

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146790.2 + phase: 0
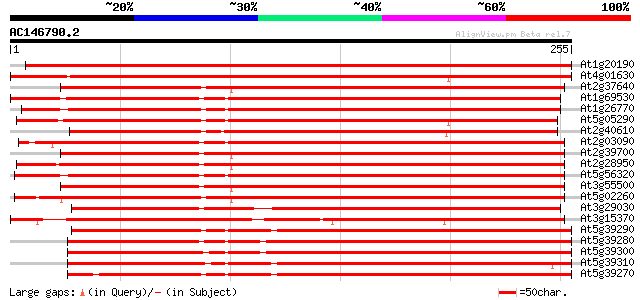
(255 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20190 expansin S2 precursor like protein 407 e-114
At4g01630 putative expansin 347 3e-96
At2g37640 putative expansin 337 4e-93
At1g69530 expansin-like protein 337 4e-93
At1g26770 expansin 10 336 6e-93
At5g05290 expansin At-EXP2 (gb|AAB38073.1) 335 1e-92
At2g40610 putative expansin 335 2e-92
At2g03090 expansin like protein 328 2e-90
At2g39700 putative expansin 327 5e-90
At2g28950 putative expansin 323 4e-89
At5g56320 expansin 322 9e-89
At3g55500 expansin-like protein 320 4e-88
At5g02260 expansin precursor - like protein 319 8e-88
At3g29030 expansin At-EXP5 309 1e-84
At3g15370 putative expansin S2 precursor 276 6e-75
At5g39290 expansin-like protein 275 2e-74
At5g39280 expansin-like protein 272 1e-73
At5g39300 expansin-like protein 271 2e-73
At5g39310 expansin-like protein 271 3e-73
At5g39270 expansin-like protein 267 5e-72
>At1g20190 expansin S2 precursor like protein
Length = 252
Score = 407 bits (1047), Expect = e-114
Identities = 186/248 (75%), Positives = 209/248 (84%)
Query: 8 LGTLIGLCCFTINSYAFKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAAL 67
L L L I AF+ SG TN HATFYGGSDA GTMGGACGYG+LYS GYGT TAAL
Sbjct: 5 LAGLAVLAALFIAVDAFRPSGLTNGHATFYGGSDASGTMGGACGYGDLYSAGYGTMTAAL 64
Query: 68 STALFNDGASCGQCYKIICDYKTDPRWCIKGRSITITATNFCPPNFDLPNDDGGWCNPPL 127
STALFNDGASCG+CY+I CD+ D RWC+KG S+ ITATNFCPPNF LPN++GGWCNPPL
Sbjct: 65 STALFNDGASCGECYRITCDHAADSRWCLKGASVVITATNFCPPNFALPNNNGGWCNPPL 124
Query: 128 KHFDMAQPAWEKIGIYRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAGSI 187
KHFDMAQPAWEKIGIYRGGIVPV+FQRV C K GGVRF +NGRDYFELV I N+ GAGSI
Sbjct: 125 KHFDMAQPAWEKIGIYRGGIVPVVFQRVSCYKKGGVRFRINGRDYFELVNIQNVGGAGSI 184
Query: 188 QSVSIKGSKTDWMAMSRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGFGQ 247
+SVSIKGSKT W+AMSRNWGANWQSNAYL+GQ++SF +TTTDG TR F ++VPS+W FGQ
Sbjct: 185 KSVSIKGSKTGWLAMSRNWGANWQSNAYLDGQALSFSITTTDGATRVFLNVVPSSWSFGQ 244
Query: 248 SFSSKIQF 255
+SS +QF
Sbjct: 245 IYSSNVQF 252
>At4g01630 putative expansin
Length = 255
Score = 347 bits (891), Expect = 3e-96
Identities = 162/256 (63%), Positives = 197/256 (76%), Gaps = 2/256 (0%)
Query: 1 MTKVMFILGTLIGLCCFTINSYAFKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGY 60
MTK+ +L +I F + + A GW AHATFYGGSDA GTMGGACGYGNLY+ GY
Sbjct: 1 MTKIFSLLVAMIFSTMFFMKISSVSA-GWLQAHATFYGGSDASGTMGGACGYGNLYTDGY 59
Query: 61 GTRTAALSTALFNDGASCGQCYKIICDYKTDPRWCIKGRSITITATNFCPPNFDLPNDDG 120
T TAALSTALFNDG SCG CY+I+CD P+WC+KG+SITITATNFCPPNF +D+G
Sbjct: 60 KTNTAALSTALFNDGKSCGGCYQILCDATKVPQWCLKGKSITITATNFCPPNFAQASDNG 119
Query: 121 GWCNPPLKHFDMAQPAWEKIGIYRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISN 180
GWCNPP HFDMAQPA+ I Y+ GIVP+L+++V C++ GG+RF++NGR+YFELVLISN
Sbjct: 120 GWCNPPRPHFDMAQPAFLTIAKYKAGIVPILYKKVGCRRSGGMRFTINGRNYFELVLISN 179
Query: 181 LAGAGSIQSVSIKGSKTD-WMAMSRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIV 239
+AG G I V IKGSK++ W MSRNWGAN+QSN YLNGQS+SFKV +DG + ++V
Sbjct: 180 VAGGGEISKVWIKGSKSNKWETMSRNWGANYQSNTYLNGQSLSFKVQLSDGSIKAALNVV 239
Query: 240 PSNWGFGQSFSSKIQF 255
PSNW FGQSF S + F
Sbjct: 240 PSNWRFGQSFKSNVNF 255
>At2g37640 putative expansin
Length = 262
Score = 337 bits (864), Expect = 4e-93
Identities = 153/230 (66%), Positives = 182/230 (78%), Gaps = 3/230 (1%)
Query: 24 FKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAALSTALFNDGASCGQCYK 83
+ W NAHATFYGGSDA GTMGGACGYGNLYSQGYG TAALSTALFN+G SCG C++
Sbjct: 31 YSGGPWQNAHATFYGGSDASGTMGGACGYGNLYSQGYGVNTAALSTALFNNGFSCGACFE 90
Query: 84 IICDYKTDPRWCIKGR-SITITATNFCPPNFDLPNDDGGWCNPPLKHFDMAQPAWEKIGI 142
I C DPRWC+ G SI +TATNFCPPNF P+DDGGWCNPP +HFD+A P + KIG+
Sbjct: 91 IKCT--DDPRWCVPGNPSILVTATNFCPPNFAQPSDDGGWCNPPREHFDLAMPMFLKIGL 148
Query: 143 YRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAGSIQSVSIKGSKTDWMAM 202
YR GIVPV ++RVPC+K GG+RF+VNG YF LVL++N+AGAG I VS+KGSKTDW+ M
Sbjct: 149 YRAGIVPVSYRRVPCRKIGGIRFTVNGFRYFNLVLVTNVAGAGDINGVSVKGSKTDWVRM 208
Query: 203 SRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGFGQSFSSK 252
SRNWG NWQSNA L GQS+SF+VT +D + T ++ P+ W FGQ+FS K
Sbjct: 209 SRNWGQNWQSNAVLIGQSLSFRVTASDRRSSTSWNVAPATWQFGQTFSGK 258
>At1g69530 expansin-like protein
Length = 250
Score = 337 bits (864), Expect = 4e-93
Identities = 152/250 (60%), Positives = 188/250 (74%), Gaps = 5/250 (2%)
Query: 1 MTKVMFILGTLIGLCCFTINSYAFKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGY 60
M V F+ +G +N YA GW NAHATFYGG DA GTMGGACGYGNLYSQGY
Sbjct: 1 MALVTFLFIATLGAMTSHVNGYA--GGGWVNAHATFYGGGDASGTMGGACGYGNLYSQGY 58
Query: 61 GTRTAALSTALFNDGASCGQCYKIICDYKTDPRWCIKGRSITITATNFCPPNFDLPNDDG 120
GT TAALSTALFN+G SCG C++I C + D +WC+ G SI +TATNFCPPN LPN+ G
Sbjct: 59 GTNTAALSTALFNNGLSCGACFEIRC--QNDGKWCLPG-SIVVTATNFCPPNNALPNNAG 115
Query: 121 GWCNPPLKHFDMAQPAWEKIGIYRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISN 180
GWCNPP +HFD++QP +++I YR GIVPV ++RVPC + GG+RF++NG YF LVLI+N
Sbjct: 116 GWCNPPQQHFDLSQPVFQRIAQYRAGIVPVAYRRVPCVRRGGIRFTINGHSYFNLVLITN 175
Query: 181 LAGAGSIQSVSIKGSKTDWMAMSRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVP 240
+ GAG + S +KGS+T W AMSRNWG NWQSN+YLNGQS+SFKVTT+DG T ++
Sbjct: 176 VGGAGDVHSAMVKGSRTGWQAMSRNWGQNWQSNSYLNGQSLSFKVTTSDGQTIVSNNVAN 235
Query: 241 SNWGFGQSFS 250
+ W FGQ+F+
Sbjct: 236 AGWSFGQTFT 245
>At1g26770 expansin 10
Length = 249
Score = 336 bits (862), Expect = 6e-93
Identities = 149/245 (60%), Positives = 192/245 (77%), Gaps = 6/245 (2%)
Query: 6 FILGTLIGLCCFTINSYAFKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTA 65
F++ ++G+ +++ Y GW NAHATFYGG DA GTMGGACGYGNLYSQGYGT TA
Sbjct: 6 FLVMIMVGVMASSVSGYG---GGWINAHATFYGGGDASGTMGGACGYGNLYSQGYGTSTA 62
Query: 66 ALSTALFNDGASCGQCYKIICDYKTDPRWCIKGRSITITATNFCPPNFDLPNDDGGWCNP 125
ALSTALFN+G SCG C++I C+ D +WC+ G SI +TATNFCPPN L N++GGWCNP
Sbjct: 63 ALSTALFNNGLSCGSCFEIRCE--NDGKWCLPG-SIVVTATNFCPPNNALANNNGGWCNP 119
Query: 126 PLKHFDMAQPAWEKIGIYRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAG 185
PL+HFD+AQP +++I YR GIVPV ++RVPC++ GG+RF++NG YF LVLI+N+ GAG
Sbjct: 120 PLEHFDLAQPVFQRIAQYRAGIVPVSYRRVPCRRRGGIRFTINGHSYFNLVLITNVGGAG 179
Query: 186 SIQSVSIKGSKTDWMAMSRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGF 245
+ S +IKGS+T W AMSRNWG NWQSN+YLNGQ++SFKVTT+DG T + P+ W +
Sbjct: 180 DVHSAAIKGSRTVWQAMSRNWGQNWQSNSYLNGQALSFKVTTSDGRTVVSFNAAPAGWSY 239
Query: 246 GQSFS 250
GQ+F+
Sbjct: 240 GQTFA 244
>At5g05290 expansin At-EXP2 (gb|AAB38073.1)
Length = 255
Score = 335 bits (860), Expect = 1e-92
Identities = 154/247 (62%), Positives = 192/247 (77%), Gaps = 6/247 (2%)
Query: 4 VMFILGTLIGLCCFTINSYAFKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTR 63
++F+ + C ++INS GW HATFYGG+DA GTMGGACGYGNL+SQGYG +
Sbjct: 9 ILFLSLCTLNFCLYSINSD--DNGGWERGHATFYGGADASGTMGGACGYGNLHSQGYGLQ 66
Query: 64 TAALSTALFNDGASCGQCYKIICDYKTDPRWCIKGRSITITATNFCPPNFDLPNDDGGWC 123
TAALSTALFN G CG C+++ C+ DP WCI G SI ++ATNFCPPNF L ND+GGWC
Sbjct: 67 TAALSTALFNSGQKCGACFELQCE--DDPEWCIPG-SIIVSATNFCPPNFALANDNGGWC 123
Query: 124 NPPLKHFDMAQPAWEKIGIYRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAG 183
NPPLKHFD+A+PA+ +I YR GIVPV F+RVPC+K GG+RF++NG YF+LVLI+N+ G
Sbjct: 124 NPPLKHFDLAEPAFLQIAQYRAGIVPVAFRRVPCEKGGGIRFTINGNPYFDLVLITNVGG 183
Query: 184 AGSIQSVSIKGSKTD-WMAMSRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSN 242
AG I++VS+KGSKTD W +MSRNWG NWQSN YL GQS+SF+VT +DG T D+VP +
Sbjct: 184 AGDIRAVSLKGSKTDQWQSMSRNWGQNWQSNTYLRGQSLSFQVTDSDGRTVVSYDVVPHD 243
Query: 243 WGFGQSF 249
W FGQ+F
Sbjct: 244 WQFGQTF 250
>At2g40610 putative expansin
Length = 253
Score = 335 bits (858), Expect = 2e-92
Identities = 151/223 (67%), Positives = 180/223 (80%), Gaps = 4/223 (1%)
Query: 28 GWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAALSTALFNDGASCGQCYKIICD 87
GW HATFYGG DA GTMGGACGYGNLY QGYGT TAALSTALFN+G +CG CY++ C+
Sbjct: 29 GWQGGHATFYGGEDASGTMGGACGYGNLYGQGYGTNTAALSTALFNNGLTCGACYEMKCN 88
Query: 88 YKTDPRWCIKGRSITITATNFCPPNFDLPNDDGGWCNPPLKHFDMAQPAWEKIGIYRGGI 147
DPRWC+ G +IT+TATNFCPPN L ND+GGWCNPPL+HFD+A+PA+ +I YR GI
Sbjct: 89 --DDPRWCL-GSTITVTATNFCPPNPGLSNDNGGWCNPPLQHFDLAEPAFLQIAQYRAGI 145
Query: 148 VPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAGSIQSVSIKGSKT-DWMAMSRNW 206
VPV F+RVPC K GG+RF++NG YF LVLISN+ GAG + +VSIKGSKT W AMSRNW
Sbjct: 146 VPVSFRRVPCMKKGGIRFTINGHSYFNLVLISNVGGAGDVHAVSIKGSKTQSWQAMSRNW 205
Query: 207 GANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGFGQSF 249
G NWQSN+Y+N QS+SF+VTT+DG T D+ PSNW FGQ++
Sbjct: 206 GQNWQSNSYMNDQSLSFQVTTSDGRTLVSNDVAPSNWQFGQTY 248
>At2g03090 expansin like protein
Length = 253
Score = 328 bits (841), Expect = 2e-90
Identities = 150/250 (60%), Positives = 191/250 (76%), Gaps = 7/250 (2%)
Query: 5 MFILGTLIGLCCFT--INSYAFKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGT 62
M +LG I L CF + S +GW NAHATFYGGSDA GTMGGACGYGNLYSQGYGT
Sbjct: 6 MGLLG--IALFCFAAMVCSVHGYDAGWVNAHATFYGGSDASGTMGGACGYGNLYSQGYGT 63
Query: 63 RTAALSTALFNDGASCGQCYKIICDYKTDPRWCIKGRSITITATNFCPPNFDLPNDDGGW 122
TAALSTALFN+G SCG C++I C ++D WC+ G +I +TATNFCPPN LPN+ GGW
Sbjct: 64 NTAALSTALFNNGLSCGACFEIKC--QSDGAWCLPG-AIIVTATNFCPPNNALPNNAGGW 120
Query: 123 CNPPLKHFDMAQPAWEKIGIYRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLA 182
CNPPL HFD++QP +++I Y+ G+VPV ++RVPC + GG+RF++NG YF LVL++N+
Sbjct: 121 CNPPLHHFDLSQPVFQRIAQYKAGVVPVSYRRVPCMRRGGIRFTINGHSYFNLVLVTNVG 180
Query: 183 GAGSIQSVSIKGSKTDWMAMSRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSN 242
GAG + SV++KGS+T W MSRNWG NWQSN LNGQ++SFKVT +DG T +I P++
Sbjct: 181 GAGDVHSVAVKGSRTRWQQMSRNWGQNWQSNNLLNGQALSFKVTASDGRTVVSNNIAPAS 240
Query: 243 WGFGQSFSSK 252
W FGQ+F+ +
Sbjct: 241 WSFGQTFTGR 250
>At2g39700 putative expansin
Length = 257
Score = 327 bits (837), Expect = 5e-90
Identities = 149/230 (64%), Positives = 179/230 (77%), Gaps = 3/230 (1%)
Query: 24 FKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAALSTALFNDGASCGQCYK 83
+ W NAHATFYGGSDA GTMGGACGYGNLYSQGYGT TAALSTALFN+G SCG C++
Sbjct: 26 YSGGAWQNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGMSCGACFE 85
Query: 84 IICDYKTDPRWCIKGR-SITITATNFCPPNFDLPNDDGGWCNPPLKHFDMAQPAWEKIGI 142
+ C DP+WC G SI ITATNFCPPN P+D+GGWCNPP +HFD+A P + KI
Sbjct: 86 LKC--ANDPQWCHSGSPSILITATNFCPPNLAQPSDNGGWCNPPREHFDLAMPVFLKIAQ 143
Query: 143 YRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAGSIQSVSIKGSKTDWMAM 202
YR GIVPV ++RVPC+K GG+RF++NG YF LVLI+N+AGAG I S+KGS+T WM++
Sbjct: 144 YRAGIVPVSYRRVPCRKRGGIRFTINGHRYFNLVLITNVAGAGDIVRASVKGSRTGWMSL 203
Query: 203 SRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGFGQSFSSK 252
SRNWG NWQSNA L GQ++SF+VT +D T T ++VPSNW FGQ+F K
Sbjct: 204 SRNWGQNWQSNAVLVGQALSFRVTGSDRRTSTSWNMVPSNWQFGQTFVGK 253
>At2g28950 putative expansin
Length = 257
Score = 323 bits (829), Expect = 4e-89
Identities = 151/250 (60%), Positives = 187/250 (74%), Gaps = 4/250 (1%)
Query: 4 VMFILGTLIGLCCFTINSYAFKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTR 63
V+ +L T++ L I + GW AHATFYGGSDA GTMGGACGYGNLYSQGYG
Sbjct: 7 VLSVLTTILALSEARIPG-VYNGGGWETAHATFYGGSDASGTMGGACGYGNLYSQGYGVN 65
Query: 64 TAALSTALFNDGASCGQCYKIICDYKTDPRWCIKGR-SITITATNFCPPNFDLPNDDGGW 122
TAALSTALFN+G SCG C+++ C +DP+WC G SI ITATNFCPPNF P+D+GGW
Sbjct: 66 TAALSTALFNNGFSCGACFELKC--ASDPKWCHSGSPSIFITATNFCPPNFAQPSDNGGW 123
Query: 123 CNPPLKHFDMAQPAWEKIGIYRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLA 182
CNPP HFD+A P + KI YR GIVPV F+RVPC+K GG+RF++NG YF LVL++N+A
Sbjct: 124 CNPPRPHFDLAMPMFLKIAEYRAGIVPVSFRRVPCRKRGGIRFTINGFRYFNLVLVTNVA 183
Query: 183 GAGSIQSVSIKGSKTDWMAMSRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSN 242
GAG+I + +KG+ T WM MSRNWG NWQSN+ L GQS+SF+VT++D + T +I P+N
Sbjct: 184 GAGNIVRLGVKGTHTSWMTMSRNWGQNWQSNSVLVGQSLSFRVTSSDRRSSTSWNIAPAN 243
Query: 243 WGFGQSFSSK 252
W FGQ+F K
Sbjct: 244 WKFGQTFMGK 253
>At5g56320 expansin
Length = 255
Score = 322 bits (826), Expect = 9e-89
Identities = 148/250 (59%), Positives = 189/250 (75%), Gaps = 6/250 (2%)
Query: 3 KVMFILGTLIGLCCFTINSYAFKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGT 62
K++ L ++ + +++ Y+ SGW NA ATFYGG+DA GTMGGACGYGNLYSQGYGT
Sbjct: 6 KMIISLSLMMMIMWKSVDGYS---SGWVNARATFYGGADASGTMGGACGYGNLYSQGYGT 62
Query: 63 RTAALSTALFNDGASCGQCYKIICDYKTDPRWCIKGRSITITATNFCPPNFDLPNDDGGW 122
TAALSTALFN G SCG C++I C DP+WCI G +IT+T TNFCPPNF N+ GGW
Sbjct: 63 NTAALSTALFNGGQSCGACFQIKC--VDDPKWCIGG-TITVTGTNFCPPNFAQANNAGGW 119
Query: 123 CNPPLKHFDMAQPAWEKIGIYRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLA 182
CNPP HFD+AQP + +I Y+ G+VPV ++RV C++ GG+RF++NG YF LVLI+N+A
Sbjct: 120 CNPPQHHFDLAQPIFLRIAQYKAGVVPVQYRRVACRRKGGIRFTINGHSYFNLVLITNVA 179
Query: 183 GAGSIQSVSIKGSKTDWMAMSRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSN 242
GAG + SVSIKG+ T W +MSRNWG NWQSNA L+GQ++SFKVTT+DG T + P N
Sbjct: 180 GAGDVISVSIKGTNTRWQSMSRNWGQNWQSNAKLDGQALSFKVTTSDGRTVISNNATPRN 239
Query: 243 WGFGQSFSSK 252
W FGQ+++ K
Sbjct: 240 WSFGQTYTGK 249
>At3g55500 expansin-like protein
Length = 260
Score = 320 bits (821), Expect = 4e-88
Identities = 145/230 (63%), Positives = 175/230 (76%), Gaps = 3/230 (1%)
Query: 24 FKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAALSTALFNDGASCGQCYK 83
F W AHATFYGG+DA GTMGGACGYGNLYSQGYGT TAALST+LFN G SCG C++
Sbjct: 29 FSGGSWQTAHATFYGGNDASGTMGGACGYGNLYSQGYGTNTAALSTSLFNSGQSCGACFE 88
Query: 84 IICDYKTDPRWCIKGR-SITITATNFCPPNFDLPNDDGGWCNPPLKHFDMAQPAWEKIGI 142
I C DP+WC G S+ +TATNFCPPN P+D+GGWCNPP HFD+A P + KI
Sbjct: 89 IKC--VNDPKWCHPGNPSVFVTATNFCPPNLAQPSDNGGWCNPPRSHFDLAMPVFLKIAE 146
Query: 143 YRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAGSIQSVSIKGSKTDWMAM 202
YR GIVP+ ++RV C+K GG+RF++NG YF LVLI+N+AGAG I S+KGSKT WM++
Sbjct: 147 YRAGIVPISYRRVACRKSGGIRFTINGHRYFNLVLITNVAGAGDIARTSVKGSKTGWMSL 206
Query: 203 SRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGFGQSFSSK 252
+RNWG NWQSNA L GQS+SF+VT++D T T +I PSNW FGQ+F K
Sbjct: 207 TRNWGQNWQSNAVLVGQSLSFRVTSSDRRTSTSWNIAPSNWQFGQTFVGK 256
>At5g02260 expansin precursor - like protein
Length = 258
Score = 319 bits (818), Expect = 8e-88
Identities = 147/254 (57%), Positives = 188/254 (73%), Gaps = 7/254 (2%)
Query: 3 KVMFILGTLIGLCCFTINSY---AFKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQG 59
KV+ + ++ + FT N+ + W NAHATFYG +DA GTMGGACGYGNLYSQG
Sbjct: 4 KVITFMAVMV-VTAFTANAKIPGVYTGGPWINAHATFYGEADASGTMGGACGYGNLYSQG 62
Query: 60 YGTRTAALSTALFNDGASCGQCYKIICDYKTDPRWCIKGR-SITITATNFCPPNFDLPND 118
YG TAALSTALFN+G SCG C+++ C DP WC+ G SI ITATNFCPPNF+ +D
Sbjct: 63 YGVNTAALSTALFNNGLSCGSCFELKCI--NDPGWCLPGNPSILITATNFCPPNFNQASD 120
Query: 119 DGGWCNPPLKHFDMAQPAWEKIGIYRGGIVPVLFQRVPCKKHGGVRFSVNGRDYFELVLI 178
+GGWCNPP +HFD+A P + I Y+ GIVPV ++R+PC+K GG+RF++NG YF LVL+
Sbjct: 121 NGGWCNPPREHFDLAMPMFLSIAKYKAGIVPVSYRRIPCRKKGGIRFTINGFKYFNLVLV 180
Query: 179 SNLAGAGSIQSVSIKGSKTDWMAMSRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQDI 238
+N+AGAG + VS+KGS T W+ +SRNWG NWQSNA L GQS+SF+V T+DG + T +I
Sbjct: 181 TNVAGAGDVIKVSVKGSNTQWLDLSRNWGQNWQSNALLVGQSLSFRVKTSDGRSSTSNNI 240
Query: 239 VPSNWGFGQSFSSK 252
PSNW FGQ++S K
Sbjct: 241 APSNWQFGQTYSGK 254
>At3g29030 expansin At-EXP5
Length = 255
Score = 309 bits (791), Expect = 1e-84
Identities = 136/222 (61%), Positives = 172/222 (77%), Gaps = 10/222 (4%)
Query: 29 WTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAALSTALFNDGASCGQCYKIICDY 88
W NAHATFYGG DA GTMGGACGYGNLYSQGYG TAALSTALF+ G SCG C++++C
Sbjct: 38 WINAHATFYGGGDASGTMGGACGYGNLYSQGYGLETAALSTALFDQGLSCGACFELMC-- 95
Query: 89 KTDPRWCIKGRSITITATNFCPPNFDLPNDDGGWCNPPLKHFDMAQPAWEKIGIYRGGIV 148
DP+WCIKGRSI +TATNFCPP GG C+PP HFD++QP +EKI +Y+ GI+
Sbjct: 96 VNDPQWCIKGRSIVVTATNFCPP--------GGACDPPNHHFDLSQPIYEKIALYKSGII 147
Query: 149 PVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAGSIQSVSIKGSKTDWMAMSRNWGA 208
PV+++RV CK+ GG+RF++NG YF LVL++N+ GAG + SVS+KGS+T W MSRNWG
Sbjct: 148 PVMYRRVRCKRSGGIRFTINGHSYFNLVLVTNVGGAGDVHSVSMKGSRTKWQLMSRNWGQ 207
Query: 209 NWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGFGQSFS 250
NWQSN+YLNGQS+SF VTT+D + ++ P W FGQ+++
Sbjct: 208 NWQSNSYLNGQSLSFVVTTSDRRSVVSFNVAPPTWSFGQTYT 249
>At3g15370 putative expansin S2 precursor
Length = 252
Score = 276 bits (707), Expect = 6e-75
Identities = 129/256 (50%), Positives = 181/256 (70%), Gaps = 20/256 (7%)
Query: 1 MTKVMFILGTL-IGLCCFTINSYAFKASGWTNAHATFYGGSDAEGTMGGACGYGNLYSQG 59
+ V+ ++ TL +G+C ++GW AHAT+YG +D+ ++GGACGY N Y G
Sbjct: 8 LVTVILLVSTLSVGMC----------SNGWIRAHATYYGVNDSPASLGGACGYDNPYHAG 57
Query: 60 YGTRTAALSTALFNDGASCGQCYKIICDYKTDPRWCIKGRSITITATNFCPPNFDLPNDD 119
+G TAALS LF G SCG CY++ CD+ DP+WC++G ++T+TATNFCP N++
Sbjct: 58 FGAHTAALSGELFRSGESCGGCYQVRCDFPADPKWCLRGAAVTVTATNFCP-----TNNN 112
Query: 120 GGWCNPPLKHFDMAQPAWEKIGIYRG--GIVPVLFQRVPCKKHGGVRFSVNGRDYFELVL 177
GWCN P HFDM+ PA+ +I RG GIVPV ++RV CK+ GGVRF++ G+ F +V+
Sbjct: 113 NGWCNLPRHHFDMSSPAFFRIA-RRGNEGIVPVFYRRVGCKRRGGVRFTMRGQGNFNMVM 171
Query: 178 ISNLAGAGSIQSVSIKGSK-TDWMAMSRNWGANWQSNAYLNGQSMSFKVTTTDGVTRTFQ 236
ISN+ G GS++SV+++GSK W+ M+RNWGANWQS+ L GQ +SFKVT TD T+TF
Sbjct: 172 ISNVGGGGSVRSVAVRGSKGKTWLQMTRNWGANWQSSGDLRGQRLSFKVTLTDSKTQTFL 231
Query: 237 DIVPSNWGFGQSFSSK 252
++VPS+W FGQ+FSS+
Sbjct: 232 NVVPSSWWFGQTFSSR 247
>At5g39290 expansin-like protein
Length = 263
Score = 275 bits (702), Expect = 2e-74
Identities = 124/227 (54%), Positives = 159/227 (69%), Gaps = 5/227 (2%)
Query: 29 WTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAALSTALFNDGASCGQCYKIICDY 88
W +A ATFYG T GACGYGNL+ QGYG TAALSTALFNDG +CG CY+I+C
Sbjct: 42 WYDARATFYGDIHGGDTQQGACGYGNLFRQGYGLATAALSTALFNDGYTCGACYEIMCT- 100
Query: 89 KTDPRWCIKGRSITITATNFCPPNFDLPNDDGGWCNPPLKHFDMAQPAWEKIGIYRGGIV 148
DP+WC+ G S+ ITATNFCP N+ D WCNPP KHFD++ + KI Y+ G+V
Sbjct: 101 -RDPQWCLPG-SVKITATNFCPANYSKTTDL--WCNPPQKHFDLSLAMFLKIAKYKAGVV 156
Query: 149 PVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAGSIQSVSIKGSKTDWMAMSRNWGA 208
PV ++R+PC K GGV+F G YF +VLI N+ GAG I+ V +K +KT W+ M +NWG
Sbjct: 157 PVRYRRIPCSKTGGVKFETKGNPYFLMVLIYNVGGAGDIKYVQVKENKTGWITMKKNWGQ 216
Query: 209 NWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGFGQSFSSKIQF 255
NW ++ L GQ +SF+VTTTDG+T+ F +++P NWGFGQ+F KI F
Sbjct: 217 NWTTSTVLTGQGLSFRVTTTDGITKDFWNVMPKNWGFGQTFDGKINF 263
>At5g39280 expansin-like protein
Length = 259
Score = 272 bits (695), Expect = 1e-73
Identities = 120/229 (52%), Positives = 161/229 (69%), Gaps = 5/229 (2%)
Query: 27 SGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAALSTALFNDGASCGQCYKIIC 86
S W +A ATFYG T GACGYG+L+ QGYG TAALSTALFN+G +CG CY+I+C
Sbjct: 36 SSWYDARATFYGDIHGGETQQGACGYGDLFKQGYGLETAALSTALFNEGYTCGACYQIMC 95
Query: 87 DYKTDPRWCIKGRSITITATNFCPPNFDLPNDDGGWCNPPLKHFDMAQPAWEKIGIYRGG 146
DP+WC+ G S+ ITATNFCPP++ +G WCNPP KHFD++ P + KI Y+ G
Sbjct: 96 --VNDPQWCLPG-SVKITATNFCPPDYS--KTEGVWCNPPQKHFDLSLPMFLKIAQYKAG 150
Query: 147 IVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAGSIQSVSIKGSKTDWMAMSRNW 206
+VPV ++R+ C + GGV+F G YF ++L N+ GAG I+ + +KG KT W+ M +NW
Sbjct: 151 VVPVKYRRISCARTGGVKFETKGNPYFLMILPYNVGGAGDIKLMQVKGDKTGWITMQKNW 210
Query: 207 GANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGFGQSFSSKIQF 255
G NW + L GQ +SF+VTT+DGVT+ F +++P+NWGFGQ+F KI F
Sbjct: 211 GQNWTTGVNLTGQGISFRVTTSDGVTKDFNNVMPNNWGFGQTFDGKINF 259
>At5g39300 expansin-like protein
Length = 260
Score = 271 bits (694), Expect = 2e-73
Identities = 120/229 (52%), Positives = 162/229 (70%), Gaps = 5/229 (2%)
Query: 27 SGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAALSTALFNDGASCGQCYKIIC 86
S W +A ATFYG T GACGYG+L+ QGYG TAALSTALFN+G +CG CY+I+C
Sbjct: 37 SSWYDARATFYGDIHGGETQQGACGYGDLFKQGYGLETAALSTALFNEGYTCGACYQIMC 96
Query: 87 DYKTDPRWCIKGRSITITATNFCPPNFDLPNDDGGWCNPPLKHFDMAQPAWEKIGIYRGG 146
+ DP+WC+ G +I ITATNFCPP++ +G WCNPP KHFD++ P + KI Y+ G
Sbjct: 97 VH--DPQWCLPG-TIKITATNFCPPDYS--KTEGVWCNPPQKHFDLSLPMFLKIAQYKAG 151
Query: 147 IVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAGSIQSVSIKGSKTDWMAMSRNW 206
+VPV ++R+ C + GGV+F G YF ++L N+ GAG I+ + +KG KT W+ M +NW
Sbjct: 152 VVPVKYRRISCARTGGVKFETKGNPYFLMILPYNVGGAGDIKLMQVKGDKTGWITMQKNW 211
Query: 207 GANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGFGQSFSSKIQF 255
G NW + L GQ +SF+VTT+DGVT+ F +++P+NWGFGQ+F KI F
Sbjct: 212 GQNWTTGVNLTGQGISFRVTTSDGVTKDFNNVMPNNWGFGQTFDGKINF 260
>At5g39310 expansin-like protein
Length = 296
Score = 271 bits (692), Expect = 3e-73
Identities = 122/230 (53%), Positives = 166/230 (72%), Gaps = 6/230 (2%)
Query: 27 SGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAALSTALFNDGASCGQCYKIIC 86
SGW + ATFYG + T GACGYG+L+ QGYG TAALSTALFN+G+ CG CY+I+C
Sbjct: 72 SGWGHGRATFYGDINGGETQQGACGYGDLHKQGYGLETAALSTALFNNGSRCGACYEIMC 131
Query: 87 DYKTDPRWCIKGRSITITATNFCPPNFDLPNDDGGWCNPPLKHFDMAQPAWEKIGIYRGG 146
++ P+WC+ G SI ITATNFCPP+F PND+ WCNPP KHFD++QP + KI Y+ G
Sbjct: 132 EHA--PQWCLPG-SIKITATNFCPPDFTKPNDN--WCNPPQKHFDLSQPMFLKIAKYKAG 186
Query: 147 IVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAGSIQSVSIKGSKTDWMAMSRNW 206
+VPV F+RVPC K GGV+F + G +F ++L N+ GAG+++++ IKG++T W+AM +NW
Sbjct: 187 VVPVKFRRVPCAKIGGVKFEIKGNPHFLMILPYNVGGAGAVRAMQIKGTRTQWIAMKKNW 246
Query: 207 GANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGF-GQSFSSKIQF 255
G W + L GQ +SF++TT+DGV + F D+ P +W GQSF K+ F
Sbjct: 247 GQIWSTGVVLTGQCLSFRLTTSDGVMKEFIDVTPPDWKCNGQSFDGKVNF 296
>At5g39270 expansin-like protein
Length = 261
Score = 267 bits (682), Expect = 5e-72
Identities = 122/229 (53%), Positives = 159/229 (69%), Gaps = 7/229 (3%)
Query: 27 SGWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAALSTALFNDGASCGQCYKIIC 86
+ W +A ATFYG D G CGYGNL+ QGYG TAALSTALFNDG +CG CY+I+C
Sbjct: 40 TNWYDARATFYG--DIHGGDTQPCGYGNLFRQGYGLATAALSTALFNDGYTCGACYEIMC 97
Query: 87 DYKTDPRWCIKGRSITITATNFCPPNFDLPNDDGGWCNPPLKHFDMAQPAWEKIGIYRGG 146
DP+WC+ G S+ ITATNFCP N+ D WCNPP KHFD++ + KI Y+ G
Sbjct: 98 T--RDPQWCLPG-SVKITATNFCPANYSKTTDL--WCNPPQKHFDLSLAMFLKIAKYKAG 152
Query: 147 IVPVLFQRVPCKKHGGVRFSVNGRDYFELVLISNLAGAGSIQSVSIKGSKTDWMAMSRNW 206
+VPV ++R+PC K GGV+F G YF +VLI N+ GAG I+ V +KG+KT W+ M +NW
Sbjct: 153 VVPVRYRRIPCSKTGGVKFETKGNPYFLMVLIYNVGGAGDIKYVQVKGNKTGWITMKKNW 212
Query: 207 GANWQSNAYLNGQSMSFKVTTTDGVTRTFQDIVPSNWGFGQSFSSKIQF 255
G NW + L GQ +SF+VTT+DG+T+ F +++P NWGFGQ+F +I F
Sbjct: 213 GQNWTTITVLTGQGLSFRVTTSDGITKDFWNVMPKNWGFGQTFDGRINF 261
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,312,423
Number of Sequences: 26719
Number of extensions: 286568
Number of successful extensions: 751
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 637
Number of HSP's gapped (non-prelim): 57
length of query: 255
length of database: 11,318,596
effective HSP length: 97
effective length of query: 158
effective length of database: 8,726,853
effective search space: 1378842774
effective search space used: 1378842774
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146790.2


