

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146790.4 - phase: 0
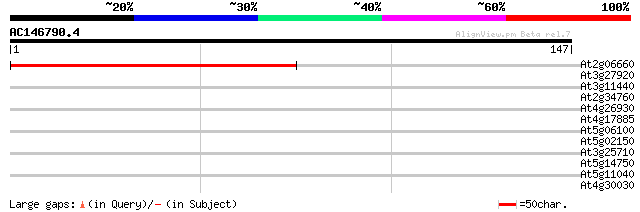
(147 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g06660 putative CENP-B/ARS binding protein-like protein 138 1e-33
At3g27920 MYB family transcription factor (GL1) 31 0.27
At3g11440 transcription factor like protein (MYB65) 28 1.3
At2g34760 unknown protein 28 1.3
At4g26930 putative myb-related protein 27 3.9
At4g17885 unknown protein 27 5.1
At5g06100 transcription factor MYB33 - like protein 26 6.7
At5g02150 unknown protein (At5g02150) 26 6.7
At3g25710 putative bHLH transcription factor (bHLH032) 26 6.7
At5g14750 myb transcription factor werewolf (WER)/ MYB66 26 8.7
At5g11040 putative protein 26 8.7
At4g30030 putative protein 26 8.7
>At2g06660 putative CENP-B/ARS binding protein-like protein
Length = 283
Score = 138 bits (347), Expect = 1e-33
Identities = 63/75 (84%), Positives = 69/75 (92%)
Query: 1 MTSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVR 60
MTSKIQPCDAGIIRAFKMHYRRRFYR+ILEGYE+GQSDPG INVLDAI+ A+ WTI+VR
Sbjct: 208 MTSKIQPCDAGIIRAFKMHYRRRFYREILEGYELGQSDPGTINVLDAISFAVSAWTINVR 267
Query: 61 KETIANCFRHCKIRS 75
+ETI NCFRHCKI S
Sbjct: 268 RETILNCFRHCKIHS 282
>At3g27920 MYB family transcription factor (GL1)
Length = 228
Score = 30.8 bits (68), Expect = 0.27
Identities = 22/80 (27%), Positives = 39/80 (48%), Gaps = 3/80 (3%)
Query: 18 MHYRRRFYRKILEGYEVGQSDPGKINVLD--AINLAIPTWTIDVRKETIANCFRHCKIRS 75
M RRR ++ E Y+ G + N+L +N W VRK + C + C++R
Sbjct: 1 MRIRRRDEKENQE-YKKGLWTVEEDNILMDYVLNHGTGQWNRIVRKTGLKRCGKSCRLRW 59
Query: 76 ASDVVGNLDESTFDEETQDL 95
+ + N+++ F E+ +DL
Sbjct: 60 MNYLSPNVNKGNFTEQEEDL 79
>At3g11440 transcription factor like protein (MYB65)
Length = 553
Score = 28.5 bits (62), Expect = 1.3
Identities = 11/41 (26%), Positives = 22/41 (52%)
Query: 55 WTIDVRKETIANCFRHCKIRSASDVVGNLDESTFDEETQDL 95
W + ++A C + C++R A+ + NL + F +E + L
Sbjct: 66 WNAVQKHTSLARCGKSCRLRWANHLRPNLKKGAFSQEEEQL 106
>At2g34760 unknown protein
Length = 613
Score = 28.5 bits (62), Expect = 1.3
Identities = 15/59 (25%), Positives = 30/59 (50%), Gaps = 6/59 (10%)
Query: 77 SDVVGNLDESTFDEETQDLQTMINQCG------YRNKMDIDNLMNYPGENEACSEVQSL 129
++V+ N DE + +++ DL +N+ ++NKM+ D L+ P + +SL
Sbjct: 555 AEVIVNGDEDSDEDDEADLDYALNKMSITPKHSFKNKMERDRLLRMPSRIRPSTSPESL 613
>At4g26930 putative myb-related protein
Length = 389
Score = 26.9 bits (58), Expect = 3.9
Identities = 12/41 (29%), Positives = 22/41 (53%)
Query: 55 WTIDVRKETIANCFRHCKIRSASDVVGNLDESTFDEETQDL 95
W +K +A C + C++R A+ + NL + +F E + L
Sbjct: 44 WNSVQKKTWLARCGKSCRLRWANHLRPNLRKGSFTPEEERL 84
>At4g17885 unknown protein
Length = 413
Score = 26.6 bits (57), Expect = 5.1
Identities = 17/61 (27%), Positives = 28/61 (45%), Gaps = 3/61 (4%)
Query: 61 KETIANCFRHCKIRSASDVVGNLDESTFDEETQDLQTMINQCGYRNKMDIDNLMNYPGEN 120
K T+ I SA + D+S D DL IN+ ++ + D+ +L+N GE
Sbjct: 337 KATLQKFAGSASISSADFYGHDQDDSNIDITASDL---INRLSFQAQQDLSSLVNIAGET 393
Query: 121 E 121
+
Sbjct: 394 K 394
>At5g06100 transcription factor MYB33 - like protein
Length = 520
Score = 26.2 bits (56), Expect = 6.7
Identities = 10/41 (24%), Positives = 21/41 (50%)
Query: 55 WTIDVRKETIANCFRHCKIRSASDVVGNLDESTFDEETQDL 95
W + ++ C + C++R A+ + NL + F +E + L
Sbjct: 57 WNAVQKHTSLFRCGKSCRLRWANHLRPNLKKGAFSQEEEQL 97
>At5g02150 unknown protein (At5g02150)
Length = 324
Score = 26.2 bits (56), Expect = 6.7
Identities = 20/68 (29%), Positives = 31/68 (45%), Gaps = 6/68 (8%)
Query: 78 DVVGNLDESTFDEETQDLQTMINQCGYRNKMDIDNLMNYPGENEACSEVQSLEDIVCTII 137
D+ G LDE E+ D+ ++ G + L+ Y + A +S D+V TI+
Sbjct: 74 DIEGLLDELQEHVESIDMANDLHSVG-----GLVPLLGYLKNSNANIRAKSA-DVVSTIV 127
Query: 138 ENNAEDDE 145
ENN E
Sbjct: 128 ENNPRSQE 135
>At3g25710 putative bHLH transcription factor (bHLH032)
Length = 344
Score = 26.2 bits (56), Expect = 6.7
Identities = 18/61 (29%), Positives = 28/61 (45%), Gaps = 12/61 (19%)
Query: 89 DEETQDLQTMINQCGYRNKMDIDNLMNYPGENEACSEVQSLEDIVCTIIE----NNAEDD 144
DEE D YR D D++ +Y E + V S+E+ + +IE NN E +
Sbjct: 266 DEEDHD--------SYRRNFDGDDVEDYDEERMMNNRVSSIEEALKAVIEKCVHNNDESN 317
Query: 145 E 145
+
Sbjct: 318 D 318
>At5g14750 myb transcription factor werewolf (WER)/ MYB66
Length = 203
Score = 25.8 bits (55), Expect = 8.7
Identities = 10/41 (24%), Positives = 20/41 (48%)
Query: 55 WTIDVRKETIANCFRHCKIRSASDVVGNLDESTFDEETQDL 95
W +K + C + C++R + + N+ F E+ +DL
Sbjct: 41 WNRIAKKTGLKRCGKSCRLRWMNYLSPNVKRGNFTEQEEDL 81
>At5g11040 putative protein
Length = 1186
Score = 25.8 bits (55), Expect = 8.7
Identities = 11/33 (33%), Positives = 21/33 (63%)
Query: 27 KILEGYEVGQSDPGKINVLDAINLAIPTWTIDV 59
KI ++ G++ G++++ DAI A+ T +DV
Sbjct: 999 KIKVRWQSGRNSSGELDIKDAIQTALQTTVMDV 1031
>At4g30030 putative protein
Length = 424
Score = 25.8 bits (55), Expect = 8.7
Identities = 17/52 (32%), Positives = 21/52 (39%), Gaps = 5/52 (9%)
Query: 60 RKETIANCFRHCKIRSASDVVGNLDESTFDEETQD-----LQTMINQCGYRN 106
R E NC H + R S+ G L E ET D Q ++ CG N
Sbjct: 144 RDEKTGNCQYHLRYRDFSNTRGILAEEKLTFETSDDGLISKQNIVFGCGQDN 195
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,283,730
Number of Sequences: 26719
Number of extensions: 128151
Number of successful extensions: 326
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 319
Number of HSP's gapped (non-prelim): 12
length of query: 147
length of database: 11,318,596
effective HSP length: 90
effective length of query: 57
effective length of database: 8,913,886
effective search space: 508091502
effective search space used: 508091502
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146790.4


