPVAEEIAANQEQVIN- 829
Query: 810 SNAIHTRSKSGIHKPKLPYIGITETYKDTVEPANVKEALTRTLWKEAMQKEFQALMSNKT 869
S+A+ TRSK+GI KP Y IT +T EP + A+ W EA+ +E + T
Sbjct: 830 SHAMTTRSKAGIQKPNTRYALITSRM-NTAEPKTLASAMKHPGWNEAVHEEINRVHMLHT 888
Query: 870 WILVPYQDQENIVDSKWVFKTKYKSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVS 929
W LVP D NI+ SKWVFKTK DGSI++ KARLVAKGF Q G+DY ETFSPVV+ +
Sbjct: 889 WSLVPPTDDMNILSSKWVFKTKLHPDGSIDKLKARLVAKGFDQEEGVDYLETFSPVVRTA 948
Query: 930 TVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYG 989
T+R++L ++ W ++QLD++NAFL+G L+E VFM+QP GF+DP KP H+C+L+KAIYG
Sbjct: 949 TIRLVLDVSTSKGWPIKQLDVSNAFLHGELQEPVFMYQPSGFIDPQKPTHVCRLTKAIYG 1008
Query: 990 LKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQ 1049
LKQAPRAWFD+ LL++GF +KSDPSLF+ I +LL+YVDDI++TGS + L+
Sbjct: 1009 LKQAPRAWFDTFSNFLLDYGFVCSKSDPSLFVCHQDGKILYLLLYVDDILLTGSDQSLLE 1068
Query: 1050 AFIKQLNDVFSLKDLGRLHYFLGIEVQRDASGMYLKQSKYIGDLLKKFKMENASPCPTPM 1109
++ L + FS+KDLG YFLGI+++ A+G++L Q+ Y D+L++ M + +P PTP
Sbjct: 1069 DLLQALKNRFSMKDLGPPRYFLGIQIEDYANGLFLHQTAYATDILQQAGMSDCNPMPTP- 1127
Query: 1110 ITGRHFTVEGEKLKDPTVFRQAIGGLQYLTHTRPDIAFSVNKLSQYMSSPTTDHWQGIKR 1169
+ + + E +PT FR G LQYLT TRPDI F+VN + Q M SPTT + +KR
Sbjct: 1128 LPQQLDNLNSELFAEPTYFRSLAGKLQYLTITRPDIQFAVNFICQRMHSPTTSDFGLLKR 1187
Query: 1170 ILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDDRKSMAGQCVFLGETLISWSSRKQ 1229
ILRY++GTI L IK ++ L ++ +SD+D A + R+S G C+ LG LISWS+++Q
Sbjct: 1188 ILRYIKGTIGMGLPIKRNSTLTLSAYSDSDHAGCKNTRRSTTGFCILLGSNLISWSAKRQ 1247
Query: 1230 KVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSAKALASNPVLHA 1289
VS SSTE+EYRAL A EI WI LL +L +P ++CDNLSA L++NP LH
Sbjct: 1248 PTVSNSSTEAEYRALTYAAREITWISFLLRDLGIPQYLPTQVYCDNLSAVYLSANPALHN 1307
Query: 1290 RSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQLRDKLGVIHSP 1347
RSKH + D HYIR+QV + ++ T Q+AD TK L F LR KLGV SP
Sbjct: 1308 RSKHFDTDYHYIREQVALGLIETQHISATFQLADVFTKSLPRRAFVDLRSKLGVSGSP 1365
>At1g53810
Length = 1522
Score = 959 bits (2480), Expect = 0.0
Identities = 554/1434 (38%), Positives = 801/1434 (55%), Gaps = 108/1434 (7%)
Query: 11 DLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEF--ITSSD-SSKSNNP 67
++ + V+V L++ NY LWKS + G L G++ G+ P + +T ++ +S+ NP
Sbjct: 10 NISNCVTVTLNQQNYILWKSQFESFLSGQGLLGFVTGSISAPAQTRSVTHNNVTSEEPNP 69
Query: 68 AFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKFQF 127
F W DQ + W+L S A ++ + +++C TS Q+W + + S++ L+ +
Sbjct: 70 EFYTWHQTDQVVKSWLLGSFAEDILSVVVNCFTSHQVWLTLANHFNRVSSSRLFELQRRL 129
Query: 128 HSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSNLIIQTLNGLDSEYNPIVVKLSD--- 184
++ K + ME +L +K++ D+L G+P+ I LNGL EY PI + +
Sbjct: 130 QTLEKKDNTMEVFLKDLKHICDQLASVGSPVPEKMKIFSALNGLGREYEPIKTTIENSVD 189
Query: 185 -HTTLSWVDLQAQLLTFESRIEQLNTLTNLNLNATANVANKFDHRDNRFNSNNNWRGSNF 243
+ +LS ++ ++L ++ R++ T ++ + NV H D+ + NNN RG
Sbjct: 190 SNPSLSLDEVASKLRGYDDRLQSYVTEPTISPHVAFNVT----HSDSGYYHNNN-RGKGR 244
Query: 244 RGWRGG------RGRG-------------RSSKAPCQVYGKTNHTAINCFHRFDKNYSRS 284
G RGRG +S CQ+ GK H A+ C+HRFD +Y
Sbjct: 245 SNSGSGKSSFSTRGRGFHQQISPTSGSQAGNSGLVCQICGKAGHHALKCWHRFDNSYQHE 304
Query: 285 NYSADSDKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGN 344
+ I ++W DS AS HVT+ Q +HG +S +V +
Sbjct: 305 DLP-----MALATMRITDVTDHHGHEWIPDSAASAHVTNNRHVLQQSQPYHGSDSIMVAD 359
Query: 345 GDKLEIVATGSSKLKS----LNLDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVK 400
G+ L I TGS + S + L +VL P+I K+LLSVSKL +D VEFD + +
Sbjct: 360 GNFLPITHTGSGSIASSSGKIPLKEVLVCPDIVKSLLSVSKLTSDYPCSVEFDADSVRIN 419
Query: 401 EKLTGKVILKGLLKNGLYQLSDTKGNPYAFV----SVKESWHRRLGHPNNKVLDKVLKSC 456
+K T K+++ G ++GLY L + K + E WHRRLGH N +VL ++ S
Sbjct: 420 DKATKKLLVMGRNRDGLYSLEEPKLQVLYSTRQNSASSEVWHRRLGHANAEVLHQLASSK 479
Query: 457 NVKVPPSDNFSFCEACQYGKMHLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYV 516
++ + + CEAC GK LPF S+ +A PLE +H D+WGP+P S GF+YYV
Sbjct: 480 SIIIINKVVKTVCEACHLGKSTRLPFMLSTFNASRPLERIHCDLWGPSPTSSVQGFRYYV 539
Query: 517 HFIDDFSRFTWIYPLKQKSETVQAFT-----TENQFNKRIKVIQCDGGGEYKPVQ--KLA 569
FID +SRFTW YPLK KS+ F ENQ +IK+ QCDGGGE+ Q K
Sbjct: 540 VFIDHYSRFTWFYPLKLKSDFFSTFVMFQKLVENQLGHKIKIFQCDGGGEFISSQFLKHL 599
Query: 570 IDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRL 629
D GIQ MSCPYT QQNG AERKHRHI E GL+++ Q+++PL YW E+F TA ++IN L
Sbjct: 600 QDHGIQQNMSCPYTPQQNGMAERKHRHIVELGLSMIFQSKLPLKYWLESFFTANFVINLL 659
Query: 630 P-SQVTQNESPYSLIFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSH 688
P S + NESPY ++ K P Y L+ FGCACYP L+ Y K + +CVFLGY+ +
Sbjct: 660 PTSSLDNNESPYQKLYGKAPEYSALRVFGCACYPTLRDYASTKFDPRSLKCVFLGYNEKY 719
Query: 689 KGYKCLNSH-GRTFISRHVIFNEDLFPFHEGFLNTRSPLKT-------------TINNPS 734
KGY+CL GR +ISRHV+F+E+ PF + + KT T P
Sbjct: 720 KGYRCLYPPTGRIYISRHVVFDENTHPFESIYSHLHPQDKTPLLEAWFKSFHHVTPTQPD 779
Query: 735 TS-FPLCSAGN----SINDASIPIIEEENQDETNEEDSQG------VTSDTEQTD--NGS 781
S +P+ S ++ A + E +++ SQ V+ E+T + +
Sbjct: 780 QSRYPVSSIPQPETTDLSAAPASVAAETAGPNASDDTSQDNETISVVSGSPERTTGLDSA 839
Query: 782 SEGDTTHEETLDI------------------VQQQNVGESSLDTNTSNAIHTRSKSGIHK 823
S GD+ H T D +Q + +A+ TR K GI K
Sbjct: 840 SIGDSYHSPTADSSHPSPARSSPASSPQGSPIQMAPAQQVQAPVTNEHAMVTRGKEGISK 899
Query: 824 PKLPYIGITETYKDTVEPANVKEALTRTLWKEAMQKEFQALMSNKTWILVPYQDQENIVD 883
P Y+ +T EP V EAL W AMQ+E +TW LVPY N++
Sbjct: 900 PNKRYVLLTHKVS-IPEPKTVTEALKHPGWNNAMQEEMGNCKETETWTLVPYSPNMNVLG 958
Query: 884 SKWVFKTKYKSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVRVILSIAVHLNW 943
S WVF+TK +DGS+++ KARLVAKGF+Q GIDY ET+SPVV+ TVR+IL +A L W
Sbjct: 959 SMWVFRTKLHADGSLDKLKARLVAKGFKQEEGIDYLETYSPVVRTPTVRLILHVATVLKW 1018
Query: 944 EVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKT 1003
E++Q+D+ NAFL+G L ETV+M QP GFVD +KP+H+C L K++YGLKQ+PRAWFD
Sbjct: 1019 ELKQMDVKNAFLHGDLTETVYMRQPAGFVDKSKPDHVCLLHKSLYGLKQSPRAWFDRFSN 1078
Query: 1004 ALLNWGFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKD 1063
LL +GF + DPSLF+ + + LL+YVDD+++TG+++ L + LN F +KD
Sbjct: 1079 FLLEFGFICSLFDPSLFVYSSNNDVILLLLYVDDMVITGNNSQSLTHLLAALNKEFRMKD 1138
Query: 1064 LGRLHYFLGIEVQRDASGMYLKQSKYIGDLLKKFKMENASPCPTPM-ITGRHFTVEGEKL 1122
+G++HYFLGI++Q G+++ Q KY DLL M N SP PTP+ + + + E
Sbjct: 1139 MGQVHYFLGIQIQTYDGGLFMSQQKYAEDLLITASMANCSPMPTPLPLQLDRVSNQDEVF 1198
Query: 1123 KDPTVFRQAIGGLQYLTHTRPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCL 1182
DPT FR G LQYLT TRPDI F+VN + Q M P+ + +KRILRY++GT++ +
Sbjct: 1199 SDPTYFRSLAGKLQYLTLTRPDIQFAVNFVCQKMHQPSVSDFNLLKRILRYIKGTVSMGI 1258
Query: 1183 HIKPS---------TDLDITGFSDADWATSIDDRKSMAGQCVFLGETLISWSSRKQKVVS 1233
+ +D D++ +SD+D+A + R+S+ G C F+G+ +ISWSS+KQ VS
Sbjct: 1259 QYNSNSSSVVSAYESDYDLSAYSDSDYANCKETRRSVGGYCTFMGQNIISWSSKKQPTVS 1318
Query: 1234 RSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSAKALASNPVLHARSKH 1293
RSSTE+EYR+L + A+EI W+ S+L E+ + LP P L+CDNLSA L +NP HAR+KH
Sbjct: 1319 RSSTEAEYRSLSETASEIKWMSSILREIGVSLPDTPELFCDNLSAVYLTANPAFHARTKH 1378
Query: 1294 IEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQLRDKLGVIHSP 1347
++D HYIR++V +VV ++P Q+AD TK L F++LR KLGV P
Sbjct: 1379 FDVDHHYIRERVALKTLVVKHIPGHLQLADIFTKSLPFEAFTRLRFKLGVDFPP 1432
>At1g59265 polyprotein, putative
Length = 1466
Score = 911 bits (2354), Expect = 0.0
Identities = 543/1462 (37%), Positives = 790/1462 (53%), Gaps = 127/1462 (8%)
Query: 7 NNKNDLPSSVS--VKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKS 64
NN + L ++S KL NY +W V + G +L G++ G+ P I +D++
Sbjct: 11 NNTSILNVNMSNVTKLTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATI-GTDAAPR 69
Query: 65 NNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLK 124
NP + W+ D+ + +L +++ + + T+ Q+W+ + + + + L+
Sbjct: 70 VNPDYTRWKRQDKLIYSAVLGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLR 129
Query: 125 FQFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSNLIIQTLNGLDSEYNPIVVKLS- 183
Q KG ++DY+ + D+L L G P+ + + + L L EY P++ +++
Sbjct: 130 TQLKQWTKGTKTIDDYMQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQIAA 189
Query: 184 DHTTLSWVDLQAQLLTFESRIEQLNTLTNLNLNATA------------NVANKFDHRDNR 231
T + ++ +LL ES+I +++ T + + A A N N+ + DNR
Sbjct: 190 KDTPPTLTEIHERLLNHESKILAVSSATVIPITANAVSHRNTTTTNNNNNGNRNNRYDNR 249
Query: 232 FNSNNN--WRGSNFRGWRGGRGRGRSSKAPCQVYGKTNHTAINCF---HRFDKNYSRSNY 286
N+NN+ W+ S+ + + + CQ+ G H+A C H S+
Sbjct: 250 NNNNNSKPWQQSS-TNFHPNNNQSKPYLGKCQICGVQGHSAKRCSQLQHFLSSVNSQQPP 308
Query: 287 SADSDKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGD 346
S + Q N + S S + W DSGA++H+T + + G + +V +G
Sbjct: 309 SPFTPWQPRANLALGSPYSSNN--WLLDSGATHHITSDFNNLSLHQPYTGGDDVMVADGS 366
Query: 347 KLEIVATGSSKLKS----LNLDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEK 402
+ I TGS+ L + LNL ++LYVPNI KNL+SV +L N + VEF VK+
Sbjct: 367 TIPISHTGSTSLSTKSRPLNLHNILYVPNIHKNLISVYRLCNANGVSVEFFPASFQVKDL 426
Query: 403 LTGKVILKGLLKNGLYQLSDTKGNPYAFVS------VKESWHRRLGHPNNKVLDKVLKSC 456
TG +L+G K+ LY+ P + + SWH RLGHP +L+ V+ +
Sbjct: 427 NTGVPLLQGKTKDELYEWPIASSQPVSLFASPSSKATHSSWHARLGHPAPSILNSVISNY 486
Query: 457 NVKV-PPSDNFSFCEACQYGKMHLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYY 515
++ V PS F C C K + +PF S+ ++ PLE +++DVW +PI+S ++YY
Sbjct: 487 SLSVLNPSHKFLSCSDCLINKSNKVPFSQSTINSTRPLEYIYSDVWS-SPILSHDNYRYY 545
Query: 516 VHFIDDFSRFTWIYPLKQKSETVQAFTT-----ENQFNKRIKVIQCDGGGEYKPVQKLAI 570
V F+D F+R+TW+YPLKQKS+ + F T EN+F RI D GGE+ + +
Sbjct: 546 VIFVDHFTRYTWLYPLKQKSQVKETFITFKNLLENRFQTRIGTFYSDNGGEFVALWEYFS 605
Query: 571 DVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLP 630
GI S P+T + NG +ERKHRHI E GLTLL+ A +P YW AF+ AVYLINRLP
Sbjct: 606 QHGISHLTSPPHTPEHNGLSERKHRHIVETGLTLLSHASIPKTYWPYAFAVAVYLINRLP 665
Query: 631 SQVTQNESPYSLIFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKG 690
+ + Q ESP+ +F PNY L+ FGCACYP L+PYNQHKL + +CVFLGYS +
Sbjct: 666 TPLLQLESPFQKLFGTSPNYDKLRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSA 725
Query: 691 YKCLNSH-GRTFISRHVIFNEDLFPFHEGFLNTRSPLK---------------------- 727
Y CL+ R +ISRHV F+E+ FPF +L T SP++
Sbjct: 726 YLCLHLQTSRLYISRHVRFDENCFPF-SNYLATLSPVQEQRRESSCVWSPHTTLPTRTPV 784
Query: 728 -------------TTINNPSTSFPLCSAGNSINDASI---------PIIEEEN--QDETN 763
T ++PS F +S D+S P +N Q T
Sbjct: 785 LPAPSCSDPHHAATPPSSPSAPFRNSQVSSSNLDSSFSSSFPSSPEPTAPRQNGPQPTTQ 844
Query: 764 EEDSQGVTSDTEQT--DNGSSEGDTTHEETLDIVQQQNVGESSLDTNTSNA--------- 812
+Q T ++ T +N ++E + ++L Q + S T+ S++
Sbjct: 845 PTQTQTQTHSSQNTSQNNPTNESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSI 904
Query: 813 -IH-----------------------TRSKSGIHKPKLPYIGITETYKDTVEPANVKEAL 848
IH TR+K+GI KP P + + EP +AL
Sbjct: 905 LIHPPPPLAQIVNNNNQAPLNTHSMGTRAKAGIIKPN-PKYSLAVSLAAESEPRTAIQAL 963
Query: 849 TRTLWKEAMQKEFQALMSNKTWILVPYQDQE-NIVDSKWVFKTKYKSDGSIERRKARLVA 907
W+ AM E A + N TW LVP IV +W+F KY SDGS+ R KARLVA
Sbjct: 964 KDERWRNAMGSEINAQIGNHTWDLVPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARLVA 1023
Query: 908 KGFQQTAGIDYEETFSPVVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQ 967
KG+ Q G+DY ETFSPV+K +++R++L +AV +W +RQLD+NNAFL G L + V+M Q
Sbjct: 1024 KGYNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQ 1083
Query: 968 PEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLKGKDH 1027
P GF+D +PN++CKL KA+YGLKQAPRAW+ L+ LL GF N+ SD SLF+L+
Sbjct: 1084 PPGFIDKDRPNYVCKLRKALYGLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKS 1143
Query: 1028 ITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASGMYLKQS 1087
I ++L+YVDDI++TG+ L + L+ FS+KD LHYFLGIE +R +G++L Q
Sbjct: 1144 IVYMLVYVDDILITGNDPTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTGLHLSQR 1203
Query: 1088 KYIGDLLKKFKMENASPCPTPMITGRHFTV-EGEKLKDPTVFRQAIGGLQYLTHTRPDIA 1146
+YI DLL + M A P TPM ++ G KL DPT +R +G LQYL TRPDI+
Sbjct: 1204 RYILDLLARTNMITAKPVTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDIS 1263
Query: 1147 FSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDD 1206
++VN+LSQ+M PT +H Q +KRILRYL GT N+ + +K L + +SDADWA DD
Sbjct: 1264 YAVNRLSQFMHMPTEEHLQALKRILRYLAGTPNHGIFLKKGNTLSLHAYSDADWAGDKDD 1323
Query: 1207 RKSMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLP 1266
S G V+LG ISWSS+KQK V RSSTE+EYR++ + ++E+ WI SLL EL + L
Sbjct: 1324 YVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLT 1383
Query: 1267 RKPILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLT 1326
R P+++CDN+ A L +NPV H+R KHI ID H+IR+QV + V +V T DQ+AD LT
Sbjct: 1384 RPPVIYCDNVGATYLCANPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLT 1443
Query: 1327 KPLSHTRFSQLRDKLGVIHSPP 1348
KPLS T F K+GV PP
Sbjct: 1444 KPLSRTAFQNFASKIGVTRVPP 1465
>At4g10690 retrotransposon like protein
Length = 1515
Score = 901 bits (2328), Expect = 0.0
Identities = 538/1406 (38%), Positives = 782/1406 (55%), Gaps = 119/1406 (8%)
Query: 42 DGYMLGTKKCPEE--FITSSD-SSKSNNPAFEEWQANDQRLLGWMLNSMATEMATQLLHC 98
+G++ G P +T D S+ N F +W DQ + W+ S++ E ++
Sbjct: 39 NGFVTGATPRPASTIIVTKDDIQSEEANQEFLKWTRIDQLVKAWIFGSLSEEALKVVIGL 98
Query: 99 ETSKQLWDEAQSLAGAHTRSQIIYLKFQFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPI 158
+++++W + ++ L+ + + K M+ YL ++KN+ D+L G P+
Sbjct: 99 NSAQEVWLGLARRFNRFSTTRKYDLQKRLGTCSKAGKTMDAYLSEVKNICDQLDSIGFPV 158
Query: 159 SNSNLIIQTLNGLDSEYNPIVVKLSDHTTL----SWVDLQAQLLTFESRIEQLNTLTNLN 214
+ I LNGL EY I + + + D+ +L TF+ ++L+T T N
Sbjct: 159 TEQEKIFGVLNGLGKEYESIATVIEHSLDVYPGPCFDDVVYKLTTFD---DKLSTYT-AN 214
Query: 215 LNATANVANKFDHRDNRFNSNNNWRGSNFRGWRG-------------------GRGRGRS 255
T ++A D + NNN RG + +RG G G
Sbjct: 215 SEVTPHLAFYTD-KSYSSRGNNNSRGGRYGNFRGRGSYSSRGRGFHQQFGSGSNNGSGNG 273
Query: 256 SKAPCQVYGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAF----IASQNSVEDYDW 311
SK CQ+ K H+A C+ RF++NY + NAF ++ QN ++W
Sbjct: 274 SKPTCQICRKYGHSAFKCYTRFEENYLPEDLP---------NAFAAMRVSDQNQASSHEW 324
Query: 312 YFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEIVATGSSKLK----SLNLDDVL 367
DS A+ H+T+ TD Q+ + G +S +VGNGD L I G+ L +L L+DVL
Sbjct: 325 LPDSAATAHITNTTDGLQNSQTYSGDDSVIVGNGDFLPITHIGTIPLNISQGTLPLEDVL 384
Query: 368 YVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVILKGLLKNGLYQLSDTKGNP 427
P ITK+LLSVSKL D FD + +K+K T +++ +G GLY L D
Sbjct: 385 VCPGITKSLLSVSKLTDDYPCSFTFDSDSVVIKDKRTQQLLTQGNKHKGLYVLKDVPFQT 444
Query: 428 YAFV----SVKESWHRRLGHPNNKVLDKVLKSCNVKVPPSDNFSFCEACQYGKMHLLPFK 483
Y S E WH+RLGHPN +VL ++K+ + V + + + CEACQ GK+ LPF
Sbjct: 445 YYSTRQQSSDDEVWHQRLGHPNKEVLQHLIKTKAIVVNKTSS-NMCEACQMGKVCRLPFV 503
Query: 484 SSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLKQKSETVQAFT- 542
+S + PLE +H D+WGPAP+ S+ GF+YYV FID++SRFTW YPLK KS+ F
Sbjct: 504 ASEFVSSRPLERIHCDLWGPAPVTSAQGFQYYVIFIDNYSRFTWFYPLKLKSDFFSVFVL 563
Query: 543 ----TENQFNKRIKVIQCDGGGE---YKPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHR 595
ENQ+ +I + QCDGGGE YK V LA GI+ +SCP+T QQNG AER+HR
Sbjct: 564 FQQLVENQYQHKIAMFQCDGGGEFVSYKFVAHLA-SCGIKQLISCPHTPQQNGIAERRHR 622
Query: 596 HIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQ-VTQNESPYSLIFHKEPNYKLLK 654
++ E GL+L+ +++P W EAF T+ +L N LPS ++ N+SPY ++ P Y L+
Sbjct: 623 YLTELGLSLMFHSKVPHKLWVEAFFTSNFLSNLLPSSTLSDNKSPYEMLHGTPPVYTALR 682
Query: 655 PFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKCLN-SHGRTFISRHVIFNEDLF 713
FG ACYP L+PY ++K + CVFLGY+N +KGY+CL+ G+ +I RHV+F+E F
Sbjct: 683 VFGSACYPYLRPYAKNKFDPKSLLCVFLGYNNKYKGYRCLHPPTGKVYICRHVLFDERKF 742
Query: 714 PFHEGFLN----TRSPLKT-----------TINNPSTS-----FPLCSAGNSINDASIPI 753
P+ + + + SPL T + PST+ FP + +S+ P
Sbjct: 743 PYSDIYSQFQTISGSPLFTAWQKGFSSTALSRETPSTNVEDIIFPSATVSSSVPTGCAPN 802
Query: 754 IEE---------------------------ENQDETNEEDSQGVTSDTEQTDNGSSEGDT 786
I E Q E + D ++D+E + + +
Sbjct: 803 IAETATAPDVDVAAAHDMVVPPSPITSTSLPTQPEESTSDQNHYSTDSETAISSAMTPQS 862
Query: 787 THE---ETLDIVQQQNV-GESSLDTNTSNAIHTRSKSGIHKPKLPYIGITETYKDTVEPA 842
+ E D Q+V ++ TS+ + TR+KSGI KP P + + EP
Sbjct: 863 INVSLFEDSDFPPLQSVISSTTAAPETSHPMITRAKSGITKPN-PKYALFSVKSNYPEPK 921
Query: 843 NVKEALTRTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKYKSDGSIERRK 902
+VKEAL W AM +E + TW LVP + + ++ KWVFKTK SDGS++R K
Sbjct: 922 SVKEALKDEGWTNAMGEEMGTMHETDTWDLVPPEMVDRLLGCKWVFKTKLNSDGSLDRLK 981
Query: 903 ARLVAKGFQQTAGIDYEETFSPVVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKET 962
ARLVA+G++Q G+DY ET+SPVV+ +TVR IL +A W ++QLD+ NAFL+ LKET
Sbjct: 982 ARLVARGYEQEEGVDYVETYSPVVRSATVRSILHVATINKWSLKQLDVKNAFLHDELKET 1041
Query: 963 VFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFL- 1021
VFM QP GF DP++P+++CKL KAIY LKQAPRAWFD + LL +GF + SDPSLF+
Sbjct: 1042 VFMTQPPGFEDPSRPDYVCKLKKAIYDLKQAPRAWFDKFSSYLLKYGFICSFSDPSLFVY 1101
Query: 1022 LKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASG 1081
LKG+D + FLL+YVDD+I+TG+++ LQ + L+ F +KD+G LHYFLGI+ G
Sbjct: 1102 LKGRD-VMFLLLYVDDMILTGNNDVLLQQLLNILSTEFRMKDMGALHYFLGIQAHYHNDG 1160
Query: 1082 MYLKQSKYIGDLLKKFKMENASPCPTPMITGRHFTVEGEKLKDPTVFRQAIGGLQYLTHT 1141
++L Q KY DLL M + S PTP+ + +PT FR+ G LQYLT T
Sbjct: 1161 LFLSQEKYTSDLLVNAGMSDCSSMPTPLQLDL-LQGNNKPFPEPTYFRRLAGKLQYLTLT 1219
Query: 1142 RPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWA 1201
RPDI F+VN + Q M +PT + +KRIL YL+GT+ +++ +TD + +SD+DWA
Sbjct: 1220 RPDIQFAVNFVCQKMHAPTMSDFHLLKRILHYLKGTMTMGINLSSNTDSVLRCYSDSDWA 1279
Query: 1202 TSIDDRKSMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFEL 1261
D R+S G C FLG +ISWS+++ VS+SSTE+EYR L A+E++WI LL E+
Sbjct: 1280 GCKDTRRSTGGFCTFLGYNIISWSAKRHPTVSKSSTEAEYRTLSFAASEVSWIGFLLQEI 1339
Query: 1262 KLPLPRKPILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQI 1321
LP + P ++CDNLSA L++NP LH+RSKH ++D +Y+R++V + V ++P + Q+
Sbjct: 1340 GLPQQQIPEMYCDNLSAVYLSANPALHSRSKHFQVDYYYVRERVALGALTVKHIPASQQL 1399
Query: 1322 ADCLTKPLSHTRFSQLRDKLGVIHSP 1347
AD TK L F LR KLGV+ P
Sbjct: 1400 ADIFTKSLPQAPFCDLRFKLGVVLPP 1425
>At1g44510 polyprotein, putative
Length = 1459
Score = 896 bits (2315), Expect = 0.0
Identities = 531/1435 (37%), Positives = 767/1435 (53%), Gaps = 113/1435 (7%)
Query: 19 KLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFEEWQANDQR 78
KL NY +W + ++ G L GY+ + P E T+ +S S NP+F W+ D+
Sbjct: 32 KLTSTNYLMWSIQIHALLDGYDLAGYLDNSVVIPPE-TTTINSVVSANPSFTLWKRQDKL 90
Query: 79 LLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKFQFHSIRKGEMKME 138
+ ++ +++ + + + S Q+W + + I L+ Q + KG ++
Sbjct: 91 IFSALIGAISPAVQSLVSRATNSSQIWSTLNNTYAKPSYGHIKQLRQQIQRLTKGTKTID 150
Query: 139 DYLIKMKNLADKLKLAGNPISNSNLIIQTLNGLDSEYNPIVVKLSDH-TTLSWVDLQAQL 197
+Y+ D+L + G P+ + + L GL EY +V ++ T + ++ +L
Sbjct: 151 EYVQSHTTRLDQLAILGKPMEHEEQVEHILKGLPEEYKTVVDQIEGKDNTPTITEIHERL 210
Query: 198 LTFESRI------------EQLNTLTNLNLNATANV---ANKFDHRDNRFNSNNNWRGSN 242
+ ES++ N + N N N N++ + N+N N + S
Sbjct: 211 INHESKLLSDEVPPSSSFPMSANAVQQRNFNNNCNQNQHKNRYQGNTHNNNTNTNSQPST 270
Query: 243 FRGWRGGRGRGRSSKAPCQVYGKTNHTAINC--FHRFDKNYSRSNYSADSDKQGSHNAFI 300
+ + G+ + CQ+ H+A C S S +S + Q N I
Sbjct: 271 YN--KSGQRTFKPYLGKCQICSVQGHSARRCPQLQAMQLPASSSAHSPFTPWQPRANLAI 328
Query: 301 ASQNSVEDYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEIVATGSSKLKS 360
S + W DSGA++H+T + ++G ++ +G L I TGS+ L S
Sbjct: 329 GSPYAANP--WLLDSGATHHITSDLNALSLHQPYNGGEYVMIADGTGLTIKQTGSTFLPS 386
Query: 361 LNLD----DVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVILKGLLKNG 416
N D VLYVP+I KNL+SV +L N + VEF VK+ TG ++L+G K+
Sbjct: 387 QNRDLALHKVLYVPDIRKNLISVYRLCNTNQVSVEFFPASFQVKDLNTGTLLLQGRTKDD 446
Query: 417 LYQLSDTKGNPYAFV--------SVKESWHRRLGHPNNKVLDKVLKSCNVKVP-PSDNFS 467
LY+ T NP A + SWH RLGHP+ +L+ +L ++ V S N +
Sbjct: 447 LYEWPVT--NPPATALFTSPSPKTTLSSWHSRLGHPSASILNTLLSKFSLPVSVASSNKT 504
Query: 468 FCEACQYGKMHLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTW 527
C C K H LPF +SS H+ PLE + TDVW +PI+S +KYY+ +D ++R+TW
Sbjct: 505 SCSDCLINKSHKLPFATSSIHSSSPLEYIFTDVW-TSPIISHDNYKYYLVLVDHYTRYTW 563
Query: 528 IYPLKQKSETVQAFTT-----ENQFNKRIKVIQCDGGGEYKPVQKLAIDVGIQFRMSCPY 582
+YPL+QKS+ F EN+F +I+ + D GGE+ ++ + GI S P+
Sbjct: 564 LYPLQQKSQVKATFIAFKALVENRFQAKIRTLYSDNGGEFIALRDFLVSNGISHLTSPPH 623
Query: 583 TFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYSL 642
T + NG +ERKHRHI E GLTLL QA +P YW AF+TAVYLINR+P+ V +SP+
Sbjct: 624 TPEHNGLSERKHRHIVETGLTLLTQASVPREYWTYAFATAVYLINRMPTPVLCLQSPFQK 683
Query: 643 IFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKCLN-SHGRTF 701
+F PNY+ L+ FGC C+P L+PY ++KL+ + RCVFLGYS + Y CL+ + R +
Sbjct: 684 LFGSSPNYQRLRVFGCLCFPWLRPYTRNKLEERSKRCVFLGYSLTQTAYLCLDVDNNRLY 743
Query: 702 ISRHVIFNEDLFPF-------------------------HEGF----LNTRSPLKTTINN 732
SRHV+F+E +PF + GF L +SP ++
Sbjct: 744 TSRHVMFDESTYPFAASIREQSQSSLVTPPESSSSSSPANSGFPCSVLRLQSPPASSPET 803
Query: 733 PS--------------TSFPLCSAGNSINDASIPIIEEENQDETNEEDSQGVTSDTEQTD 778
PS T P S + + D+++ + E G + +
Sbjct: 804 PSPPQQQNDSPVSPRQTGSPTPSHHSQVRDSTLSPSPSVSNSEPTAPHENGPEPEAQSNP 863
Query: 779 N----GSSEGDTTHEETLDIVQQQNVGESS---LDTN------TSNA----------IHT 815
N G ++Q+ V +S+ L N TSN+ + T
Sbjct: 864 NSPFIGPLPNPNPETNPSSSIEQRPVDKSTTTALPPNQTTIAATSNSRSQPPKNNHQMKT 923
Query: 816 RSKSGIHKPKLPY-IGITETYKDTVEPANVKEALTRTLWKEAMQKEFQALMSNKTWILVP 874
RSK+ I KPK + + T EP V +AL W+ AM EF A N TW LVP
Sbjct: 924 RSKNNITKPKTKTSLTVALTQPHLSEPNTVTQALKDKKWRFAMSDEFDAQQRNHTWDLVP 983
Query: 875 YQDQENIVDSKWVFKTKYKSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVRVI 934
+++V +WVFK KY +G I++ KARLVAKGF Q G+DY ETFSPV+K +T+RV+
Sbjct: 984 PNPTQHLVGCRWVFKLKYLPNGLIDKYKARLVAKGFNQQYGVDYAETFSPVIKATTIRVV 1043
Query: 935 LSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAP 994
L +AV NW ++QLD+NNAFL G L E V+M QP GFVD +P+H+C+L KAIYGLKQAP
Sbjct: 1044 LDVAVKKNWPLKQLDVNNAFLQGTLTEEVYMAQPPGFVDKDRPSHVCRLRKAIYGLKQAP 1103
Query: 995 RAWFDSLKTALLNWGFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQ 1054
RAW+ LK LLN GF N+ +D SLF+ + +LL+YVDDIIVTGS + + A +
Sbjct: 1104 RAWYMELKQHLLNIGFVNSLADTSLFIYSHGTTLLYLLVYVDDIIVTGSDHKSVSAVLSS 1163
Query: 1055 LNDVFSLKDLGRLHYFLGIEVQRDASGMYLKQSKYIGDLLKKFKMENASPCPTPMITGRH 1114
L + FS+KD LHYFLGIE R +G++L Q KY+ DLL K M +A P TP+ T
Sbjct: 1164 LAERFSIKDPTDLHYFLGIEATRTNTGLHLMQRKYMTDLLAKHNMLDAKPVATPLPTSPK 1223
Query: 1115 FTVE-GEKLKDPTVFRQAIGGLQYLTHTRPDIAFSVNKLSQYMSSPTTDHWQGIKRILRY 1173
T+ G KL D + +R +G LQYL TRPDIAF+VN+LSQ+M PT+DHWQ KR+LRY
Sbjct: 1224 LTLHGGTKLNDASEYRSVVGSLQYLAFTRPDIAFAVNRLSQFMHQPTSDHWQAAKRVLRY 1283
Query: 1174 LQGTINYCLHIKPSTDLDITGFSDADWATSIDDRKSMAGQCVFLGETLISWSSRKQKVVS 1233
L GT + + + S+ + + FSDADWA D S ++LG ISWSS+KQ+ VS
Sbjct: 1284 LAGTTTHGIFLNSSSPIHLHAFSDADWAGDSADYVSTNAYVIYLGRNPISWSSKKQRGVS 1343
Query: 1234 RSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSAKALASNPVLHARSKH 1293
RSSTESEYRA+ + A+EI W+ SLL EL + LP P ++CDN+ A + +NPV H+R KH
Sbjct: 1344 RSSTESEYRAVANAASEIRWLCSLLTELHIRLPHGPTIFCDNIGATYICANPVFHSRMKH 1403
Query: 1294 IEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQLRDKLGVIHSPP 1348
I +D H++R + + V++V T DQ+AD LTK LS F R K+GV PP
Sbjct: 1404 IALDYHFVRGMIQSRALRVSHVSTNDQLADALTKSLSRPHFLSARSKIGVRQLPP 1458
>At4g16870 retrotransposon like protein
Length = 1474
Score = 893 bits (2308), Expect = 0.0
Identities = 525/1454 (36%), Positives = 782/1454 (53%), Gaps = 121/1454 (8%)
Query: 11 DLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFE 70
++ +S KL NNY +W + ++ G +L G++ G+ + P +T+++ S NP +
Sbjct: 25 NINTSNVTKLTSNNYLMWSLQIHALLDGYELAGHLDGSIETPAPTLTTNNVV-SANPQYT 83
Query: 71 EWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKFQFHSI 130
W+ D+ + ++ +++ + + + Q+W + + I L+ Q +
Sbjct: 84 LWKRQDRLIFSALIGAISPPVQPLVSRATKASQIWKTLTNTYAKSSYDHIKQLRTQIKQL 143
Query: 131 RKGEMKMEDYLIKMKNLADKLKLAGNPISNSNLIIQTLNGLDSEYNPIVVKLSDH-TTLS 189
+KG +++Y++ L D+L + G P+ + + + L GL +Y +V ++ T S
Sbjct: 144 KKGTKTIDEYVLSHTTLLDQLAILGKPMEHEEQVERILEGLPEDYKTVVDQIEGKDNTPS 203
Query: 190 WVDLQAQLLTFESRIEQLNTLTNLNLNATANVANKFDHRDNRFNSNNNWR--GSNFRGWR 247
++ +L+ E+++ L++ +L +ANVA + H +NR N+ N R G+ +
Sbjct: 204 ITEIHERLINHEAKLLSTAALSSSSLPMSANVAQQRHHNNNRNNNQNKNRTQGNTYTNNW 263
Query: 248 GGRGRGRSSKAP-------CQVYGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFI 300
+S + P CQ+ H+A C + S+ SA + A +
Sbjct: 264 QPSANNKSGQRPFKPYLGKCQICNVQGHSARRC-PQLQAMQPSSSSSASTFTPWQPRANL 322
Query: 301 ASQNSVEDYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEIVATGSSKLKS 360
A +W DSGA++H+T + L + + + ++ +G L+I TGS+ L S
Sbjct: 323 AMGAPYTANNWLLDSGATHHITSDLNALA-LHQPYNGDDVMIADGTSLKITKTGSTFLPS 381
Query: 361 ----LNLDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVILKGLLKNG 416
L L+ VLYVP+I KNL+SV +L N + VEF VK+ TG ++L+G K+
Sbjct: 382 NARDLTLNKVLYVPDIQKNLVSVYRLCNTNQVSVEFFPASFQVKDLNTGTLLLQGRTKDE 441
Query: 417 LYQLSDTKGNPYAFVSVKE------SWHRRLGHPNNKVLDKVLKSCNVKVPPS-DNFSFC 469
LY+ T A + SWH RLGHP++ +L+ ++ ++ V S N C
Sbjct: 442 LYEWPVTNPKATALFTTPSPKTTLSSWHSRLGHPSSSILNTLISKFSLPVSVSASNKLAC 501
Query: 470 EACQYGKMHLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIY 529
C K H LPF SS + PLE + +DVW +PI+S +KYY+ +D +R+TW+Y
Sbjct: 502 SDCFINKSHKLPFSISSIKSTSPLEYIFSDVW-MSPILSPDNYKYYLVLVDHHTRYTWLY 560
Query: 530 PLKQKSETVQAFTT-----ENQFNKRIKVIQCDGGGEYKPVQKLAIDVGIQFRMSCPYTF 584
PL+QKS+ F EN+F +I+ + D GGE+ +++ + GI S P+T
Sbjct: 561 PLQQKSQVKSTFIAFKALVENRFQAKIRTLYSDNGGEFIALREFLVSNGISHLTSPPHTP 620
Query: 585 QQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYSLIF 644
+ NG +ERKHRHI E GLTLL QA +P YW AF+ AVYLINR+P+ V ESP+ +F
Sbjct: 621 EHNGLSERKHRHIVETGLTLLTQASVPREYWPYAFAAAVYLINRMPTPVLSMESPFQKLF 680
Query: 645 HKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKCLN-SHGRTFIS 703
+PNY+ L+ FGC C+P L+PY +KL+ + RCVFLGYS + Y C + H R + S
Sbjct: 681 GSKPNYERLRVFGCLCFPWLRPYTHNKLEERSRRCVFLGYSLTQTAYLCFDVEHKRLYTS 740
Query: 704 RHVIFNEDLFPFHE----------GFLNTRSPLKTTINNPSTSFPLC-SAGNSINDASIP 752
RHV+F+E FPF F + SPL T I + S+ P C S+ ++ P
Sbjct: 741 RHVVFDEASFPFSNLTSQNSLPTVTFEQSSSPLVTPILSSSSVLPSCLSSPCTVLHQQQP 800
Query: 753 IIEEENQDETNE------------------------------EDSQGVTSD-TEQTDNGS 781
+ N +++ S + S+ T +NG
Sbjct: 801 PVTTPNSPHSSQPTTSPAPLSPHRSTTMDFQVPQVRSSSPLLSSSSSLNSEPTAPNENGP 860
Query: 782 SE----------GDTTHEETLDIVQQQNVGESS--------------LDTNTSNAIHTRS 817
+ THE + + N ++ T T+ T
Sbjct: 861 EPEAQSPPIGPLSNPTHEAFIGPLPNPNRNPTNEIEPTPAPHPKPVKPTTTTTTPNRTTV 920
Query: 818 KSGIHKPKLP---YIGITETYKDTVEPANVKEALTRTL-------------------WKE 855
H+P P + K+ ++ N K +LT TL W+
Sbjct: 921 SDASHQPTAPQQNQHNMKTRAKNNIKKPNTKFSLTATLPNRSPSEPTNVTQALKDKKWRF 980
Query: 856 AMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKYKSDGSIERRKARLVAKGFQQTAG 915
AM EF A N TW LVP++ Q +V KWVFK KY +G+I++ KARLVAKGF Q G
Sbjct: 981 AMSDEFDAQQRNHTWDLVPHESQ-LLVGCKWVFKLKYLPNGAIDKYKARLVAKGFNQQYG 1039
Query: 916 IDYEETFSPVVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQPEGFVDPT 975
+DY ETFSPV+K +T+R++L +AV +WE++QLD+NNAFL G L E V+M QP GF+D
Sbjct: 1040 VDYAETFSPVIKSTTIRLVLDVAVKKDWEIKQLDVNNAFLQGTLTEEVYMAQPPGFIDKD 1099
Query: 976 KPNHICKLSKAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLKGKDHITFLLIYV 1035
+P H+C+L KAIYGLKQAPRAW+ LK L N GF N+ SD SLF+ ++L+YV
Sbjct: 1100 RPTHVCRLRKAIYGLKQAPRAWYMELKQHLFNIGFVNSLSDASLFIYCHGTTFVYVLVYV 1159
Query: 1036 DDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASGMYLKQSKYIGDLLK 1095
DDIIVTGS + + A + L + FS+KD LHYFLGIE R G++L Q KYI DLL
Sbjct: 1160 DDIIVTGSDKSSIDAVLTSLAERFSIKDPTDLHYFLGIEATRTKQGLHLMQRKYIKDLLA 1219
Query: 1096 KFKMENASPCPTPMITGRHFTVE-GEKLKDPTVFRQAIGGLQYLTHTRPDIAFSVNKLSQ 1154
K M +A P TP+ T T+ G KL D + +R +G LQYL TRPDIA++VN+LSQ
Sbjct: 1220 KHNMADAKPVLTPLPTSPKLTLHGGTKLNDASEYRSVVGSLQYLAFTRPDIAYAVNRLSQ 1279
Query: 1155 YMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDDRKSMAGQC 1214
M PT DHWQ KR+LRYL GT + + + ++ L++ FSDADWA DD S
Sbjct: 1280 LMPQPTEDHWQAAKRVLRYLAGTSTHGIFLDTTSPLNLHAFSDADWAGDSDDYVSTNAYV 1339
Query: 1215 VFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPILWCD 1274
++LG+ ISWSS+KQ+ V+RSSTESEYRA+ + A+E+ W+ SLL +L + LP +P ++CD
Sbjct: 1340 IYLGKNPISWSSKKQRGVARSSTESEYRAVANAASEVKWLCSLLSKLHIRLPIRPSIFCD 1399
Query: 1275 NLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRF 1334
N+ A L +NPV H+R KHI ID H++R+ + + V++V T DQ+AD LTKPLS F
Sbjct: 1400 NIGATYLCANPVFHSRMKHIAIDYHFVRNMIQSGALRVSHVSTRDQLADALTKPLSRAHF 1459
Query: 1335 SQLRDKLGVIHSPP 1348
R K+GV PP
Sbjct: 1460 QSARFKIGVRQLPP 1473
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 884 bits (2283), Expect = 0.0
Identities = 526/1462 (35%), Positives = 769/1462 (51%), Gaps = 144/1462 (9%)
Query: 6 NNNKNDLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSN 65
N N ++ S KL NY +W V + G +L G++ G+ P I +D+
Sbjct: 12 NTNILNVNMSNVTKLTSTNYLMWSRQVHALFDGYELAGFLDGSTPMPPATI-GTDAVPRV 70
Query: 66 NPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKF 125
NP + W+ D+ + +L +++ + + T+ Q+W+ + + + + L+F
Sbjct: 71 NPDYTRWRRQDKLIYSAILGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLRF 130
Query: 126 QFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSNLIIQTLNGLDSEYNPIVVKLS-D 184
D+L L G P+ + + + L L +Y P++ +++
Sbjct: 131 ITRF-------------------DQLALLGKPMDHDEQVERVLENLPDDYKPVIDQIAAK 171
Query: 185 HTTLSWVDLQAQLLTFESRIEQLNT----------LTNLNLNATANVANKFDHRDNRFNS 234
T S ++ +L+ ES++ LN+ +T+ N N N N+ D+R+ +N+
Sbjct: 172 DTPPSLTEIHERLINRESKLLALNSAEVVPITANVVTHRNTNTNRNQNNRGDNRN--YNN 229
Query: 235 NNNWRGSNFRGWRGGRGRGRSSK---APCQVYGKTNHTAINC--FHRFDKNYSRSNYSAD 289
NNN S G R R K CQ+ H+A C H+F ++ S
Sbjct: 230 NNNRSNSWQPSSSGSRSDNRQPKPYLGRCQICSVQGHSAKRCPQLHQFQSTTNQQQ-STS 288
Query: 290 SDKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLE 349
A +A + +W DSGA++H+T + + G + ++ +G +
Sbjct: 289 PFTPWQPRANLAVNSPYNANNWLLDSGATHHITSDFNNLSFHQPYTGGDDVMIADGSTIP 348
Query: 350 IVATGSSKL----KSLNLDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTG 405
I TGS+ L +SL+L+ VLYVPNI KNL+SV +L N + VEF VK+ TG
Sbjct: 349 ITHTGSASLPTSSRSLDLNKVLYVPNIHKNLISVYRLCNTNRVSVEFFPASFQVKDLNTG 408
Query: 406 KVILKGLLKNGLYQLSDTKGNPYAFVS------VKESWHRRLGHPNNKVLDKVLKSCNVK 459
+L+G K+ LY+ + + SWH RLGHP+ +L+ V+ + ++
Sbjct: 409 VPLLQGKTKDELYEWPIASSQAVSMFASPCSKATHSSWHSRLGHPSLAILNSVISNHSLP 468
Query: 460 V-PPSDNFSFCEACQYGKMHLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVHF 518
V PS C C K H +PF +S+ + +PLE +++DVW +PI+S ++YYV F
Sbjct: 469 VLNPSHKLLSCSDCFINKSHKVPFSNSTITSSKPLEYIYSDVWS-SPILSIDNYRYYVIF 527
Query: 519 IDDFSRFTWIYPLKQKSETVQAFT-----TENQFNKRIKVIQCDGGGEYKPVQKLAIDVG 573
+D F+R+TW+YPLKQKS+ F EN+F RI + D GGE+ ++ G
Sbjct: 528 VDHFTRYTWLYPLKQKSQVKDTFIIFKSLVENRFQTRIGTLYSDNGGEFVVLRDYLSQHG 587
Query: 574 IQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQV 633
I S P+T + NG +ERKHRHI E GLTLL+ A +P YW AFS AVYLINRLP+ +
Sbjct: 588 ISHFTSPPHTPEHNGLSERKHRHIVEMGLTLLSHASVPKTYWPYAFSVAVYLINRLPTPL 647
Query: 634 TQNESPYSLIFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKC 693
Q +SP+ +F + PNY+ LK FGCACYP L+PYN+HKL+ + +C F+GYS + Y C
Sbjct: 648 LQLQSPFQKLFGQPPNYEKLKVFGCACYPWLRPYNRHKLEDKSKQCAFMGYSLTQSAYLC 707
Query: 694 LN-SHGRTFISRHVIFNEDLFPFHEGFLNTRSP--------------------------- 725
L+ GR + SRHV F+E FPF +
Sbjct: 708 LHIPTGRLYTSRHVQFDERCFPFSTTNFGVSTSQEQRSDSAPNWPSHTTLPTTPLVLPAP 767
Query: 726 ------LKTTINNPSTSFPLCSAGNSINDASIPIIEEENQDETNEEDSQGV--TSDTEQT 777
L T+ PS+ PLC+ S ++ I + E G T+ QT
Sbjct: 768 PCLGPHLDTSPRPPSSPSPLCTTQVSSSNLPSSSISSPSSSEPTAPSHNGPQPTAQPHQT 827
Query: 778 DNGSSEG---------------------------DTTHEETLDIVQQQNVGESSLDTNT- 809
N +S + H T + SS T+T
Sbjct: 828 QNSNSNSPILNNPNPNSPSPNSPNQNSPLPQSPISSPHIPTPSTSISEPNSPSSSSTSTP 887
Query: 810 ---------------------SNAIHTRSKSGIHKPKLPYIGITETYKDTVEPANVKEAL 848
++++ TR+K GI KP Y T ++ EP +A+
Sbjct: 888 PLPPVLPAPPIIQVNAQAPVNTHSMATRAKDGIRKPNQKYSYATSLAANS-EPRTAIQAM 946
Query: 849 TRTLWKEAMQKEFQALMSNKTWILVPYQDQE-NIVDSKWVFKTKYKSDGSIERRKARLVA 907
W++AM E A + N TW LVP IV +W+F K+ SDGS+ R KARLVA
Sbjct: 947 KDDRWRQAMGSEINAQIGNHTWDLVPPPPPSVTIVGCRWIFTKKFNSDGSLNRYKARLVA 1006
Query: 908 KGFQQTAGIDYEETFSPVVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQ 967
KG+ Q G+DY ETFSPV+K +++R++L +AV +W +RQLD+NNAFL G L + V+M Q
Sbjct: 1007 KGYNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDEVYMSQ 1066
Query: 968 PEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLKGKDH 1027
P GFVD +P+++C+L KAIYGLKQAPRAW+ L+T LL GF N+ SD SLF+L+
Sbjct: 1067 PPGFVDKDRPDYVCRLRKAIYGLKQAPRAWYVELRTYLLTVGFVNSISDTSLFVLQRGRS 1126
Query: 1028 ITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASGMYLKQS 1087
I ++L+YVDDI++TG+ L+ + L+ FS+K+ LHYFLGIE +R G++L Q
Sbjct: 1127 IIYMLVYVDDILITGNDTVLLKHTLDALSQRFSVKEHEDLHYFLGIEAKRVPQGLHLSQR 1186
Query: 1088 KYIGDLLKKFKMENASPCPTPMITGRHFTV-EGEKLKDPTVFRQAIGGLQYLTHTRPDIA 1146
+Y DLL + M A P TPM T T+ G KL DPT +R +G LQYL TRPD++
Sbjct: 1187 RYTLDLLARTNMLTAKPVATPMATSPKLTLHSGTKLPDPTEYRGIVGSLQYLAFTRPDLS 1246
Query: 1147 FSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDD 1206
++VN+LSQYM PT DHW +KR+LRYL GT ++ + +K L + +SDADWA DD
Sbjct: 1247 YAVNRLSQYMHMPTDDHWNALKRVLRYLAGTPDHGIFLKKGNTLSLHAYSDADWAGDTDD 1306
Query: 1207 RKSMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLP 1266
S G V+LG ISWSS+KQK V RSSTE+EYR++ + ++E+ WI SLL EL + L
Sbjct: 1307 YVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSELQWICSLLTELGIQLS 1366
Query: 1267 RKPILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLT 1326
P+++CDN+ A L +NPV H+R KHI +D H+IR+QV + V +V T DQ+AD LT
Sbjct: 1367 HPPVIYCDNVGATYLCANPVFHSRMKHIALDYHFIRNQVQSGALRVVHVSTHDQLADTLT 1426
Query: 1327 KPLSHTRFSQLRDKLGVIHSPP 1348
KPLS F K+GVI PP
Sbjct: 1427 KPLSRVAFQNFSRKIGVIKVPP 1448
>At4g27210 putative protein
Length = 1318
Score = 848 bits (2191), Expect = 0.0
Identities = 499/1289 (38%), Positives = 715/1289 (54%), Gaps = 157/1289 (12%)
Query: 111 LAGAHTRSQIIYLKFQFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSNLIIQTLNG 170
L+ A ++ L+ + ++ K + M+ YL +KN+ D+L G+P++ I LNG
Sbjct: 38 LSTATLHMRLFELQRRLQNVSKRDKTMDAYLNDLKNICDQLASVGSPVTEKMKIFAALNG 97
Query: 171 LDSEYNPIVVKLSDHTTLSWVDLQAQLLTFESRIEQLNTLTNLNLNATANVANKFDHRDN 230
L EY PI TT+ N++ + NV K D+
Sbjct: 98 LGREYEPI------KTTIE------------------NSMDTQPGPSLENVIPKLTGYDD 133
Query: 231 RFNSNNNWRGSNFRGWRGGRGRGRSSKAPCQVYGKTNHTAINCFHRFDKNYSRSNYSADS 290
R +G + Y N T D +Y R++Y+
Sbjct: 134 RL-------------------QGYLEETTISPYVAFNITTS------DDSY-RNSYNRGK 167
Query: 291 DKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEI 350
D H +W DS A+ HVT+ Q +HG ++ +V +G+ L I
Sbjct: 168 DVTDQHGN-----------EWLPDSAATAHVTNSPRSLQQSQPYHGTDAIMVDDGNYLPI 216
Query: 351 VATGSSKLKSLN----LDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGK 406
TGS+ L S + L DVL P+ITK+LLS+SKL D VEF+ + V +K T K
Sbjct: 217 THTGSTNLASSSGTVPLTDVLVCPSITKSLLSMSKLTQDFPCTVEFEYDGVRVNDKATKK 276
Query: 407 VILKGLLKNGLYQLSDTKGNPYAFVSVK------ESWHRRLGHPNNKVLDKVLKSCNVKV 460
++L G ++GLY L D K AF S + E WHRRLGHP+ ++L
Sbjct: 277 LLLMGSNRDGLYCLKDDK-QFQAFFSTRQRSASDEVWHRRLGHPHPQIL----------- 324
Query: 461 PPSDNFSFCEACQYGKMHLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFID 520
+PLE VH D+WGP I S GF+YY FID
Sbjct: 325 ------------------------------QPLERVHCDLWGPTTITSVQGFRYYAVFID 354
Query: 521 DFSRFTWIYPLKQKSETVQAFTT-----ENQFNKRIKVIQCDGGGEY---KPVQKLAIDV 572
+SRF+WIYPLK KS+ F ENQ +++I V QCDGGGE+ K +Q L
Sbjct: 355 HYSRFSWIYPLKLKSDFYNIFLAFHKLVENQLSQKISVFQCDGGGEFVSHKFLQHLQSH- 413
Query: 573 GIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLP-S 631
GIQ ++SCP+T QQNG AERKHRH+ E GL++L Q+ +P +W EAF TA +LIN LP S
Sbjct: 414 GIQQQLSCPHTPQQNGLAERKHRHLVELGLSMLFQSHVPHKFWVEAFFTANFLINLLPTS 473
Query: 632 QVTQNESPYSLIFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGY 691
+ ++ SPY ++ K+P+Y L+ FG AC+P L+ Y ++K + +CVFLGY+ +KGY
Sbjct: 474 ALKESISPYEKLYDKKPDYTSLRSFGSACFPTLRDYAENKFNPCSLKCVFLGYNEKYKGY 533
Query: 692 KCLNSH-GRTFISRHVIFNEDLFPFHEGFLNTR----------------SPLKTTINNPS 734
+CL GR +ISRHVIF+E ++PF + + SP +T +PS
Sbjct: 534 RCLYPPTGRLYISRHVIFDESVYPFSHTYKHLHPQPRTPLLAAWLRSSDSPAPSTSTSPS 593
Query: 735 TSFPLCSAGN---------SINDASIPIIEEENQDETNEEDSQGVTSDTEQTDNGSSEGD 785
+ PL ++ + + +PI + + S S+ + +S GD
Sbjct: 594 SRSPLFTSADFPPLPQRKTPLLPTLVPISSVSHASNITTQQSPDFDSERTTDFDSASIGD 653
Query: 786 TTHEETLDIVQQQNVGESSLD---TNTSNAIH---TRSKSGIHKPKLPYIGITETYKDTV 839
++H ++ + ++S++ T+ S +H TR+K GI KP Y+ ++
Sbjct: 654 SSHSSQAGSDSEETIQQASVNVHQTHASTNVHPMVTRAKVGISKPNPRYVFLSHKVSYP- 712
Query: 840 EPANVKEALTRTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKYKSDGSIE 899
EP V AL W AM +E +TW LVPY+ +++ SKWVF+TK +DG++
Sbjct: 713 EPKTVTAALKHPGWTGAMTEEIGNCSETQTWSLVPYKSDMHVLGSKWVFRTKLHADGTLN 772
Query: 900 RRKARLVAKGFQQTAGIDYEETFSPVVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGYL 959
+ KAR+VAKGF Q GIDY ET+SPVV+ TVR++L +A LNW+++Q+D+ NAFL+G L
Sbjct: 773 KLKARIVAKGFLQEEGIDYLETYSPVVRTPTVRLVLHLATALNWDIKQMDVKNAFLHGDL 832
Query: 960 KETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSL 1019
KETV+M QP GFVDP+KP+H+C L K+IYGLKQ+PRAWFD T LL +GF +KSDPSL
Sbjct: 833 KETVYMTQPAGFVDPSKPDHVCLLHKSIYGLKQSPRAWFDKFSTFLLEFGFFCSKSDPSL 892
Query: 1020 FLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDA 1079
F+ +++ LL+YVDD+++TG+S+ L + + LN F + D+G+LHYFLGI+VQR
Sbjct: 893 FIYAHNNNLILLLLYVDDMVITGNSSQTLTSLLAALNKEFRMTDMGQLHYFLGIQVQRQQ 952
Query: 1080 SGMYLKQSKYIGDLLKKFKMENASPCPTPMITGRHFTVEGEKL-KDPTVFRQAIGGLQYL 1138
+G+++ Q KY DLL ME+ +P PTP+ E+L DPT FR G LQYL
Sbjct: 953 NGLFMSQQKYAEDLLIAASMEHCTPLPTPLPVQLDRVPHQEELFSDPTYFRSIAGKLQYL 1012
Query: 1139 THTRPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDA 1198
T TRPDI F+VN + Q M PT + +KRILRY++GTI + + + +SD+
Sbjct: 1013 TLTRPDIQFAVNFVCQKMHQPTISDFHLLKRILRYIKGTITMGISYSRDSPTLLQAYSDS 1072
Query: 1199 DWATSIDDRKSMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLL 1258
DW R+S+ G C F+G L+SWSS+K VSRSSTE+EY++L D A+EI W+ +LL
Sbjct: 1073 DWGNCKQTRRSVGGLCTFMGTNLVSWSSKKHPTVSRSSTEAEYKSLSDAASEILWLSTLL 1132
Query: 1259 FELKLPLPRKPILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTT 1318
EL++PLP P L+CDNLSA L +NP HAR+KH +ID H++R++V +VV ++P +
Sbjct: 1133 RELRIPLPDTPELFCDNLSAVYLTANPAFHARTKHFDIDFHFVRERVALKALVVKHIPGS 1192
Query: 1319 DQIADCLTKPLSHTRFSQLRDKLGVIHSP 1347
+QIAD TK L + F LR KLGV SP
Sbjct: 1193 EQIADIFTKSLPYEAFIHLRGKLGVTLSP 1221
>At4g28900 putative protein
Length = 1415
Score = 845 bits (2183), Expect = 0.0
Identities = 522/1406 (37%), Positives = 756/1406 (53%), Gaps = 169/1406 (12%)
Query: 16 VSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPE--EFITSSDS-SKSNNPAFEEW 72
V++KL NY LWK + +L G++ G CP I + D +++ NP F W
Sbjct: 17 VTLKLSTANYLLWKIQFETWLNNQRLLGFVTGANPCPNATRSIRNGDQVTEATNPDFLTW 76
Query: 73 QANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKFQFHSIRK 132
NDQ+++GW+L S++ + + TS+++W + S+ L+ + + + K
Sbjct: 77 VQNDQKIMGWLLGSLSEDALRSVYGLHTSREVWFSLAKKYNRVSASRKSDLQRRLNPVSK 136
Query: 133 GEMKMEDYLIKMKNLADKLKLAGNPISNSNLIIQTLNGLDSEYNPIVVKLS---DHTTLS 189
E M +YL +K + D+L G P+ + I LNGL EY + + D +S
Sbjct: 137 NEKSMLEYLNCVKQICDQLDSIGCPVPENEKIFGVLNGLGQEYMLVSTMIKGSMDTYPMS 196
Query: 190 WVDLQAQLLTFESRIEQLNTLTNLNLNATANVANKFDHRDNRFNSNNNWRGSNF-RGWRG 248
+ D+ +L+ F+ +++ + NR +N +G F +
Sbjct: 197 FEDVVFKLINFDDKLQNGQS------------------GGNRGRNNYTTKGRGFPQQISS 238
Query: 249 GRGRGRSSKAPCQVYGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFIASQNSVED 308
G ++ CQ+ K H+A C+ RFD + ++S AF A + S +
Sbjct: 239 GSPSDSGTRPTCQICNKYGHSAYKCWKRFDHAFQSEDFS---------KAFAAMRVSDQK 289
Query: 309 YD-WYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEIVATGSSKLKS----LNL 363
+ W DSGA++H+T+ T + Q + G++S +VGN D L I GS+ L S L L
Sbjct: 290 SNPWVTDSGATSHITNSTSQLQSAQPYSGEDSVIVGNSDFLPITHIGSAVLTSNQGNLPL 349
Query: 364 DDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVILKGLLKNGLYQLSDT 423
DVL PNITK+LLSVSKL +D +EFD + VK+KLT +++ KG N LY L +
Sbjct: 350 RDVLVCPNITKSLLSVSKLTSDYPCVIEFDSDGVIVKDKLTKQLLTKGTRHNDLYLLENP 409
Query: 424 KG----NPYAFVSVKESWHRRLGHPNNKVLDKVLKSCNVKVPPSDNFSFCEACQYGKMHL 479
K + + E WH RLGHPN VL ++L++ + + + + S C+ACQ GK+
Sbjct: 410 KFMACYSSRQQATSDEVWHMRLGHPNQDVLQQLLRNKAIVISKTSH-SLCDACQMGKICK 468
Query: 480 LPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLKQKSETVQ 539
LPF SS + LE VH D+WGPAP++SS GF+YYV FID++SRFTW YPL+ KS+
Sbjct: 469 LPFASSDFVSSRLLERVHCDLWGPAPVVSSQGFRYYVIFIDNYSRFTWFYPLRLKSDFFS 528
Query: 540 AFTT-----ENQFNKRIKVIQCDGGGEYKPVQKLA--IDVGIQFRMSCPYTFQQNGRAER 592
F T ENQ ++I QCDGGGE+ Q ++ + GI+ +SCPYT QQNG AER
Sbjct: 529 VFLTFQKMVENQCQQKIASFQCDGGGEFISNQFVSHLAECGIRQLISCPYTPQQNGIAER 588
Query: 593 KHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQVTQNE-SPYSLIFHKEPNYK 651
KHRHI E G +++ Q ++P W EAF T+ +L N LPS V +++ SPY ++ K P Y
Sbjct: 589 KHRHITELGSSMMFQGKVPQFLWVEAFYTSNFLCNLLPSSVLKDQKSPYEVLMGKAPVYT 648
Query: 652 LLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKCLN-SHGRTFISRHVIFNE 710
L+ FGCACYP L+PY +K + CVF GY+ +KGYKC + G+ +I+RHV+F+E
Sbjct: 649 SLRVFGCACYPNLRPYASNKFDPKSLLCVFTGYNEKYKGYKCFHPPTGKIYINRHVLFDE 708
Query: 711 DLFPFHEGFLNTRSPLKTT----------------------INNPSTSFP---------- 738
F F + + + S +T I+N + SF
Sbjct: 709 SKFLFSDIYSDKVSGTNSTLVSAWQSNFLPKSIPATPEVLDISNTAASFSDEQGEFSGAV 768
Query: 739 -----LCSA-------GNSINDASIPIIEEENQDETNEEDSQGVTSDTEQTDNGSSEGDT 786
C+A GNS+ S P+ ++ + S G +D E D+ SE
Sbjct: 769 GGGGCGCTADLDSVPIGNSL--PSSPVTQQNSPQPETPISSAGSGNDAE--DSELSENSE 824
Query: 787 THEETLDIVQQQNVGESSLDTNTSNAIH---TRSKSGIHKPKLPYIGITETYKDTVEPAN 843
E + V + E+ NT++ H TRSKSGI KP Y T V P
Sbjct: 825 NSESS---VFSEATTETEAADNTNDQSHPMITRSKSGIFKPNPKYAMFTVKSNYPV-PKT 880
Query: 844 VKEALTRTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKYKSDGSIERRKA 903
VK AL W +AM +E+ + TW LVP + +WVFKTK K+DG+++R KA
Sbjct: 881 VKTALKDPGWTDAMGEEYDSFEETHTWDLVPPDSFITPLGCRWVFKTKLKADGTLDRLKA 940
Query: 904 RLVAKGFQQTAGIDYEETFSPVVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKETV 963
RLVAKG++Q G+DY ET+SPVV+ +TVR IL +A WE++QLD+ NAFL+G LKETV
Sbjct: 941 RLVAKGYEQEEGVDYMETYSPVVRTATVRTILHVATINKWEIKQLDVKNAFLHGDLKETV 1000
Query: 964 FMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFL-L 1022
+M+QP GF + +P+++CKL+KAIYGLKQAPRAWFD T LL +GF T SDPSLF+ L
Sbjct: 1001 YMYQPPGFENQDRPDYVCKLNKAIYGLKQAPRAWFDKFSTFLLEFGFICTYSDPSLFVFL 1060
Query: 1023 KGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASGM 1082
KG+D + FLL+Y+DD+++TG++
Sbjct: 1061 KGRD-LMFLLLYMDDMLLTGNN-------------------------------------- 1081
Query: 1083 YLKQSKYIGDLLKKFKMENASPCPTPM-ITGRHFTVEGEKLKDPTVFRQAIGGLQYLTHT 1141
KY DLL M + +P PTP+ + + E DPT FR
Sbjct: 1082 ----KKYAMDLLVAAGMADCAPMPTPLPLQLDKVPGQQESFADPTYFR------------ 1125
Query: 1142 RPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWA 1201
+ +VN + Q M SPT + +KR+LRYL+G + L++ +TD+ + +SD+DWA
Sbjct: 1126 ----SLAVNLVCQKMHSPTVADFNLLKRVLRYLKGKVQMGLNLHNNTDITLRAYSDSDWA 1181
Query: 1202 TSIDDRKSMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFEL 1261
+ R+S+ G C FLG +ISWS+++ VSRSSTE+EYR L A E+ WI SLL E+
Sbjct: 1182 NCKETRRSVGGFCTFLGTNIISWSAKRHPTVSRSSTEAEYRTLSIAATEVKWISSLLREI 1241
Query: 1262 KLPLPRKPILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQI 1321
+ P P L+CDNLSA L +NP +H RSK ++D HY+R++V +VV +VP + Q+
Sbjct: 1242 GIYQPAPPELYCDNLSAVYLTANPAMHNRSKAFDVDFHYVRERVALGALVVKHVPASHQL 1301
Query: 1322 ADCLTKPLSHTRFSQLRDKLGVIHSP 1347
AD TK L F LR KLGV+ P
Sbjct: 1302 ADIFTKSLPQRPFFDLRYKLGVVLPP 1327
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 660 bits (1702), Expect = 0.0
Identities = 382/1066 (35%), Positives = 581/1066 (53%), Gaps = 61/1066 (5%)
Query: 302 SQNSVEDYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEIVATGSSKLKS- 360
+++++ W DSGA++HV+H F L + ++ + G ++I G+ KL
Sbjct: 422 ARHTLSSATWVIDSGATHHVSHDRSLFSSL-DTSVLSAVNLPTGPTVKISGVGTLKLNDD 480
Query: 361 LNLDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVILKGLLKNGLYQL 420
+ L +VL++P NL+S+S L D V FDKN C +++ + G+++ +G LY L
Sbjct: 481 ILLKNVLFIPEFRLNLISISSLTDDIGSRVIFDKNSCEIQDLIKGRMLGQGRRVANLYLL 540
Query: 421 S--DTKGNPYAFVSVKESWHRRLGHPNNKVLDKVLKSCNVKVPPSDNFSFCEACQYGKMH 478
D + A V + WHRRLGH + + LD + S + FC C K
Sbjct: 541 DVGDQSISVNAVVDIS-MWHRRLGHASLQRLDAISDSLGTTRHKNKGSDFCHVCHLAKQR 599
Query: 479 LLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLKQKSETV 538
L F +S+ +E +L+H DVWGP + + G+KY++ +DD SR TW+Y LK KSE +
Sbjct: 600 KLSFPTSNKVCKEIFDLLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWMYLLKTKSEVL 659
Query: 539 QAFTT-----ENQFNKRIKVIQCDGGGEYKPVQKLAIDVGIQFRMSCPYTFQQNGRAERK 593
F ENQ+ ++K ++ D E K A + GI SCP T +QN ERK
Sbjct: 660 TVFPAFIQQVENQYKVKVKAVRSDNAPELKFTSFYA-EKGIVSFHSCPETPEQNSVVERK 718
Query: 594 HRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYSLIFHKEPNYKLL 653
H+HI L+ Q+Q+PL W + TAV+LINR PSQ+ N++PY ++ P Y+ L
Sbjct: 719 HQHILNVARALMFQSQVPLSLWGDCVLTAVFLINRTPSQLLMNKTPYEILTGTAPVYEQL 778
Query: 654 KPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKCLNSHGRT-FISRHVIFNEDL 712
+ FGC CY P +HK Q + C+FLGY + +KGYK ++ T FISR+V F+E++
Sbjct: 779 RTFGCLCYSSTSPKQRHKFQPRSRACLFLGYPSGYKGYKLMDLESNTVFISRNVQFHEEV 838
Query: 713 FPFHEGFLNTRSPLKTTINNPSTSFPLCSAGNSINDASIPIIEEENQDETNEEDSQGVTS 772
FP + NP + L + +P+ S G+ S
Sbjct: 839 FPLAK--------------NPGSESSL-----KLFTPMVPV-------------SSGIIS 866
Query: 773 DTEQTDNG--SSEGDTTHEETLDIVQQQNVGESSLDTNTSNAIH------TRSKSGIHKP 824
DT + + S D + + V++ + NT + H T S S I
Sbjct: 867 DTTHSPSSLPSQISDLPPQISSQRVRKPPAHLNDYHCNTMQSDHKYPISSTISYSKISPS 926
Query: 825 KLPYIGITETYKDTVEPANVKEALTRTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDS 884
+ YI P N EA W EA+ E A+ TW + + V
Sbjct: 927 HMCYIN---NITKIPIPTNYAEAQDTKEWCEAVDAEIGAMEKTNTWEITTLPKGKKAVGC 983
Query: 885 KWVFKTKYKSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVRVILSIAVHLNWE 944
KWVF K+ +DG++ER KARLVAKG+ Q G+DY +TFSPV K++T++++L ++ W
Sbjct: 984 KWVFTLKFLADGNLERYKARLVAKGYTQKEGLDYTDTFSPVAKMTTIKLLLKVSASKKWF 1043
Query: 945 VRQLDINNAFLNGYLKETVFMHQPEGFVDPT----KPNHICKLSKAIYGLKQAPRAWFDS 1000
++QLD++NAFLNG L+E +FM PEG+ + N + +L ++IYGLKQA R WF
Sbjct: 1044 LKQLDVSNAFLNGELEEEIFMKIPEGYAERKGIVLPSNVVLRLKRSIYGLKQASRQWFKK 1103
Query: 1001 LKTALLNWGFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFS 1060
++LL+ GF+ T D +LFL +L+YVDDI++ +S ++L+ F
Sbjct: 1104 FSSSLLSLGFKKTHGDHTLFLKMYDGEFVIVLVYVDDIVIASTSEAAAAQLTEELDQRFK 1163
Query: 1061 LKDLGRLHYFLGIEVQRDASGMYLKQSKYIGDLLKKFKMENASPCPTPMITGRHFTVE-G 1119
L+DLG L YFLG+EV R +G+ + Q KY +LL+ M P PMI + G
Sbjct: 1164 LRDLGDLKYFLGLEVARTTAGISICQRKYALELLQSTGMLACKPVSVPMIPNLKMRKDDG 1223
Query: 1120 EKLKDPTVFRQAIGGLQYLTHTRPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTIN 1179
+ ++D +R+ +G L YLT TRPDI F+VNKL Q+ S+P T H R+L+Y++GT+
Sbjct: 1224 DLIEDIEQYRRIVGKLMYLTITRPDITFAVNKLCQFSSAPRTTHLTAAYRVLQYIKGTVG 1283
Query: 1180 YCLHIKPSTDLDITGFSDADWATSIDDRKSMAGQCVFLGETLISWSSRKQKVVSRSSTES 1239
L S+DL + GF+D+DWA+ D R+S +F+G++LISW S+KQ VSRSS E+
Sbjct: 1284 QGLFYSASSDLTLKGFADSDWASCQDSRRSTTSFTMFVGDSLISWRSKKQHTVSRSSAEA 1343
Query: 1240 EYRALVDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSAKALASNPVLHARSKHIEIDVH 1299
EYRAL E+ W+ +LL L+ P PIL+ D+ +A +A+NPV H R+KHI++D H
Sbjct: 1344 EYRALALATCEMVWLFTLLVSLQAS-PPVPILYSDSTAAIYIATNPVFHERTKHIKLDCH 1402
Query: 1300 YIRDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQLRDKLGVIH 1345
+R+++ ++ + +V T DQ+AD LTKPL +F L+ K+ +++
Sbjct: 1403 TVRERLDNGELKLLHVRTEDQVADILTKPLFPYQFEHLKSKMSILN 1448
Score = 48.1 bits (113), Expect = 3e-05
Identities = 31/136 (22%), Positives = 58/136 (41%), Gaps = 10/136 (7%)
Query: 16 VSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFEEWQAN 75
+S +LD NY W +L + G++ GT P E SD + F W
Sbjct: 75 ISHRLDETNYGDWSVAMLISLDAKNKTGFIDGTLSRPLE----SDLN------FRLWSRC 124
Query: 76 DQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKFQFHSIRKGEM 135
+ + W+LNS++ ++ +L + +W + S + L + R+G +
Sbjct: 125 NSMVKSWLLNSVSPQIYRSILRMNDASDIWRDLNSRFNVTNLPRTYNLTQEIQDFRQGTL 184
Query: 136 KMEDYLIKMKNLADKL 151
+ +Y ++K L D+L
Sbjct: 185 SLSEYYTRLKTLWDQL 200
>At1g70010 hypothetical protein
Length = 1315
Score = 659 bits (1701), Expect = 0.0
Identities = 412/1323 (31%), Positives = 662/1323 (49%), Gaps = 78/1323 (5%)
Query: 65 NNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLK 124
++P + W+ + + W+LNS++ E+ T +L+ T+ +W + + + ++ L+
Sbjct: 24 DDPYCKIWRRCNSMVKSWLLNSVSKEIYTSILYFPTAAAIWKDLYTRFHKSSLPRLYKLR 83
Query: 125 FQFHSIRKGEMKMEDYLIKMKNLADKL-KLAGNPIS--------NSNLIIQTLNGLDSEY 175
Q HS+R+G + + Y + + L ++L L P + +N +I L GL+ Y
Sbjct: 84 QQIHSLRQGNLDLSSYHTRTQTLWEELTSLQAVPRTVEDLLIERETNRVIDFLMGLNDCY 143
Query: 176 NPIVVKLSDHTTLSWVDLQAQLLTFESRIEQLNTLTNLNLNATANVANKFDHRDNRFNSN 235
+ + ++ TL L + I+Q T + ++ T + + N+ +S
Sbjct: 144 DTVRSQILMKKTLP------SLSEVFNMIDQDETQRSARISTTPGMTSSVFPVSNQ-SSQ 196
Query: 236 NNWRGSNFRGWRGGRGRGRSSKAPCQVYGKTNHTAINCFHRFDKNYSRSNYSADSDKQGS 295
+ G ++ + + C + H C+ + S + S
Sbjct: 197 SALNGDTYQ---------KKERPVCSYCSRPGHVEDTCYKKHGYPTSFKSKQKFVKPSIS 247
Query: 296 HNAFIASQNSVEDYDW----YFDSGASNHVTHQTDKFQDLT-----EHHGKNSQVVGNGD 346
NA I S+ V + S V+ + K Q + E H + +
Sbjct: 248 ANAAIGSEEVVNNTSVSTGDLTTSQIQQLVSFLSSKLQPPSTPVQPEVHSISVSSDPSSS 307
Query: 347 KLEIVATGSSKL-KSLNLDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTG 405
+GS L + L L+DVL++P NLLSVS L + FD+ C +++
Sbjct: 308 STVCPISGSVHLGRHLILNDVLFIPQFKFNLLSVSSLTKSMGCRIWFDETSCVLQDATRE 367
Query: 406 KVILKGLLKNGLYQLS-DTKGNP-------YAFVSVKESWHRRLGHPNNKVLDKVLKSCN 457
++ G LY + D+ +P A V+ + WH+RLGHP+ + L + +
Sbjct: 368 LMVGMGKQVANLYIVDLDSLSHPGTDSSITVASVTSHDLWHKRLGHPSVQKLQPMSSLLS 427
Query: 458 VKVPPSDNFSFCEACQYGKMHLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVH 517
++ C C K LPF S ++ + P +L+H D WGP + + G++Y++
Sbjct: 428 FPKQKNNTDFHCRVCHISKQKHLPFVSHNNKSSRPFDLIHIDTWGPFSVQTHDGYRYFLT 487
Query: 518 FIDDFSRFTWIYPLKQKSETVQAFTT-----ENQFNKRIKVIQCDGGGEYKPVQKLAIDV 572
+DD+SR TW+Y L+ KS+ + T ENQF IK ++ D E Q
Sbjct: 488 IVDDYSRATWVYLLRNKSDVLTVIPTFVTMVENQFETTIKGVRSDNAPELNFTQ-FYHSK 546
Query: 573 GIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQ 632
GI SCP T QQN ERKH+HI +L Q+ +P+ YW + TAVYLINRLP+
Sbjct: 547 GIVPYHSCPETPQQNSVVERKHQHILNVARSLFFQSHIPISYWGDCILTAVYLINRLPAP 606
Query: 633 VTQNESPYSLIFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYK 692
+ +++ P+ ++ P Y +K FGC CY P ++HK C F+GY + KGYK
Sbjct: 607 ILEDKCPFEVLTKTVPTYDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIGYPSGFKGYK 666
Query: 693 CLNSHGRTFI-SRHVIFNEDLFPFHEGFLNTRSPLKTTINNPSTSFPLCSAGNSINDASI 751
L+ + I SRHV+F+E+LFPF L+ NP+
Sbjct: 667 LLDLETHSIIVSRHVVFHEELFPFLGSDLSQEEQNFFPDLNPT----------------- 709
Query: 752 PIIEEENQDETNEEDSQG---VTSDTEQTDNGSSEG-DTTHEETLDIVQQQNVGESSLDT 807
P ++ ++ D N DS + T+N T+H + Q+ S+ +
Sbjct: 710 PPMQRQSSDHVNPSDSSSSVEILPSANPTNNVPEPSVQTSHRKAKKPAYLQDYYCHSVVS 769
Query: 808 NTSNAIHTRSKSGIHKPKLPYIGITETYKDTVEPANVKEALTRTLWKEAMQKEFQALMSN 867
+T + I R + PY+ T EP+N EA +W++AM EF L
Sbjct: 770 STPHEI--RKFLSYDRINDPYLTFLACLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGT 827
Query: 868 KTWILVPYQDQENIVDSKWVFKTKYKSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVK 927
TW + + + +W+FK KY SDGS+ER KARLVA+G+ Q GIDY ETFSPV K
Sbjct: 828 HTWEVCSLPADKRCIGCRWIFKIKYNSDGSVERYKARLVAQGYTQKEGIDYNETFSPVAK 887
Query: 928 VSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQPEGFV----DPTKPNHICKL 983
+++V+++L +A + QLDI+NAFLNG L E ++M P+G+ D PN +C+L
Sbjct: 888 LNSVKLLLGVAARFKLSLTQLDISNAFLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRL 947
Query: 984 SKAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGS 1043
K++YGLKQA R W+ + LL GF + D + FL +L+Y+DDII+ +
Sbjct: 948 KKSLYGLKQASRQWYLKFSSTLLGLGFIQSYCDHTCFLKISDGIFLCVLVYIDDIIIASN 1007
Query: 1044 SNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASGMYLKQSKYIGDLLKKFKMENAS 1103
++ + Q+ F L+DLG L YFLG+E+ R G+++ Q KY DLL +
Sbjct: 1008 NDAAVDILKSQMKSFFKLRDLGELKYFLGLEIVRSDKGIHISQRKYALDLLDETGQLGCK 1067
Query: 1104 PCPTPMITGRHFTVE-GEKLKDPTVFRQAIGGLQYLTHTRPDIAFSVNKLSQYMSSPTTD 1162
P PM F + G + +R+ IG L YL TRPDI F+VNKL+Q+ +P
Sbjct: 1068 PSSIPMDPSMVFAHDSGGDFVEVGPYRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKA 1127
Query: 1163 HWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDDRKSMAGQCVFLGETLI 1222
H Q + +IL+Y++GTI L +++L + +++AD+ + D R+S +G C+FLG++LI
Sbjct: 1128 HLQAVYKILQYIKGTIGQGLFYSATSELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLI 1187
Query: 1223 SWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSAKALA 1282
W SRKQ VVS+SS E+EYR+L E+ W+ + L EL++PL + +L+CDN +A +A
Sbjct: 1188 CWKSRKQDVVSKSSAEAEYRSLSVATDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIA 1247
Query: 1283 SNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQLRDKLG 1342
+N V H R+KHIE D H +R+++L+ + ++ T QIAD TKPL + F +L K+G
Sbjct: 1248 NNHVFHERTKHIESDCHSVRERLLKGLFELYHINTELQIADPFTKPLYPSHFHRLISKMG 1307
Query: 1343 VIH 1345
+++
Sbjct: 1308 LLN 1310
>At1g57640
Length = 1444
Score = 615 bits (1587), Expect = e-176
Identities = 374/1087 (34%), Positives = 571/1087 (52%), Gaps = 89/1087 (8%)
Query: 311 WYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEIVATGSSKLKS-LNLDDVLYV 369
W D+GAS+H+T + D+ ++ +G+K V+ G+ +L S L L V YV
Sbjct: 398 WILDTGASHHMTGNLELLSDMRSM-SPVLIILADGNKRVAVSEGTVRLGSHLILKSVFYV 456
Query: 370 PNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVILKGLLKNGLYQLSDTKGNPYA 429
+ +L+SV ++ D+N C N + K P+
Sbjct: 457 KELESDLISVGQM---------MDENHCV----------------NAAAVHTSVKA-PF- 489
Query: 430 FVSVKESWHRRLGHPNNKV---LDKVLKSCNVKVPPSDNFSFCEACQYGKMHLLPFKSSS 486
+ WHRRLGH ++K+ L + L S ++ + C+ C K F S
Sbjct: 490 -----DLWHRRLGHASDKIVNLLPRELLSSGKEILEN----VCDTCMRAKQTRDTFPLSD 540
Query: 487 SHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLKQKSETVQAFT---- 542
+ + + +L+H DVWGP S SG +Y++ +DD+SR W+Y + KSET +
Sbjct: 541 NRSMDSFQLIHCDVWGPYRAPSYSGARYFLTIVDDYSRGVWVYLMTDKSETQKHLKDFIA 600
Query: 543 -TENQFNKRIKVIQCDGGGEYKPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFG 601
E QF+ IK+++ D G E+ +++ + GI SC T QNGR ERKHRHI
Sbjct: 601 LVERQFDTEIKIVRSDNGTEFLCMREYFLHKGIAHETSCVGTPHQNGRVERKHRHILNIA 660
Query: 602 LTLLAQAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYSLIFHKEPNYKLLKPFGCACY 661
L Q+ +P+ +W E +A YLINR PS + Q +SPY +++ P Y L+ FG CY
Sbjct: 661 RALRFQSYLPIQFWGECILSAAYLINRTPSMLLQGKSPYEMLYKTAPKYSHLRVFGSLCY 720
Query: 662 PCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKCLN-SHGRTFISRHVIFNEDLFPFHEGFL 720
+ + K + RCVF+GY + KG++ + + F+SR VIF E FP+ +
Sbjct: 721 AHNQNHKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIFQETEFPYSKMSC 780
Query: 721 NTRSP--LKTTINNP---STSFPLCSAGNSINDASI-------PIIEEENQDETNEEDSQ 768
N L + P P G +I +A++ PII E NQ+ ++ +
Sbjct: 781 NEEDERVLVDCVGPPFIEEAIGPRTIIGRNIGEATVGPNVATGPIIPEINQESSSPSEFV 840
Query: 769 GVTS-------DTEQTDNGSSEGDTTHEETLDIVQQQNVGESSLDTNTSNAIHTRS---- 817
++S T QT + T L +Q L +N + S
Sbjct: 841 SLSSLDPFLASSTVQTADLPLSSTTPAPIQLRRSSRQTQKPMKLKNFVTNTVSVESISPE 900
Query: 818 -KSGIHKPKLPYIG---ITETYKDTV-------EPANVKEALTRTLWKEAMQKEFQALMS 866
S P Y+ T ++K + EP EA+ W+EAM E ++L
Sbjct: 901 ASSSSLYPIEKYVDCHRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRV 960
Query: 867 NKTWILVPYQDQENIVDSKWVFKTKYKSDGSIERRKARLVAKGFQQTAGIDYEETFSPVV 926
N+T+ +V + + +KWV+K KY+SDG+IER KARLV G Q G+DY+ETF+PV
Sbjct: 961 NQTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVA 1020
Query: 927 KVSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQPEGFV--DPTKPNHICKLS 984
K+STVR+ L +A +W V Q+D++NAFL+G LKE V+M P+GF DP+K +C+L
Sbjct: 1021 KMSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSK---VCRLH 1077
Query: 985 KAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSS 1044
K++YGLKQAPR WF L +AL +GF + SD SLF +L+YVDD+I++GS
Sbjct: 1078 KSLYGLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGIFVHVLVYVDDLIISGSC 1137
Query: 1045 NNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASGMYLKQSKYIGDLLKKFKMENASP 1104
+ + F L F +KDLG L YFLGIEV R+A G YL Q KY+ D++ + + A P
Sbjct: 1138 PDAVAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGARP 1197
Query: 1105 CPTPMITGRHFTVEGEKL-KDPTVFRQAIGGLQYLTHTRPDIAFSVNKLSQYMSSPTTDH 1163
P+ ++ L D + +R+ +G L YL TRP++++SV+ L+Q+M +P DH
Sbjct: 1198 SAFPLEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLVVTRPELSYSVHTLAQFMQNPRQDH 1257
Query: 1164 WQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDDRKSMAGQCVFLGETLIS 1223
W R++RYL+ + + ++ L I G+ D+D+A R+S+ G V LG+T IS
Sbjct: 1258 WNAAIRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQLGDTPIS 1317
Query: 1224 WSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSAKALAS 1283
W ++KQ VSRSS E+EYRA+ L E+ W+ +L++L + + ++ D+ SA AL+
Sbjct: 1318 WKTKKQPTVSRSSAEAEYRAMAFLTQELMWLKRVLYDLGVSHVQAMRIFSDSKSAIALSV 1377
Query: 1284 NPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQLRDKLGV 1343
NPV H R+KH+E+D H+IRD +L + ++VP+ Q+AD LTK L KLG+
Sbjct: 1378 NPVQHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILTKALGEKEVRYFLRKLGI 1437
Query: 1344 --IHSPP 1348
+H+PP
Sbjct: 1438 LDVHAPP 1444
>At1g26990 polyprotein, putative
Length = 1436
Score = 596 bits (1536), Expect = e-170
Identities = 370/1055 (35%), Positives = 543/1055 (51%), Gaps = 65/1055 (6%)
Query: 311 WYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEIVATGSSKLK-SLNLDDVLYV 369
W DSGAS+HVTH+ + + + + + NG ++I TG +L +L+L +VL++
Sbjct: 421 WVIDSGASHHVTHERNLYHTY-KALDRTFVRLPNGHTVKIEGTGFIQLTDALSLHNVLFI 479
Query: 370 PNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVILKGLLKNGLYQLS------DT 423
P NLLSVS L V F + C ++ ++ KG LY L+ D
Sbjct: 480 PEFKFNLLSVSVLTKTLQSKVSFTSDECMIQALTKELMLGKGSQVGNLYILNLDKSLVDV 539
Query: 424 KGNPYAFV--SVK---ESWHRRLGHPNNKVLDKVLKSCNV-KVPPSDNFSFCEACQYGKM 477
P V SVK E WH+RLGHP+ +D + + K + + S C C K
Sbjct: 540 SSFPGKSVCSSVKNESEMWHKRLGHPSFAKIDTLSDVLMLPKQKINKDSSHCHVCHLSKQ 599
Query: 478 HLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLKQKSET 537
LPFKS + ++ ELVH D WGP + + ++Y++ +DDFSR TWIY LKQKS+
Sbjct: 600 KHLPFKSVNHIREKAFELVHIDTWGPFSVPTVDSYRYFLTIVDDFSRATWIYLLKQKSDV 659
Query: 538 VQAFTT-----ENQFNKRIKVIQCDGGGEYKPVQKLAIDVGIQFRMSCPYTFQQNGRAER 592
+ F + E Q++ ++ ++ D E K +L GI+ CP T +QN ER
Sbjct: 660 LTVFPSFLKMVETQYHTKVCSVRSDNAHELK-FNELFAKEGIKADHPCPETPEQNFVVER 718
Query: 593 KHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYSLIFHKEPNYKL 652
KH+H+ L+ Q+ +PL YW + TAV+LINRL S V NE+PY + +P+Y
Sbjct: 719 KHQHLLNVARALMFQSGIPLEYWGDCVLTAVFLINRLLSPVINNETPYERLTKGKPDYSS 778
Query: 653 LKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKCLNSHGRTF-ISRHVIFNED 711
LK FGC CY P ++ K C+FLGY +KGYK L+ + ISRHVIF ED
Sbjct: 779 LKAFGCLCYCSTSPKSRTKFDPRAKACIFLGYPMGYKGYKLLDIETYSVSISRHVIFYED 838
Query: 712 LFPFHEGFLNTRSPLKTTINNPSTSFPLCSAGNSINDASIPIIEEENQDETNEEDSQGVT 771
+FPF + + FP ND +P+++ + N ++S +
Sbjct: 839 IFPFASSNITDAA---------KDFFPHIYLPAPNNDEHLPLVQSSSDAPHNHDESSSMI 889
Query: 772 SDTEQTDNGSSEGDTTHEETLDIVQQQNVGESSLDTNTSNAIHTRSKSGIHKPKLPYIGI 831
+ + +H + + +N I S S + +P +I I
Sbjct: 890 FVPSEPKSTRQRKLPSHLQDFHCYNNTPTTTKTSPYPLTNYI---SYSYLSEPFGAFINI 946
Query: 832 TETYKDTVEPANVKEALTRTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTK 891
K P EA +W +AM KE A + TW + + V KW+ K
Sbjct: 947 ITATK---LPQKYSEARLDKVWNDAMGKEISAFVRTGTWSICDLPAGKVAVGCKWIITIK 1003
Query: 892 YKSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVRVILSIAVHLNWEVRQLDIN 951
+ +DGSIER KARLVAKG+ Q GID+ TFSPV K+ TV+V+LS+A + W + QLDI+
Sbjct: 1004 FLADGSIERHKARLVAKGYTQQEGIDFFNTFSPVAKMVTVKVLLSLAPKMKWYLHQLDIS 1063
Query: 952 NAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKTALLNWGFQ 1011
NA LNG L+E ++M P G+ + I G + +P A
Sbjct: 1064 NALLNGDLEEEIYMKLPPGYSE-------------IQGQEVSPNA--------------- 1095
Query: 1012 NTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFL 1071
D +LF+ +L+YVDDI++ ++ QL+ F L+DLG +FL
Sbjct: 1096 KCHGDHTLFVKAQDGFFLVVLVYVDDILIASTTEAASAELTSQLSSFFQLRDLGEPKFFL 1155
Query: 1072 GIEVQRDASGMYLKQSKYIGDLLKKFKMENASPCPTPMITGRHFTVE-GEKLKDPTVFRQ 1130
GIE+ R+A G+ L Q KY+ DLL + P PM + + + G L+D +R+
Sbjct: 1156 GIEIARNADGISLCQRKYVLDLLASSDFSDCKPSSIPMEPNQKLSKDTGTLLEDGKQYRR 1215
Query: 1131 AIGGLQYLTHTRPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDL 1190
+G LQYL TRPDI F+V+KL+QY S+PT H Q + +ILRYL+GTI L T+
Sbjct: 1216 ILGKLQYLCLTRPDINFAVSKLAQYSSAPTDIHLQALHKILRYLKGTIGQGLFYGADTNF 1275
Query: 1191 DITGFSDADWATSIDDRKSMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAE 1250
D+ GFSD+DW T D R+ + G +F+G +L+SW S+KQ VVS SS E+EYRA+ E
Sbjct: 1276 DLRGFSDSDWQTCPDTRRCVTGFAIFVGNSLVSWRSKKQDVVSMSSAEAEYRAMSVATKE 1335
Query: 1251 IAWIHSLLFELKLPLPRKPILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKV 1310
+ W+ +L K+P L+CDN +A +A+N V H R+KHIE D H +R+ + +
Sbjct: 1336 LIWLGYILTAFKIPFTHPAYLYCDNEAALHIANNSVFHERTKHIENDCHKVRECIEAGIL 1395
Query: 1311 VVAYVPTTDQIADCLTKPLSHTRFSQLRDKLGVIH 1345
+V T +Q+AD LTKPL F + KLG+++
Sbjct: 1396 KTIFVRTDNQLADTLTKPLYPKPFRENNSKLGLLN 1430
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 581 bits (1497), Expect = e-165
Identities = 372/1113 (33%), Positives = 546/1113 (48%), Gaps = 165/1113 (14%)
Query: 289 DSDKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKL 348
++D + ++ A Q + W DSGAS+HV F++L G +
Sbjct: 460 NNDLKFQNHTLSALQKFLPSDAWIIDSGASSHVCSDLAMFRELKSVSGT----------V 509
Query: 349 EIVATGSSKLKSLNLDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVI 408
I + L L +VL+VP+ NL+SVS L + F +CC ++E G +I
Sbjct: 510 HIT-------QKLILHNVLHVPDFKFNLMSVSSLVKTISCSAHFYVDCCLIQELSQGLMI 562
Query: 409 LKGLLKNGLYQLSDTKGNPY-----------AFVSVKESWHRRLGHPNNKVLDKVLKSCN 457
+G L + LY L +P + ++ WH+RLGHP++ VL K+ +
Sbjct: 563 GRGRLYHNLYILETENTSPSTSTPAACLFTGSVLNDGHLWHQRLGHPSSVVLQKLKR--- 619
Query: 458 VKVPPSDNFSFCEACQYGKMHLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVH 517
L + S ++ A P +LVH D+WGP I S GF+Y++
Sbjct: 620 ----------------------LAYISHNNLASNPFDLVHLDIWGPFSIESIEGFRYFLT 657
Query: 518 FIDDFSRFTWIYPLKQKSETVQAFT-----TENQFNKRIKVIQCDGGGEYKPVQKLAIDV 572
+DD +R TW+Y L+ K + F QFN +IK I+ D E ++ +
Sbjct: 658 VVDDCTRTTWVYMLRNKKDVSSVFPEFIKLVSTQFNAKIKAIRSDNAPELG-FTEIVKEH 716
Query: 573 GIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQ 632
G+ SC YT QQN ERKH+HI LL Q+ +P+ YW + +TAV+LINRLPS
Sbjct: 717 GMLHHFSCAYTPQQNSVVERKHQHILNVARALLFQSNIPMQYWSDCVTTAVFLINRLPSP 776
Query: 633 VTQNESPYSLIFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYK 692
+ N+SPY LI +K+P+Y LLK FGC C+ + + K CVFLGY + +KGYK
Sbjct: 777 LLNNKSPYELILNKQPDYSLLKNFGCLCFVSTNAHERTKFTPRARACVFLGYPSGYKGYK 836
Query: 693 CLN--SHGRTFISRHVIFNEDLFPFHEG---------FLNTRSPLKTTINNPSTSFPLCS 741
L+ SH T +SR+V+F E +FPF F N+ PL ++ T PL
Sbjct: 837 VLDLESHSVT-VSRNVVFKEHVFPFKTSELLNKAVDMFPNSILPLPAPLHFVET-MPLID 894
Query: 742 AGNSI---------------NDASIP-IIEEENQDETNEEDSQGVT-SDTEQTDNGSSEG 784
+ I + +++P II + ET + DS V + +++T S
Sbjct: 895 EDSLIPTTTDSRTADNHASSSSSALPSIIPPSSNTETQDIDSNAVPITRSKRTTRAPSYL 954
Query: 785 DTTHEETLDIVQQQNVGESSLDTNTSNAIHTRSKSGIHKPKLPYIGITET---------- 834
H + + +SS+ + I T S K PY T
Sbjct: 955 SEYHCSLVPSISTLPPTDSSIPIHPLPEIFTASSP---KKTTPYPISTVVSYDKYTPLCQ 1011
Query: 835 -----YKDTVEPANVKEALTRTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFK 889
Y EP +A+ W +E QA+ NKTW + +N+V KWVF
Sbjct: 1012 SYIFAYNTETEPKTFSQAMKSEKWIRVAVEELQAMELNKTWSVESLPPDKNVVGCKWVFT 1071
Query: 890 TKYKSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVRVILSIAVHLNWEVRQLD 949
KY DG++ER KARLVA+GF Q GID+ +TFSPV K+++ +++L +A W + Q+D
Sbjct: 1072 IKYNPDGTVERYKARLVAQGFTQQEGIDFLDTFSPVAKLTSAKMMLGLAAITGWTLTQMD 1131
Query: 950 INNAFLNGYLKETVFMHQPEGFVDPT----KPNHICKLSKAIYGLKQAPRAWFDSLKTAL 1005
+++AFL+G L E +FM P+G+ P PN +C+L K+IYGLKQA R W+ AL
Sbjct: 1132 VSDAFLHGDLDEEIFMSLPQGYTPPAGTILPPNPVCRLLKSIYGLKQASRQWYKRFVAAL 1191
Query: 1006 LNWGFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLG 1065
+Y+DDI++ +++ ++ L F +KDLG
Sbjct: 1192 ---------------------------VYIDDIMIASNNDAEVENLKALLRSEFKIKDLG 1224
Query: 1066 RLHYFLGIEVQRDASGMYLKQSKYIGDLLKKFKMENASPCPTPMITGRHFTVE-GEKLKD 1124
+FLG+ P PM H + G L +
Sbjct: 1225 PARFFLGLL--------------------------GCKPSSIPMDPTLHLVRDMGTPLPN 1258
Query: 1125 PTVFRQAIGGLQYLTHTRPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHI 1184
PT +R+ IG L YLT TRPDI ++V++LSQ++S+P+ H Q ++LRY++ L
Sbjct: 1259 PTAYRKLIGRLLYLTITRPDITYAVHQLSQFISAPSDIHLQAAHKVLRYIKANPGQGLMY 1318
Query: 1185 KPSTDLDITGFSDADWATSIDDRKSMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRAL 1244
++ + GFSDADWA D R+S++G C++LG +LISW S+KQ V SRSSTESEYR++
Sbjct: 1319 SADYEICLNGFSDADWAACKDTRRSISGFCIYLGTSLISWKSKKQAVASRSSTESEYRSM 1378
Query: 1245 VDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQ 1304
EI W+ LL +L +PL L+CDN SA + NPV H R+KHIEID H +RDQ
Sbjct: 1379 AQATCEIIWLQQLLKDLHIPLTCPAKLFCDNKSALHSSLNPVFHERTKHIEIDCHTVRDQ 1438
Query: 1305 VLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQL 1337
+ + +VPT +Q AD LTK L F L
Sbjct: 1439 IKAGNLKALHVPTENQHADILTKALHPGPFHHL 1471
Score = 32.3 bits (72), Expect = 1.9
Identities = 23/125 (18%), Positives = 51/125 (40%), Gaps = 10/125 (8%)
Query: 23 NNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFEEWQANDQRLLGW 82
+++ W+ +L + G++ GT P E ++ F W + + W
Sbjct: 55 SDFHSWRRSILMALNVRNKLGFINGTITKPPE----------DHRDFGAWSRCNDIVSTW 104
Query: 83 MLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKFQFHSIRKGEMKMEDYLI 142
++NS+ ++ LL+ T + +W+ S +I ++ + I +G M + Y
Sbjct: 105 LMNSVDKKIGQSLLYIATVQGIWNNLLSRFKQDDAPRIFDIEQKLSKIEQGSMDISTYYT 164
Query: 143 KMKNL 147
+ L
Sbjct: 165 ALLTL 169
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 567 bits (1460), Expect = e-161
Identities = 402/1335 (30%), Positives = 655/1335 (48%), Gaps = 122/1335 (9%)
Query: 66 NPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWD--EAQSLAGAHTRS---QI 120
+P ++ + ND + S+ Q+ +TSK +W+ + ++L + Q
Sbjct: 15 DPGSDDMEKNDMAR-ALLFQSVPESTILQVGKHKTSKAMWEAIKTRNLGAERVKEAKLQT 73
Query: 121 IYLKFQFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSNLIIQTLNGLD-SEYNPIV 179
+ +F +++ E +++++ ++ ++ K + G I S ++ + L L +Y I+
Sbjct: 74 LMAEFDRLNMKDNET-IDEFVGRISEISTKSESLGEEIEESKIVKKFLKSLPRKKYIHII 132
Query: 180 VKLS---DHTTLSWVDLQAQLLTFESRI-EQLNTLTNLNLNATANVANKFDHRDNRFNSN 235
L D T + D+ ++ T+E R+ ++ ++ AN + +D R R
Sbjct: 133 AALEQILDLNTTGFEDIVGRMKTYEDRVCDEDDSPEEQGKLMYANSESSYDTRGGR---- 188
Query: 236 NNWRGSNFRGWRGGRGRGR-------SSKAPCQVYGKTNHTAINCFHRF---DKNYSRSN 285
RG RG GRGRG SK C KT H A C R K +
Sbjct: 189 --GRG---RGRSSGRGRGGYGYQQRDKSKVICYRCDKTGHYASECLDRLLKLIKAQEQQQ 243
Query: 286 YSADSDKQGS---HNAFIASQNSVE--------DYDWYFDSGASNHVTHQTDKFQDLTE- 333
+ D D+ S H ++ SV+ D WY D+GASNH+T F L E
Sbjct: 244 NNEDDDEIESLMMHEVVYLNERSVKPKEFEACSDNSWYLDNGASNHMTGNLQWFSKLNEM 303
Query: 334 -----HHGKNSQVVGNGDKLEIVATGSSKLKSLNLDDVLYVPNITKNLLSVSKLA-ADNN 387
G +S++ G ++ T K+L DV ++P++ N++S+ + A +
Sbjct: 304 ITGKVRFGDDSRIDIKGKGSIVLITKGGIRKTLT--DVYFIPDLKSNIISLGQATEAGCD 361
Query: 388 IFVEFDKNCCFVKEKLTGKVILKGLL-KNGLYQLSDTKGNPYAFVSVKESWHRRLGHPNN 446
+ ++ D+ +E G ++L+ +N LY++ N + R+
Sbjct: 362 VRMKDDQLTLHDRE---GCLLLRATRSRNRLYKVDLNVENVKCLQLEAATMVRK------ 412
Query: 447 KVLDKVLKSCNVKVPPSDNFSFCEACQYGKMHLLPF-KSSSSHAQEPLELVHTDVWGPAP 505
+ V+ N+ P + + C +C GK PF K+++ A + LELVH D+ GP
Sbjct: 413 ---ELVIGISNI---PKEKET-CGSCLLGKQARQPFPKATTYRASQVLELVHGDLCGPIT 465
Query: 506 IMSSSGFKYYVHFIDDFSRFTWIYPLKQKSETVQAFT-----TENQFNKRIKVIQCDGGG 560
+++ +Y + IDD +R+ W LK+KSE + F E + +IK + D GG
Sbjct: 466 QSTTAKKRYILVLIDDHTRYMWSMLLKEKSEAFEKFRDFKTKVEQESGVKIKTFRTDKGG 525
Query: 561 EY--KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEA 618
E+ + Q GI ++ PYT QQNG ER++R + ++L +MP + W EA
Sbjct: 526 EFVSQEFQDFCAKEGINRHLTAPYTPQQNGVVERRNRTLLGMTRSILKHMKMPNYLWGEA 585
Query: 619 FSTAVYLINRLPSQVTQNESPYSLIFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTR 678
+ Y+INR+ ++ QN++PY + ++PN + L+ FGC Y ++ + KL +
Sbjct: 586 VRHSTYIINRVGTRSLQNQTPYEVFKQRKPNVEHLRVFGCIGYAKIEGPHLRKLDDRSKM 645
Query: 679 CVFLGYSNSHKGYKCLNSHGRTFISRHVIFNEDLFPFHEGFLNTRSPLKTTINNPSTSFP 738
V+LG K Y+ L+ R I + S +T + + S
Sbjct: 646 LVYLGTEPGSKAYRLLDPTNRKIIKWN-----------------NSDSETRDISGTFSLT 688
Query: 739 LCSAGNSINDASIPIIEEENQDET-NEEDSQGVTSDTEQTDNGSSEGDTTHEETLDIVQQ 797
L GN+ S I E+N +E+ N + +G EQ + E +H L ++
Sbjct: 689 LGEFGNNGIQESDDIETEKNGEESENSHEEEGENEHNEQEQIDAEETQPSHATPLPTLR- 747
Query: 798 QNVGESSLDTNTSNAIHTRSKSGIHKPKL--PYIGITETYKDTV------EPANVKEALT 849
RS + KP Y+ + E + V EP + KEA
Sbjct: 748 ------------------RSTRQVGKPNYLDDYVLMAEIEGEQVLLAINDEPWDFKEANK 789
Query: 850 RTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKYKSDGSIERRKARLVAKG 909
W++A ++E ++ NKTW L+ + ++ KWVFK K SDGSI + KARLVAKG
Sbjct: 790 LKEWRDACKEEILSIEKNKTWSLIDLPVRRKVIGLKWVFKIKRNSDGSINKYKARLVAKG 849
Query: 910 FQQTAGIDYEETFSPVVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQPE 969
+ Q GIDY+E F+ V ++ T+RVI+++A WEV LD+ AFL+G L+E V++ QPE
Sbjct: 850 YVQRHGIDYDEVFAHVARIETIRVIIALAASNGWEVHHLDVKTAFLHGELREDVYVTQPE 909
Query: 970 GFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLKGKDHIT 1029
GF + + KL KA+YGLKQAPRAW L L F +PS++ + + +
Sbjct: 910 GFTNKDNEGKVYKLHKALYGLKQAPRAWNTKLNKILQELNFVKCSKEPSVYRRQEEKKLL 969
Query: 1030 FLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASGMYLKQSKY 1089
+ IYVDD++VTGSS + + F K + F + DLG+L Y+LGIEV +G+ L+Q +Y
Sbjct: 970 IVAIYVDDLLVTGSSLDLILCFKKDMAGKFEMSDLGQLTYYLGIEVLHRKNGIILRQERY 1029
Query: 1090 IGDLLKKFKMENASPCPTPMITGRHF-TVEGEKLKDPTVFRQAIGGLQYLTHTRPDIAFS 1148
++++ M N +P PM G + EK +R+ IG L+Y+ HTRPD+++
Sbjct: 1030 AMKIIEEAGMSNCNPVLIPMAAGLELCKAQEEKCITERDYRRMIGCLRYIVHTRPDLSYC 1089
Query: 1149 VNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDDRK 1208
V LS+Y+ P H +K++LRYL+GT+++ L++K + G+SD+ + +DD K
Sbjct: 1090 VGVLSRYLQQPRESHGNALKQVLRYLKGTMSHGLYLKRGFKSGLVGYSDSSHSADLDDGK 1149
Query: 1209 SMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRK 1268
S AG +L + I+W S+KQ+VV+ SS E+E+ A + A + W+ L E+ K
Sbjct: 1150 STAGHIFYLHQCPITWCSQKQQVVALSSCEAEFMAATEAAKQAIWLQDLFAEVCGTTSEK 1209
Query: 1269 PILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKP 1328
++ DN SA AL N V H RSKHI H+IR+ V N V V +VP +Q AD LTKP
Sbjct: 1210 VMIRVDNKSAIALTKNLVFHGRSKHIHRRYHFIRECVENNLVEVDHVPGVEQRADILTKP 1269
Query: 1329 LSHTRFSQLRDKLGV 1343
L +F ++R+ +GV
Sbjct: 1270 LGRIKFREMRELVGV 1284
>At3g25450 hypothetical protein
Length = 1343
Score = 563 bits (1452), Expect = e-160
Identities = 355/1062 (33%), Positives = 552/1062 (51%), Gaps = 59/1062 (5%)
Query: 311 WYFDSGASNHVTHQTDKFQDLTE------HHGKNS--QVVGNGDKLEIVATGSSKLKSLN 362
WY D+GASNH+T F L E G +S + G G I G K+
Sbjct: 292 WYLDNGASNHMTGNRAWFCKLDEMITGKVRFGDDSCINIKGKGSIPFISKGGERKI---- 347
Query: 363 LDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVILKGLLKNGLYQLSD 422
L DV Y+P++ N+LS+ + A ++ + ++ + ++ +I +N LY++S
Sbjct: 348 LFDVYYIPDLKSNILSLGQ-ATESGCDIRMREDYLTLHDREGNLLIKAQRSRNRLYKVSL 406
Query: 423 TKGNPYAF--VSVKES--WHRRLGHPNNKVLDKVLKS---CNVKVPPSDNFSFCEACQYG 475
N + ES WH RLGH + + + ++K + C +C +G
Sbjct: 407 EVENSKCLQLTTTNESTIWHARLGHISFETIKAMIKKELVIGISSSVPQEKETCGSCLFG 466
Query: 476 KM--HLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLKQ 533
K H P K++S A + LEL+H D+ GP +++ +Y IDD SR+ W LK+
Sbjct: 467 KQARHSFP-KATSYRAAQVLELIHGDLCGPISPSTAAKKRYVFVLIDDHSRYMWSILLKE 525
Query: 534 KSETVQAFT-----TENQFNKRIKVIQCDGGGEY--KPVQKLAIDVGIQFRMSCPYTFQQ 586
KSE F E + IK + D GGE+ Q+ GI ++ PYT QQ
Sbjct: 526 KSEAFGKFKEFKALVEQECGAIIKTFRTDRGGEFLSHEFQEFCAKEGINRHLTAPYTPQQ 585
Query: 587 NGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYSLIFHK 646
NG ER++R + ++L MP + W EA + YLINR+ ++ N++PY + HK
Sbjct: 586 NGVVERRNRTLLGMTRSILKHMNMPNYLWGEAVRHSTYLINRVGTRSLSNQTPYEVFKHK 645
Query: 647 EPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKCLN-SHGRTFISRH 705
+PN + L+ FGC Y ++ N KL + V+LG K Y+ L+ + R F+SR
Sbjct: 646 KPNVEHLRVFGCVSYAKVEVPNLKKLDDRSRMLVYLGTEPGSKAYRLLDPTKRRIFVSRD 705
Query: 706 VIFNEDLFPFHEGFLNTRSPLKTTINNPSTSFPLCSAGN---SINDASIPIIEEENQDET 762
V+F+E + ++ S +T + + + L GN + ND S E E +
Sbjct: 706 VVFDE-----NRSWMWQESSSETDKESGTFTITLSEFGNNGVTENDISTEPEETEEAEIN 760
Query: 763 NEEDSQGVTSDTEQTDNGSSEGDTTHEETLDIVQQQNVGESSLDTNTSNAIHTRSKSGIH 822
E+++ ++TE+ D E +++ + + L A H
Sbjct: 761 GEDENIIEEAETEEHDQSQEEPQPVRRSQRQVIRPNYLKDYVLCAEI-EAEHL------- 812
Query: 823 KPKLPYIGITETYKDTVEPANVKEALTRTLWKEAMQKEFQALMSNKTWILVPYQDQENIV 882
+ + + EP + KEA W++A ++E Q++ N+TW LV +
Sbjct: 813 -----LLAVND------EPWDFKEANKSKEWRDACKEEIQSIEKNRTWSLVDLPVGSKAI 861
Query: 883 DSKWVFKTKYKSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVRVILSIAVHLN 942
KWVFK K+ SDGSI + KARLVAKG+ Q G+D+EE F+PV ++ TVR+I+++A
Sbjct: 862 GVKWVFKLKHNSDGSINKYKARLVAKGYVQRHGVDFEEVFAPVARIETVRLIIALAASNG 921
Query: 943 WEVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLK 1002
WE+ LD+ AFL+G L+E V++ QPEGF + + KL KA+YGL+QAPRAW L
Sbjct: 922 WEIHHLDVKTAFLHGELREDVYVSQPEGFTNKESKEKVYKLHKALYGLRQAPRAWNTKLN 981
Query: 1003 TALLNWGFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLK 1062
L F+ +PSL+ + ++I + +YVDD++VTGS+ + + F K + F +
Sbjct: 982 EILKELKFEKCHKEPSLYRKQEGENILVVAVYVDDLLVTGSNLDIILNFKKGMVGKFEMS 1041
Query: 1063 DLGRLHYFLGIEVQRDASGMYLKQSKYIGDLLKKFKMENASPCPTPMITGRHFT-VEGEK 1121
DLG+L Y+LGIEV + G+ LKQ +Y +L++ M + TPMI + + EK
Sbjct: 1042 DLGKLTYYLGIEVLQSKDGITLKQERYAKKILEEAGMSKCNTVNTPMIASLELSKAQDEK 1101
Query: 1122 LKDPTVFRQAIGGLQYLTHTRPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYC 1181
D T +R+ IG L+YL HTRPD++++V LS+Y+ P H +K+ILRYLQGT ++
Sbjct: 1102 RIDETDYRRNIGCLRYLLHTRPDLSYNVGILSRYLQEPRESHGAALKQILRYLQGTTSHG 1161
Query: 1182 LHIKPSTDLDITGFSDADWATSIDDRKSMAGQCVFLGETLISWSSRKQKVVSRSSTESEY 1241
L+ K + + G+SD+ +DD KS G +L + I+W S+KQ+VV+ SS E+E+
Sbjct: 1162 LYFKKGENAGLIGYSDSSHNVDLDDGKSTGGHIFYLNDCPITWCSQKQQVVTLSSCEAEF 1221
Query: 1242 RALVDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSAKALASNPVLHARSKHIEIDVHYI 1301
A + A + W+ LL E+ K + DN SA AL NPV H RSKHI H+I
Sbjct: 1222 MAATEAAKQAIWLQELLAEVIGTECEKVTIRVDNKSAIALTKNPVFHGRSKHIHRRYHFI 1281
Query: 1302 RDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQLRDKLGV 1343
R+ V ++ V +VP Q AD LTK L +F ++R+ +GV
Sbjct: 1282 RECVENGQIEVEHVPGVRQKADILTKALGKIKFLEMRELIGV 1323
>At3g45520 copia-like polyprotein
Length = 1363
Score = 538 bits (1386), Expect = e-153
Identities = 392/1392 (28%), Positives = 656/1392 (46%), Gaps = 115/1392 (8%)
Query: 22 RNNYPLWKSLVLPVVRGCKLDGYM------LGTKKCPEEFITSSDSSKSNNPAFEEWQAN 75
R +Y +WK +L + L + +G ++ E+ S + K E ++
Sbjct: 14 RGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEK---SDEDEKEEREKMEAFEEK 70
Query: 76 DQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKFQFHSIRKGE- 134
++ ++ S++ + ++ ++ + + L + IYLK + +S + E
Sbjct: 71 KRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQKLYSFKMSEN 130
Query: 135 MKMEDYLIKMKNLADKLKLAGNPISNSNLIIQTLNGLDSEYNPIVVKL---SDHTTLSWV 191
+ +E + + ++ L+ +S+ + I L L ++ + L S T LS
Sbjct: 131 LSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYSSGKTVLSLD 190
Query: 192 DLQAQLLTFESRIEQLNTLTNLNLNATANVANKFDHRDNRFNSNNNWRGSNFRGWRGGRG 251
++ A + + E + ++ A D +NR S +G +G R
Sbjct: 191 EVAAAIYSRELEFGSVKK----SIKGQAEGLYVKDKAENRGRSEQKDKG------KGKRS 240
Query: 252 RGRSSKAPCQVYGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFIASQNSVE---- 307
+ +S + C + G+ H C ++ + + G N S N VE
Sbjct: 241 KSKSKRG-CWICGEDGHLKSTCPNKNKPQFKNQGSNKGESSGGKGNLVEGSVNFVESAGM 299
Query: 308 -------------DYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEIVATG 354
+ +W D+G H+TH+ + +D E G S +GN + G
Sbjct: 300 FVSEALSSTDIHLEDEWIMDTGCIYHMTHKREWLEDFDEEAG-GSVRMGNKSISRVKGVG 358
Query: 355 SSKLKSLN-----LDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVIL 409
+ ++ + N L +V Y+P++ +NLLS+ + +F+ ++ K +V+L
Sbjct: 359 TVRIVNDNGLTVTLQNVRYIPDMDRNLLSLGTFEKAGH---KFESENGMLRIKSGNQVLL 415
Query: 410 KGLLKNGLYQLSDTKGNPYAFVSVKES-----WHRRLGHPNNKVLDKVLKSCNVKVPPSD 464
+G + LY L + + + WHRRL H + K + ++K +
Sbjct: 416 EGRRYDTLYILHGKPATDESLAVARANDDTVLWHRRLCHMSQKNMSLLIKKGFLDKKKVS 475
Query: 465 NFSFCEACQYGKMHLLPFKSSSSHAQEPLELVHTDVWG-PAPIMSSSGFKYYVHFIDDFS 523
CE C YG+ + F + ++ LE VH+D+WG P MS +Y++ FIDD++
Sbjct: 476 MLDTCEDCIYGRAKKIGFNLAQHDTKKKLEYVHSDLWGAPTVPMSLGNCQYFISFIDDYT 535
Query: 524 RFTWIYPLKQKSETVQAFTT-----ENQFNKRIKVIQCDGGGEY--KPVQKLAIDVGIQF 576
R W+Y LK K E + F + ENQ +R+K ++ D G E+ + + G Q
Sbjct: 536 RKVWVYFLKTKDEAFEKFVSWISLVENQSGERVKTLRTDNGLEFCNRMFDGFCEEKGFQR 595
Query: 577 RMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQVTQN 636
+C YT QQNG ER +R I E ++L + +P +W EA TAV LIN+ P
Sbjct: 596 HRTCAYTPQQNGVVERMNRTIMEKVRSMLCDSGLPKRFWAEATHTAVLLINKTPCSAINF 655
Query: 637 ESPYSLIFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKC-LN 695
E P K P Y L+ +GC + + KL + V +GY + KGYK L
Sbjct: 656 EFPDKRWSGKAPIYSYLRRYGCVTF---VHTDGGKLNLRAKKGVLIGYPSGVKGYKVWLI 712
Query: 696 SHGRTFISRHVIFNEDLFPFHEGFLNTRSPLKTTINNPSTSFPLCSAGNSINDASIPIIE 755
+ +SR+V F E+ ++ + + + ++ + S+ + +
Sbjct: 713 EEKKCVVSRNVSFQEN--AVYKDLMQRKEQVSCEEDDHAGSY-----------IDLDLEA 759
Query: 756 EENQDETNEEDSQGVTSDTEQTDNGSSEGDTTHEETLDIVQQQNVGESSLDTNTSNAIHT 815
+++ E+ VT T G+ ET D +++ +V +S L + + +
Sbjct: 760 DKDNSSGGEQSQAQVTPAT----RGAVTSTPPRYET-DDIEETDVHQSPL---SYHLVRD 811
Query: 816 RSKSGIHKPKL----PYIG---ITETYKDTVEPANVKEAL---TRTLWKEAMQKEFQALM 865
R + I P+ Y T D VEPA+ KEA+ W+ AM +E ++ +
Sbjct: 812 RERREIRAPRRFDDEDYYAEALYTTEDGDAVEPADYKEAVRDENWDKWRLAMNEEIESQL 871
Query: 866 SNKTWILVPYQDQENIVDSKWVFKTKYKSDGSIERR-KARLVAKGFQQTAGIDYEETFSP 924
N TW V +++ I+ S+W++K K G E R KARLVAKG+ Q G+DY E F+P
Sbjct: 872 KNDTWTTVTRPEKQRIIGSRWIYKYKQGIPGVEEPRFKARLVAKGYAQREGVDYHEIFAP 931
Query: 925 VVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLS 984
VVK ++R++LSI N E+ QLD+ AFL+G LKE ++M PEG K N +C L+
Sbjct: 932 VVKHVSIRILLSIVAQENLELEQLDVKTAFLHGELKEKIYMMPPEGCESLFKENEVCLLN 991
Query: 985 KAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLKGKDHIT-FLLIYVDDIIVTGS 1043
K++YGLKQAPR W + + GF+ + D + K D T +LL YVDD++V +
Sbjct: 992 KSLYGLKQAPRQWNEKFNHYMTEIGFKRSDYDSCAYTKKLSDDSTMYLLFYVDDMLVAAN 1051
Query: 1044 SNNFLQAFIKQLNDVFSLKDLGRLHYFLGIE--VQRDASGMYLKQSKYIGDLLKKFKMEN 1101
+ + A K+L+ F +KDLG LGIE + R+A ++L Q Y+ +LK F M
Sbjct: 1052 NMQAIDALKKELSIKFEMKDLGAAKKILGIEIIIDREAGVLWLSQESYLNKVLKTFNMLE 1111
Query: 1102 ASPCPTPMITGRHFTVEG---------EKLKDPTVFRQAIGGLQY-LTHTRPDIAFSVNK 1151
+ P TP+ G H ++ E+ + + A+G + Y + TRPD+A+ V
Sbjct: 1112 SKPALTPL--GAHLKMKSATEEKLSTEEEYMNSVPYSSAVGSIMYAMIGTRPDLAYPVGV 1169
Query: 1152 LSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDDRKSMA 1211
+S++MS P +HW G+K +LRY++GT++ L K ++D I G+ DAD+A +D R+S+
Sbjct: 1170 VSRFMSQPAKEHWLGVKWVLRYIKGTVDTRLCYKRNSDFSICGYCDADYAADLDKRRSIT 1229
Query: 1212 GQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPIL 1271
G LG ISW S Q+VV++SSTE EY +L + E W+ LL + + +
Sbjct: 1230 GLVFTLGGNTISWKSGLQRVVAQSSTECEYMSLTEAVKEAIWLKGLLKDFGYE-QKNVEI 1288
Query: 1272 WCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSH 1331
+CD+ SA AL+ N V H R+KHI++ H+IR+ + KV V+ + T AD TK L
Sbjct: 1289 FCDSQSAIALSKNNVHHERTKHIDVKFHFIREIIADGKVEVSKISTEKNPADIFTKVLPV 1348
Query: 1332 TRFSQLRDKLGV 1343
+F D L V
Sbjct: 1349 NKFQTALDFLRV 1360
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 533 bits (1374), Expect = e-151
Identities = 400/1335 (29%), Positives = 650/1335 (47%), Gaps = 135/1335 (10%)
Query: 86 SMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKFQFHSIRKGEMKMEDYLIKMK 145
++A ++ ++ C+T+ + W+ L + +Y + F++ + E K D +
Sbjct: 56 NVADKVLRKIELCKTAAEAWETLDRLFMIRSLPHRVYTQLSFYTFKMQENKKID-----E 110
Query: 146 NLADKLKLAGN------PISNSNLIIQTLNGLDSEYNPIVVKLSDHTTLSWVDLQAQLLT 199
N+ D LK+ + +++ I L+ L + Y+ +V T+ + + + +L
Sbjct: 111 NIDDFLKIVADLNHLQIDVTDEVQAILLLSSLPARYDGLV------ETMKYSNSREKLRL 164
Query: 200 FESRIEQLNTLTNLNLNATANVANKFDHRDNRFNSNNNWRGSNFRGWRGGRGRGRSSKAP 259
+ + + L+ N V F R + NN +G+ +G R + K
Sbjct: 165 DDVMVAARDKERELSQNNRPVVEGHFAR--GRPDGKNNNQGN--KGKNRSRSKSADGKRV 220
Query: 260 CQVYGKTNHTAINCFHRFDKNYSRSNYS--ADSDKQGSHNAF------IASQNSVEDYD- 310
C + GK H C+ ++N S+ S +S S AF +A+ ++ D
Sbjct: 221 CWICGKEGHFKKQCYKWIERNKSKQQGSDNGESSLAKSTEAFNPAMVLLATDETLVVTDS 280
Query: 311 ----WYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEIVATGSSKLKSLN---- 362
W D+G S H+T + D F+D E ++ GN + GS K+++ +
Sbjct: 281 IANEWVLDTGCSFHMTPRKDWFKDFKELSSGYVKM-GNDTYSPVKGIGSIKIRNSDGSQV 339
Query: 363 -LDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKV------ILKGLLKN 415
L DV Y+PN+T+NL+S+ L D+ C F + K+ ILKG ++
Sbjct: 340 ILTDVRYMPNMTRNLISLGTLE---------DRGCWFKSQDGILKIVKGCSTILKGQKRD 390
Query: 416 GLYQLSDT--KGNPYAFVSVKES---WHRRLGHPNNKVLDKVLKSCNVKVPPSDNFSFCE 470
LY L +G ++ VK+ WH RLGH + K ++ ++K ++ FCE
Sbjct: 391 TLYILDGVTEEGESHSSAEVKDETALWHSRLGHMSQKGMEILVKKGCLRREVIKELEFCE 450
Query: 471 ACQYGKMHLLPFKSSSSHAQEPLELVHTDVWG-PAPIMSSSGFKYYVHFIDDFSRFTWIY 529
C YGK H + F + +E L VH+D+WG P S +Y++ F+DD+SR WIY
Sbjct: 451 DCVYGKQHRVSFAPAQHVTKEKLAYVHSDLWGSPHNPASLGNSQYFISFVDDYSRKVWIY 510
Query: 530 PLKQKSETVQAFT-----TENQFNKRIKVIQCDGGGEY--KPVQKLAIDVGIQFRMSCPY 582
L++K E + F ENQ ++++K ++ D G EY +K + GI +C Y
Sbjct: 511 FLRKKDEAFEKFVEWKKMVENQSDRKVKKLRTDNGLEYCNHYFEKFCKEEGIVRHKTCAY 570
Query: 583 TFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYSL 642
T QQNG AER +R I + ++L+++ M +W EA STAVYLINR PS + P
Sbjct: 571 TPQQNGIAERLNRTIMDKVRSMLSRSGMEKKFWAEAASTAVYLINRSPSTAINFDLPEEK 630
Query: 643 IFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKC-LNSHGRTF 701
P+ L+ FGC Y +Q KL + + +F Y KGYK + +
Sbjct: 631 WTGALPDLSSLRKFGCLAYIHA---DQGKLNPRSKKGIFTSYPEGVKGYKVWVLEDKKCV 687
Query: 702 ISRHVIFNEDLFPFHEGFLNTRSPLKTTINNPSTSFPLCSAGNSINDASIPIIEEENQDE 761
ISR+VIF E + F + + + TI+ + + D + N D
Sbjct: 688 ISRNVIFREQVM-----FKDLKGDSQNTISE-----------SDLEDLRV------NPDM 725
Query: 762 TNEE--DSQGVTSDTEQTDNGSSEGDTTHEETL-------DIVQQQNVGESSLDTNTSNA 812
++E D G T D ++ SE T+H L D ++ ++ D +T
Sbjct: 726 NDQEFTDQGGATQD----NSNPSEATTSHNPVLNSPTHSQDEESEEEDSDAVEDLSTYQL 781
Query: 813 IHTRSKSGIH-KPKLP---YIGITETYKDT--VEPANVKEALTRTLWKE---AMQKEFQA 863
+ R + I PK +G +D EP + +EAL W++ AM++E +
Sbjct: 782 VRDRVRRTIKANPKYNESNMVGFAYYSEDDGKPEPKSYQEALLDPDWEKWNAAMKEEMVS 841
Query: 864 LMSNKTWILVPYQDQENIVDSKWVFKTKYKSDG-SIERRKARLVAKGFQQTAGIDYEETF 922
+ N TW LV ++ ++ +WVF K G R ARLVAKGF Q G+DY E F
Sbjct: 842 MSKNHTWDLVTKPEKVKLIGCRWVFTRKAGIPGVEAPRFIARLVAKGFTQKEGVDYNEIF 901
Query: 923 SPVVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICK 982
SPVVK ++R +LS+ VH N E++Q+D+ AFL+G+L+E ++M QPEGF N +C
Sbjct: 902 SPVVKHVSIRYLLSMVVHYNMELQQMDVKTAFLHGFLEEEIYMAQPEGFEIKRGSNKVCL 961
Query: 983 LSKAIYGLKQAPRAWFDSLKTALLNWGFQNTKS--DPSLFLLK-GKDHITFLLIYVDDII 1039
L +++YGLKQ+PR W +L+ G + T+S D ++ K D +LL+YVDD++
Sbjct: 962 LKRSLYGLKQSPRQW--NLRFDEFMRGIKYTRSAYDSCVYFKKCNGDTYIYLLLYVDDML 1019
Query: 1040 VTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEV--QRDASGMYLKQSKYIGDLLKKF 1097
+ ++ + + + L+ F +KDLG LG+E+ RDA + L Q Y+ +L+ F
Sbjct: 1020 IASANKSEVNELKQLLSREFEMKDLGDAKKILGMEISRDRDAGLLTLSQEGYVKKVLRSF 1079
Query: 1098 KMENASPCPTPM--------ITGRHFTVEGEKLKDPTVFRQAIGGLQY-LTHTRPDIAFS 1148
+M+NA P TP+ T + + + E++K + IG + Y + TRPD+A+S
Sbjct: 1080 QMDNAKPVSTPLGIHFKLKAATDKEYEEQFERMK-IVPYANTIGSIMYSMIGTRPDLAYS 1138
Query: 1149 VNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDDRK 1208
+ +S++MS P DHWQ +K +LRY++GT L + D + G+ D+D+ ++ D R+
Sbjct: 1139 LGVISRFMSKPLKDHWQAVKWVLRYMRGTEKKKLCFRKQEDFLLRGYCDSDYGSNFDTRR 1198
Query: 1209 SMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRK 1268
S+ G +G ISW S+ QKVV+ SSTE+EY AL + E W+ EL
Sbjct: 1199 SITGYVFTVGGNTISWKSKLQKVVAISSTEAEYMALTEAVKEALWLKGFAAELGHSQDYV 1258
Query: 1269 PILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKP 1328
+ D+ SA LA N V H R+KHI+I +H+IRD + + V + T A+ TK
Sbjct: 1259 EV-HSDSQSAITLAKNSVHHERTKHIDIRLHFIRDIICAGLIKVVKIATECNPANIFTKT 1317
Query: 1329 LSHTRFSQLRDKLGV 1343
+ +F + L V
Sbjct: 1318 VPLAKFEGALNMLRV 1332
>At1g37110
Length = 1356
Score = 523 bits (1347), Expect = e-148
Identities = 362/1146 (31%), Positives = 571/1146 (49%), Gaps = 81/1146 (7%)
Query: 247 RGGRGRGRS-----SKAPCQVYGKTNHTAINCFHRFDKNYSRSNYSAD--SDKQGSHNAF 299
+GG+G+GRS +K PC K H +C+ R K S A ++K A
Sbjct: 238 KGGQGKGRSRSNSKTKVPCWYCKKEGHVKKDCYSRKKKMESEGQGEAGVITEKLVFSEAL 297
Query: 300 IASQNSVEDYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEIVATGSSKLK 359
++ V+D W DSG ++H+T + D F E G + ++G+ +E G+ ++
Sbjct: 298 SVNEQMVKDL-WILDSGCTSHMTSRRDWFISFQEK-GNTTILLGDDHSVESQGQGTIRID 355
Query: 360 SLN-----LDDVLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVILKGLLK 414
+ L++V YVP++ +NL+S L + + + V+ K L+G L
Sbjct: 356 THGGTIKILENVKYVPHLRRNLISTGTL---DKLGYRHEGGEGKVRYFKNNKTALRGSLS 412
Query: 415 NGLYQLSDTKG-----NPYAFVSVKESWHRRLGHPNNKVLDKVLKSCNVKVPPSDNFSFC 469
NGLY L + N WH RLGH + L + + + FC
Sbjct: 413 NGLYVLDGSTVMSELCNAETDKVKTALWHSRLGHMSMNNLKVLAGKGLIDRKEINELEFC 472
Query: 470 EACQYGKMHLLPFKSSSSHAQEPLELVHTDVWG-PAPIMSSSGFKYYVHFIDDFSRFTWI 528
E C GK + F +++ L VH D+WG P S SG +Y++ IDD +R W+
Sbjct: 473 EHCVMGKSKKVSFNVGKHTSEDALSYVHADLWGSPNVTPSISGKQYFLSIIDDKTRKVWL 532
Query: 529 YPLKQKSETVQAFT-----TENQFNKRIKVIQCDGGGEY--KPVQKLAIDVGIQFRMSCP 581
Y LK K ET F ENQ NK++K ++ D G E+ + GI+ +C
Sbjct: 533 YFLKSKDETFDKFCEWKSLVENQVNKKVKCLRTDNGLEFCNSRFDSYCKEHGIERHRTCT 592
Query: 582 YTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYS 641
YT QQNG AER +R I E LL ++ + +W EA +TA YLINR P+ + P
Sbjct: 593 YTPQQNGVAERMNRTIMEKVRCLLNKSGVEEVFWAEAAATAAYLINRSPASAINHNVPEE 652
Query: 642 LIFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKC-LNSHGRT 700
+ +++P YK L+ FG Y +Q KL+ + FLGY KGYK L +
Sbjct: 653 MWLNRKPGYKHLRKFGSIAYV---HQDQGKLKPRALKGFFLGYPAGTKGYKVWLLEEEKC 709
Query: 701 FISRHVIFNE-----DLFPFHEGFLNTRSPLKTTINNPSTSFPLCSAGNSI---NDASIP 752
ISR+V+F E DL + N T+ F S + S P
Sbjct: 710 VISRNVVFQESVVYRDLKVKEDDTDNLNQKETTSSEVEQNKFAEASGSGGVIQLQSDSEP 769
Query: 753 IIEEENQDETNEEDSQGVTSDTEQTDNGSSEGDTTHEETLDIVQQQNVGESSLDTNTSN- 811
I E E ++ EE + T++T + G TT++ D V++ N+ + T S+
Sbjct: 770 ITEGEQSSDSEEEVEY--SEKTQETPKRT--GLTTYKLARDRVRR-NINPPTRFTEESSV 824
Query: 812 --AIHTRSKSGIHKPKLPYIGITETYKDTVEPANVKEALTRTLWKEAMQKEFQALMSNKT 869
A+ + +P ++Y++ +E + ++ W A E +LM N T
Sbjct: 825 TFALVVVENCIVQEP--------QSYQEAMESQDCEK------WDMATHDEMDSLMKNGT 870
Query: 870 WILVPYQDQENIVDSKWVFKTKYKSDGSIERR-KARLVAKGFQQTAGIDYEETFSPVVKV 928
W LV I+ +W+FK K G R KARLVAKG+ Q G+DY+E F+PVVK
Sbjct: 871 WDLVDKPKDRKIIGCRWLFKLKSGIPGVEPTRFKARLVAKGYTQREGVDYQEIFAPVVKH 930
Query: 929 STVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIY 988
++R+++S+ V + E+ Q+D+ FL+G L+E ++M QPEGFV + N +C+L K++Y
Sbjct: 931 VSIRILMSLVVDKDLELEQMDVKTTFLHGDLEEELYMEQPEGFVSDSSENKVCRLKKSLY 990
Query: 989 GLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLKGKDH-ITFLLIYVDDIIVTGSSNNF 1047
GLKQ+PR W + + F ++ D +++ +H +LL+YVDD+++ G+S
Sbjct: 991 GLKQSPRQWNKRFDRFMSSQQFIRSEHDACVYVKHVSEHDFIYLLLYVDDMLIAGASKAE 1050
Query: 1048 LQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASGMYLKQSK--YIGDLLKKFKMENASPC 1105
+ +QL+ F +KD+G LGI++ RD G LK S+ YI +L +F M A
Sbjct: 1051 INRVKEQLSTEFEMKDMGGASRILGIDIYRDRKGGVLKLSQEIYIRKVLDRFNMSGAKMT 1110
Query: 1106 PTPMITGRHFTVEGEKLKDPTV------FRQAIGGLQY-LTHTRPDIAFSVNKLSQYMSS 1158
P+ G HF + + +D V + A+G + Y + TRPD+A+++ +S+YMS
Sbjct: 1111 NAPV--GAHFKLAAVREEDECVDTDVVPYSSAVGSIMYAMLGTRPDLAYAICLISRYMSK 1168
Query: 1159 PTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDDRKSMAGQCVFLG 1218
P + HW+ +K ++RYL+G + L D +TG+ D+++A +D R+S++G +G
Sbjct: 1169 PGSMHWEAVKWVMRYLKGAQDLNLVFTKEKDFTVTGYCDSNYAADLDRRRSISGYVFTIG 1228
Query: 1219 ETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSA 1278
+SW + Q VV+ S+TE+EY AL + A E WI LL ++ + + I WCD+ SA
Sbjct: 1229 GNTVSWKASLQPVVAMSTTEAEYIALAEAAKEAMWIKGLLQDMGMQQDKVKI-WCDSQSA 1287
Query: 1279 KALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQLR 1338
L+ N V H R+KHI++ +YIRD V V V + T+ D LTK + +F +
Sbjct: 1288 ICLSKNSVYHERTKHIDVRFNYIRDVVESGDVDVLKIHTSRNPVDALTKCIPVNKF---K 1344
Query: 1339 DKLGVI 1344
LGV+
Sbjct: 1345 SALGVL 1350
>At4g27200 putative protein
Length = 819
Score = 523 bits (1346), Expect = e-148
Identities = 291/725 (40%), Positives = 419/725 (57%), Gaps = 51/725 (7%)
Query: 657 GCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKCLNSH-GRTFISRHVIFNEDLFPF 715
G AC+P L+ Y ++K + +CVFLGY+ +KGY+CL GR +ISRHVIF+E ++PF
Sbjct: 15 GSACFPTLRDYAENKFNPCSLKCVFLGYNEKYKGYRCLYPPTGRLYISRHVIFDESVYPF 74
Query: 716 HEGFLNTR----------------SPLKTTINNPSTSFPLCSAGN---------SINDAS 750
+ + SP +T +PS+ PL ++ + +
Sbjct: 75 SHTYKHLHPQPRTPLLAAWLRSSDSPAPSTSTSPSSRSPLFTSADFPPLPQRKTPLLPTL 134
Query: 751 IPIIEEENQDETNEEDSQGVTSDTEQTDNGSSEGDTTHEETLDIVQQQNVGESSLD---T 807
+PI + + S S+ + +S GD++H ++ + ++S++ T
Sbjct: 135 VPISSVSHASNITTQQSPDFDSERTTDFDSASIGDSSHSSQAGSDSEETIQQASVNVHQT 194
Query: 808 NTSNAIH---TRSKSGIHKPKLPYIGITETYKDTVEPANVKEALTRTLWKEAMQKEFQAL 864
S +H TR+K GI KP Y+ ++ EP V AL W AM +E
Sbjct: 195 PASTNVHPMVTRAKVGISKPNPRYVFLSHKVSYP-EPKTVTAALKHPGWTGAMTEEIGNC 253
Query: 865 MSNKTWILVPYQDQENIVDSKWVFKTKYKSDGSIERRKARLVAKGFQQTAGIDYEETFSP 924
+TW LVPY+ +++ SKWVF+TK +DG++ + KAR+VAKGF Q GIDY ET+SP
Sbjct: 254 SETQTWSLVPYKSDMHVLGSKWVFRTKLHADGTLNKLKARIVAKGFLQEEGIDYLETYSP 313
Query: 925 VVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLS 984
VV+ TVR++L +A LNW+++Q+D+ NAFL+G LKETV+M QP D H+C L
Sbjct: 314 VVRTPTVRLVLHLATALNWDIKQMDVKNAFLHGDLKETVYMTQPAANRD-----HVCLLH 368
Query: 985 KAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSS 1044
K+IYGLKQ+PRAWFD T LL +GF KSDPSLF+ +++ LL+
Sbjct: 369 KSIYGLKQSPRAWFDKFSTFLLEFGFFCRKSDPSLFIYAHNNNLILLLL----------- 417
Query: 1045 NNFLQAFIKQLNDVFSLKDLGRLHY-FLGIEVQRDASGMYLKQSKYIGDLLKKFKMENAS 1103
+ L + + LN F + D+G+ FLGI+VQR +G+++ Q KY DLL ME+ +
Sbjct: 418 SQTLTSLLAALNKEFRMTDMGQHSLTFLGIQVQRQQNGLFMSQQKYAEDLLIAASMEHCT 477
Query: 1104 PCPTPMITGRHFTVEGEKL-KDPTVFRQAIGGLQYLTHTRPDIAFSVNKLSQYMSSPTTD 1162
P PTP+ E+L DPT FR G LQYLT TRPDI F+VN + Q M PT
Sbjct: 478 PLPTPLPVQLDRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDIQFAVNFVCQKMHQPTIS 537
Query: 1163 HWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDDRKSMAGQCVFLGETLI 1222
+ +KRILRY++GTI + + + +SD+DW R+S+ G C F+G L+
Sbjct: 538 DFHLLKRILRYIKGTITMGISYSRDSPTLLQAYSDSDWGNCKQTRRSVGGLCTFMGTNLV 597
Query: 1223 SWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSAKALA 1282
SWSS+K VSRSSTE+EY++L D A+EI W+ +LL EL++PLP P L+CDNLSA L
Sbjct: 598 SWSSKKHPTVSRSSTEAEYKSLSDAASEILWLSTLLRELRIPLPDTPELFCDNLSAVYLT 657
Query: 1283 SNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQLRDKLG 1342
+NP HAR+KH +ID H++R++V +VV ++P ++QIAD TK L + F LR KLG
Sbjct: 658 ANPAFHARTKHFDIDFHFVRERVALKALVVKHIPGSEQIADIFTKSLPYEAFIHLRGKLG 717
Query: 1343 VIHSP 1347
V P
Sbjct: 718 VTLPP 722
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,200,559
Number of Sequences: 26719
Number of extensions: 1378724
Number of successful extensions: 5736
Number of sequences better than 10.0: 181
Number of HSP's better than 10.0 without gapping: 131
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 4735
Number of HSP's gapped (non-prelim): 384
length of query: 1349
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1238
effective length of database: 8,352,787
effective search space: 10340750306
effective search space used: 10340750306
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146790.9


