

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146784.6 - phase: 0
(617 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
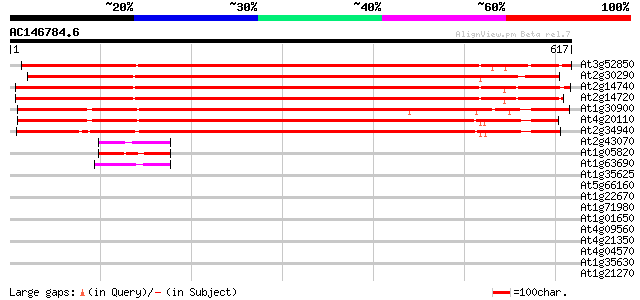
Score E
Sequences producing significant alignments: (bits) Value
At3g52850 Spot 3 protein and vacuolar sorting receptor homolog/A... 861 0.0
At2g30290 putative vacuolar sorting receptor 833 0.0
At2g14740 putative vacuolar sorting receptor 814 0.0
At2g14720 putative vacuolar sorting receptor 811 0.0
At1g30900 F17F8.23 693 0.0
At4g20110 vacuolar sorting receptor-like protein 682 0.0
At2g34940 putative vacuolar sorting receptor 650 0.0
At2g43070 unknown protein 51 2e-06
At1g05820 unknown protein 51 2e-06
At1g63690 unknown protein 49 1e-05
At1g35625 integral membrane protein, putative 39 0.007
At5g66160 ReMembR-H2 protein JR700 (gb|AAF32325.1) 38 0.015
At1g22670 putative RING zinc finger protein 37 0.042
At1g71980 unknown protein 36 0.072
At1g01650 unknown protein 34 0.21
At4g09560 putative protein 31 1.8
At4g21350 putative protein 31 2.3
At4g04570 receptor-like protein kinase 31 2.3
At1g35630 integral membrane protein like protein 30 3.0
At1g21270 putative protein 30 3.0
>At3g52850 Spot 3 protein and vacuolar sorting receptor
homolog/AtELP1
Length = 623
Score = 861 bits (2225), Expect = 0.0
Identities = 411/620 (66%), Positives = 486/620 (78%), Gaps = 23/620 (3%)
Query: 14 VLFLECCFGRFLVEKNSLRITSPKSLKGSYECAIGNFGVPQYGGTLVGSVVYPNVNQKGC 73
+L L GRF+VEKN+L++TSP S+KG YECAIGNFGVPQYGGTLVG+VVYP NQK C
Sbjct: 11 LLILNLAMGRFVVEKNNLKVTSPDSIKGIYECAIGNFGVPQYGGTLVGTVVYPKSNQKAC 70
Query: 74 KNFTDFSASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGGAAAILVADDREETLITMDT 133
K+++DF SF S PG PTFVL+DRGDCYFTLKAW AQ GAAAILVAD + E LITMDT
Sbjct: 71 KSYSDFDISFKSKPGRLPTFVLIDRGDCYFTLKAWIAQQAGAAAILVADSKAEPLITMDT 130
Query: 134 PEEGNVVNDDYIEKINIPSALISKSLGDRIKKALSDGEMVHINLDWREALPHPDDRVEYE 193
PEE + DY++ I IPSALI+K+LGD IK ALS G+MV++ LDW E++PHPD+RVEYE
Sbjct: 131 PEEDKS-DADYLQNITIPSALITKTLGDSIKSALSGGDMVNMKLDWTESVPHPDERVEYE 189
Query: 194 LWTNSNDECGPKCDNQINFVKSFKGAAQLLEKKGFTQFTPHYITWYCPKEFLLSRRCKSQ 253
LWTNSNDECG KCD QI F+K+FKGAAQ+LEK G TQFTPHYITWYCP+ F LS++CKSQ
Sbjct: 190 LWTNSNDECGKKCDTQIEFLKNFKGAAQILEKGGHTQFTPHYITWYCPEAFTLSKQCKSQ 249
Query: 254 CINHGRYCAPDPEQDFNKGYDGKDVVVQNLRQACFFKVANESGRPWQWWDYVTDFSIRCP 313
CINHGRYCAPDPEQDF KGYDGKDVVVQNLRQAC ++V N++G+PW WWDYVTDF+IRCP
Sbjct: 250 CINHGRYCAPDPEQDFTKGYDGKDVVVQNLRQACVYRVMNDTGKPWVWWDYVTDFAIRCP 309
Query: 314 MKEKKYTEECSDEVIKSLGVDLKKIKDCVGDPLADVENPVLKAEQEAQIGKESRGDVTIL 373
MKEKKYT+EC+D +IKSLG+DLKK+ C+GDP ADVENPVLKAEQE+QIGK SRGDVTIL
Sbjct: 310 MKEKKYTKECADGIIKSLGIDLKKVDKCIGDPEADVENPVLKAEQESQIGKGSRGDVTIL 369
Query: 374 PTLVINNRQYRGKLSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGCWKEKSSN 433
PTLV+NNRQYRGKL + AVLKAMC+GFQE+TEP+ICLT D+ETNECLENNGGCW++K++N
Sbjct: 370 PTLVVNNRQYRGKLEKGAVLKAMCSGFQESTEPAICLTEDLETNECLENNGGCWQDKAAN 429
Query: 434 ITACRDTFRGRVCVCPVVNNIKFVGDGYTHCEASGTLSCEFNNGGCWKASHGGRLYSACH 493
ITACRDTFRGR+C CP V +KFVGDGYTHC+ASG L C NNGGCW+ S GG YSAC
Sbjct: 430 ITACRDTFRGRLCECPTVQGVKFVGDGYTHCKASGALHCGINNGGCWRESRGGFTYSACV 489
Query: 494 DDYRKGCECPSGFRGDGVRSCEDCFNIFRILTMLQI----LMNAKKSRLASVHN------ 543
DD+ K C+CP GF+GDGV++CED + + T+ Q N S S N
Sbjct: 490 DDHSKDCKCPLGFKGDGVKNCED-VDECKEKTVCQCPECKCKNTWGSYECSCSNGLLYMR 548
Query: 544 ------ANAKIPLGVMSASAIAVCSTHEKMTRVLSGGYAFYKYRIQRYMDTEIRAIMAQY 597
+ K+ +S S + + + + GYA YKYRI+ YMD EIR IMAQY
Sbjct: 549 EHDTCIGSGKVGTTKLSWSFLWILIIGVGVAGL--SGYAVYKYRIRSYMDAEIRGIMAQY 606
Query: 598 MPLDNQPLIIPNPNQVHHDI 617
MPL++QP PN + H DI
Sbjct: 607 MPLESQP---PNTSGHHMDI 623
>At2g30290 putative vacuolar sorting receptor
Length = 625
Score = 833 bits (2151), Expect = 0.0
Identities = 388/603 (64%), Positives = 467/603 (77%), Gaps = 26/603 (4%)
Query: 20 CFGRFLVEKNSLRITSPKSLKGSYECAIGNFGVPQYGGTLVGSVVYPNVNQKGCKNFTDF 79
C GRF+VEKN+LR+TSP+S++G YECA+GNFGVPQYGG++ G+VVYP NQK CKNF DF
Sbjct: 21 CTGRFVVEKNNLRVTSPESIRGVYECALGNFGVPQYGGSMSGAVVYPKTNQKACKNFDDF 80
Query: 80 SASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGGAAAILVADDREETLITMDTPEEGNV 139
SF S PTFVLVDRGDCYFTLKAWNAQ GAA ILVAD+R E LITMD PE+
Sbjct: 81 EISFRSRVAGLPTFVLVDRGDCYFTLKAWNAQRAGAATILVADNRPEQLITMDAPED-ET 139
Query: 140 VNDDYIEKINIPSALISKSLGDRIKKALSDGEMVHINLDWREALPHPDDRVEYELWTNSN 199
+ DY++ I IPSAL+S+SLG IK A++ G+ VHI+LDWREALPHP+DRV YELWTNSN
Sbjct: 140 SDADYLQNITIPSALVSRSLGSAIKTAIAHGDPVHISLDWREALPHPNDRVAYELWTNSN 199
Query: 200 DECGPKCDNQINFVKSFKGAAQLLEKKGFTQFTPHYITWYCPKEFLLSRRCKSQCINHGR 259
DECG KCD QI F+K FKGAAQ+LEK G+T+FTPHYITWYCP+ FL SR+CK+QCIN GR
Sbjct: 200 DECGSKCDAQIRFLKRFKGAAQILEKGGYTRFTPHYITWYCPEAFLASRQCKTQCINGGR 259
Query: 260 YCAPDPEQDFNKGYDGKDVVVQNLRQACFFKVANESGRPWQWWDYVTDFSIRCPMKEKKY 319
YCAPDPEQDF++GY+GKDV++QNLRQACFF+V NESG+PW WWDYVTDF+IRCPMKE+KY
Sbjct: 260 YCAPDPEQDFSRGYNGKDVIIQNLRQACFFRVTNESGKPWLWWDYVTDFAIRCPMKEEKY 319
Query: 320 TEECSDEVIKSLGVDLKKIKDCVGDPLADVENPVLKAEQEAQIGKESRGDVTILPTLVIN 379
++C+D+VI+SLGVD+KKI C+GD A+ ENPVLK EQ AQ+GK SRGDVTILPT+VIN
Sbjct: 320 NKKCADQVIQSLGVDVKKIDKCIGDIDANAENPVLKEEQVAQVGKGSRGDVTILPTIVIN 379
Query: 380 NRQYRGKLSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGCWKEKSSNITACRD 439
NRQYRGKL R AVLKA+C+GF+ETTEP ICLT D+ETNECL+NNGGCW++K++NITACRD
Sbjct: 380 NRQYRGKLQRSAVLKALCSGFRETTEPPICLTEDIETNECLQNNGGCWEDKTTNITACRD 439
Query: 440 TFRGRVCVCPVVNNIKFVGDGYTHCEASGTLSCEFNNGGCWKASHGGRLYSACHDDYRKG 499
TFRGRVC CP+V +KF+GDGYTHCEASG L C NNGGCWK + G+ YSAC DD+ KG
Sbjct: 440 TFRGRVCQCPIVQGVKFLGDGYTHCEASGALRCGINNGGCWKQTQMGKTYSACRDDHSKG 499
Query: 500 CECPSGFRGDGVRSCED----------------CFNIFRI--LTMLQILMNAKKSRLASV 541
C+CP GF GDG++ C+D C N + + L+ ++ +
Sbjct: 500 CKCPPGFIGDGLKECKDVNECEEKTACQCRDCKCKNTWGSYECSCSGSLLYIREHDICIN 559
Query: 542 HNANAKIPLGVMSASAIAVCSTHEKMTRVLSGGYAFYKYRIQRYMDTEIRAIMAQYMPLD 601
+A GV+ + + G Y YKYRI+ YMD+EIRAIMAQYMPLD
Sbjct: 560 RDARGDFSWGVIWIIIMG-------LGAAALGAYTVYKYRIRTYMDSEIRAIMAQYMPLD 612
Query: 602 NQP 604
N P
Sbjct: 613 NNP 615
>At2g14740 putative vacuolar sorting receptor
Length = 628
Score = 814 bits (2102), Expect = 0.0
Identities = 389/624 (62%), Positives = 472/624 (75%), Gaps = 20/624 (3%)
Query: 7 PLLLCVLVLFLECCFGRFLVEKNSLRITSPKSLKGSYECAIGNFGVPQYGGTLVGSVVYP 66
P LL + +L RF+VEKNSL +TSP+S+KG+++ AIGNFG+PQYGG++ G+VVYP
Sbjct: 9 PWLLLLTLLVSPLNDARFVVEKNSLSVTSPESIKGTHDSAIGNFGIPQYGGSMAGTVVYP 68
Query: 67 NVNQKGCKNFTDFSASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGGAAAILVADDREE 126
NQK CK F+DFS SF S PG PTF+LVDRGDC+F LK WNAQ GA+A+LVAD+ +E
Sbjct: 69 KENQKSCKEFSDFSISFKSQPGALPTFLLVDRGDCFFALKVWNAQKAGASAVLVADNVDE 128
Query: 127 TLITMDTPEEGNVVNDDYIEKINIPSALISKSLGDRIKKALSDGEMVHINLDWREALPHP 186
LITMDTPEE +V + YIE I IPSAL++K G+++KKA+S G+MV++NLDWREA+PHP
Sbjct: 129 PLITMDTPEE-DVSSAKYIENITIPSALVTKGFGEKLKKAISGGDMVNLNLDWREAVPHP 187
Query: 187 DDRVEYELWTNSNDECGPKCDNQINFVKSFKGAAQLLEKKGFTQFTPHYITWYCPKEFLL 246
DDRVEYELWTNSNDECG KCD + FVK FKGAAQ+LEK GFTQF PHYITWYCP F L
Sbjct: 188 DDRVEYELWTNSNDECGVKCDMLMEFVKDFKGAAQILEKGGFTQFRPHYITWYCPHAFTL 247
Query: 247 SRRCKSQCINHGRYCAPDPEQDFNKGYDGKDVVVQNLRQACFFKVANESGRPWQWWDYVT 306
SR+CKSQCIN GRYCAPDPEQDF+ GYDGKDVVV+NLRQ C +KVANE+G+PW WWDYVT
Sbjct: 248 SRQCKSQCINKGRYCAPDPEQDFSSGYDGKDVVVENLRQLCVYKVANETGKPWVWWDYVT 307
Query: 307 DFSIRCPMKEKKYTEECSDEVIKSLGVDLKKIKDCVGDPLADVENPVLKAEQEAQIGKES 366
DF IRCPMKEKKY +EC+D VIKSLG+D KK+ C+GDP AD++NPVLK EQ+AQ+GK S
Sbjct: 308 DFQIRCPMKEKKYNKECADSVIKSLGIDSKKLDKCMGDPDADLDNPVLKEEQDAQVGKGS 367
Query: 367 RGDVTILPTLVINNRQYRGKLSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGC 426
RGDVTILPTLV+NNRQYRGKL + AVLKA+C+GF+ETTEP+ICL+ D+E+NECL+NNGGC
Sbjct: 368 RGDVTILPTLVVNNRQYRGKLEKSAVLKALCSGFEETTEPAICLSTDVESNECLDNNGGC 427
Query: 427 WKEKSSNITACRDTFRGRVCVCPVVNNIKFVGDGYTHCEASGTLSCEFNNGGCWKASHGG 486
W++KS+NITAC+DTFRGRVC CP V+ ++F GDGY+HCE SG C NNGGCW G
Sbjct: 428 WQDKSANITACKDTFRGRVCECPTVDGVQFKGDGYSHCEPSGPGRCTINNGGCWHEERDG 487
Query: 487 RLYSACHDDYRKGCECPSGFRGDGVRSCEDCFNIFRILTMLQILMNAKKSRLASVH---- 542
+SAC D CECP GF+GDG + CED N + Q + K+ S
Sbjct: 488 HAFSACVDKDSVKCECPPGFKGDGTKKCED-INECKEKKACQCPECSCKNTWGSYECSCS 546
Query: 543 ----------NANAKIPLGVMSASAIAVCSTHEKMTRVLSGGYAFYKYRIQRYMDTEIRA 592
+K V SA A AV + +G Y YKYR+++YMD+EIRA
Sbjct: 547 GDLLYIRDHDTCISKTGAQVRSAWA-AVWLIMLSLGLAAAGAYLVYKYRLRQYMDSEIRA 605
Query: 593 IMAQYMPLDNQPLIIPNPNQVHHD 616
IMAQYMPLD+QP I PN V+ +
Sbjct: 606 IMAQYMPLDSQPEI---PNHVNDE 626
>At2g14720 putative vacuolar sorting receptor
Length = 628
Score = 811 bits (2095), Expect = 0.0
Identities = 385/617 (62%), Positives = 472/617 (76%), Gaps = 18/617 (2%)
Query: 7 PLLLCVLVLFLECCFGRFLVEKNSLRITSPKSLKGSYECAIGNFGVPQYGGTLVGSVVYP 66
P LL + ++ RF+VEKNSL +TSP+S+KG+++ AIGNFG+PQYGG++ G+VVYP
Sbjct: 9 PWLLLLSLVVSPFNEARFVVEKNSLSVTSPESIKGTHDSAIGNFGIPQYGGSMAGTVVYP 68
Query: 67 NVNQKGCKNFTDFSASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGGAAAILVADDREE 126
NQK CK F+DFS SF S PG PTF+LVDRGDC+F LK WNAQ GA+A+LVAD+ +E
Sbjct: 69 KENQKSCKEFSDFSISFKSQPGALPTFLLVDRGDCFFALKVWNAQKAGASAVLVADNVDE 128
Query: 127 TLITMDTPEEGNVVNDDYIEKINIPSALISKSLGDRIKKALSDGEMVHINLDWREALPHP 186
LITMDTPEE +V + YIE I IPSAL++K G+++KKA+S G+MV++NLDWREA+PHP
Sbjct: 129 PLITMDTPEE-DVSSAKYIENITIPSALVTKGFGEKLKKAISGGDMVNLNLDWREAVPHP 187
Query: 187 DDRVEYELWTNSNDECGPKCDNQINFVKSFKGAAQLLEKKGFTQFTPHYITWYCPKEFLL 246
DDRVEYELWTNSNDECG KCD + FVK FKGAAQ+LEK GFTQF PHYITWYCP F L
Sbjct: 188 DDRVEYELWTNSNDECGVKCDMLMEFVKDFKGAAQILEKGGFTQFRPHYITWYCPHAFTL 247
Query: 247 SRRCKSQCINHGRYCAPDPEQDFNKGYDGKDVVVQNLRQACFFKVANESGRPWQWWDYVT 306
SR+CKSQCIN GRYCAPDPEQDF+ GYDGKDVVV+NLRQ C +KVANE+G+PW WWDYVT
Sbjct: 248 SRQCKSQCINKGRYCAPDPEQDFSSGYDGKDVVVENLRQLCVYKVANETGKPWVWWDYVT 307
Query: 307 DFSIRCPMKEKKYTEECSDEVIKSLGVDLKKIKDCVGDPLADVENPVLKAEQEAQIGKES 366
DF IRCPMKEKKY ++C++ VIKSLG+D +KI C+GDP AD++NPVLK EQ+AQ+GK +
Sbjct: 308 DFQIRCPMKEKKYNKDCAESVIKSLGIDSRKIDKCMGDPDADLDNPVLKEEQDAQVGKGT 367
Query: 367 RGDVTILPTLVINNRQYRGKLSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGC 426
RGDVTILPTLV+NNRQYRGKL + AVLKA+C+GF+E+TEP+ICL+ DMETNECL+NNGGC
Sbjct: 368 RGDVTILPTLVVNNRQYRGKLEKSAVLKALCSGFEESTEPAICLSTDMETNECLDNNGGC 427
Query: 427 WKEKSSNITACRDTFRGRVCVCPVVNNIKFVGDGYTHCEASGTLSCEFNNGGCWKASHGG 486
W++KS+NITAC+DTFRG+VCVCP+V+ ++F GDGY+HCE SG C NNGGCW G
Sbjct: 428 WQDKSANITACKDTFRGKVCVCPIVDGVRFKGDGYSHCEPSGPGRCTINNGGCWHEERDG 487
Query: 487 RLYSACHDDYRKGCECPSGFRGDGVRSCEDCFNIFRILTMLQILMNAKKSRLASVH---- 542
+SAC D CECP GF+GDGV+ CED N + Q + K+ S
Sbjct: 488 HAFSACVDKDSVKCECPPGFKGDGVKKCED-INECKEKKACQCPECSCKNTWGSYECSCS 546
Query: 543 ----------NANAKIPLGVMSASAIAVCSTHEKMTRVLSGGYAFYKYRIQRYMDTEIRA 592
+K V SA A AV + +G Y YKYR+++YMD+EIRA
Sbjct: 547 GDLLYMRDHDTCISKTGSQVKSAWA-AVWLIMLSLGLAAAGAYLVYKYRLRQYMDSEIRA 605
Query: 593 IMAQYMPLDNQPLIIPN 609
IMAQYMPLD+QP +PN
Sbjct: 606 IMAQYMPLDSQP-EVPN 621
>At1g30900 F17F8.23
Length = 649
Score = 693 bits (1789), Expect = 0.0
Identities = 336/647 (51%), Positives = 433/647 (65%), Gaps = 58/647 (8%)
Query: 9 LLCVLVLFLECCFGRFLVEKNSLRITSPKSLKGSYECAIGNFGVPQYGGTLVGSVVYPNV 68
L L + + FGRF+VEK+S+ I +P +++ ++ AI NFGVP YGG ++GSVVY
Sbjct: 12 LFLALTMVVNGVFGRFIVEKSSVTILNPLAMRSKHDAAIANFGVPNYGGYMIGSVVYAGQ 71
Query: 69 NQKGCKNFTD-FSASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGGAAAILVADDREET 127
GC +F F F PT +++DRG+CYF LK WN Q G AA+LVAD+ +E
Sbjct: 72 GAYGCDSFDKTFKPKFPR-----PTILIIDRGECYFALKVWNGQQSGVAAVLVADNVDEP 126
Query: 128 LITMDTPEEGNVVNDDYIEKINIPSALISKSLGDRIKKALSDGEMVHINLDWREALPHPD 187
LITMD+PEE DD+IEK+NIPSALI S + +K+AL GE V + +DW E+LPHPD
Sbjct: 127 LITMDSPEESKEA-DDFIEKLNIPSALIDFSFANTLKQALKKGEEVVLKIDWSESLPHPD 185
Query: 188 DRVEYELWTNSNDECGPKCDNQINFVKSFKGAAQLLEKKGFTQFTPHYITWYCPKEFLLS 247
+RVEYELWTN+NDECG +CD Q+NFVK+FKG AQ+LEK G++ FTPHYITW+CPK+++ S
Sbjct: 186 ERVEYELWTNTNDECGARCDEQMNFVKNFKGHAQILEKGGYSLFTPHYITWFCPKDYVSS 245
Query: 248 RRCKSQCINHGRYCAPDPEQDFNKGYDGKDVVVQNLRQACFFKVANESGRPWQWWDYVTD 307
+CKSQCIN GRYCAPDPEQDF GYDGKD+V +NLRQ C KVA E+ R W WWDYVTD
Sbjct: 246 NQCKSQCINQGRYCAPDPEQDFGDGYDGKDIVFENLRQLCVHKVAKENNRSWVWWDYVTD 305
Query: 308 FSIRCPMKEKKYTEECSDEVIKSLGVDLKKIKDCVGDPLADVENPVLKAEQEAQIGKESR 367
F IRC MKEKKY++EC++ V++SLG+ L KIK C+GDP A+VEN VLKAEQ Q+G+ R
Sbjct: 306 FHIRCSMKEKKYSKECAERVVESLGLPLDKIKKCIGDPDANVENEVLKAEQALQVGQGDR 365
Query: 368 GDVTILPTLVINNRQYRGKLSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGCW 427
GDVTILPTL++NN QYRGKL R AVLKA+C+GF+E TEP ICL+ D+ETNECLE NGGCW
Sbjct: 366 GDVTILPTLIVNNAQYRGKLERNAVLKAICSGFKERTEPGICLSGDIETNECLEANGGCW 425
Query: 428 KEKSSNITACR---------------------------DTFRGRVCVCPVVNNIKFVGDG 460
++K SN+TAC+ DTFRGRVC CPVVN +++ GDG
Sbjct: 426 EDKKSNVTACKVLRTDELKGLHFYRYLVSFIPKNGFYQDTFRGRVCECPVVNGVQYKGDG 485
Query: 461 YTHCEASGTLSCEFNNGGCWKASHGGRLYSACHDDYRKGCECPSGFRGDGVR-------- 512
YT CE G C N GGCW + G +SAC + GC CP GF+GDG++
Sbjct: 486 YTSCEPYGPARCSINQGGCWSETKKGLTFSACSNLETSGCRCPPGFKGDGLKCEACQCDG 545
Query: 513 -SCEDCFNIFRILTMLQILMNAKKSRLASVHNANAKI---PLGVMSASAIAVCSTHEKMT 568
+C++ + F L K + + + ++I P V+ A+ ++C
Sbjct: 546 CNCKNKWGGFECKCSGNRLY--MKEQDTCIERSGSRIGWFPTFVILAAVASICV------ 597
Query: 569 RVLSGGYAFYKYRIQRYMDTEIRAIMAQYMPLDNQPLIIPNPNQVHH 615
GGY FYKYR++ YMD+EI AIM+QYMPL++Q P + H
Sbjct: 598 ----GGYVFYKYRLRSYMDSEIMAIMSQYMPLESQNTTDPMTGESQH 640
>At4g20110 vacuolar sorting receptor-like protein
Length = 625
Score = 682 bits (1761), Expect = 0.0
Identities = 328/615 (53%), Positives = 418/615 (67%), Gaps = 36/615 (5%)
Query: 9 LLCVLVLFLECCFGRFLVEKNSLRITSPKSLKGSYECAIGNFGVPQYGGTLVGSVVYPNV 68
LL L + RF+VEK S+ + +P+ ++ ++ +I NFG+P YGG L+GSVVYP+
Sbjct: 13 LLAALTIIAMVVEARFVVEKESISVLNPEEMRSKHDGSIANFGLPDYGGFLIGSVVYPDS 72
Query: 69 NQKGCKNF-TDFSASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGGAAAILVADDREET 127
GC F F F PT +L+DRG CYF LKAW+AQ GAAA+LVAD+ +E
Sbjct: 73 KTDGCSAFGKTFKPKFPR-----PTILLLDRGGCYFALKAWHAQQAGAAAVLVADNVDEP 127
Query: 128 LITMDTPEEGNVVNDDYIEKINIPSALISKSLGDRIKKALSDGEMVHINLDWREALPHPD 187
L+TMD+PEE D +IEK+ IPS LI KS GD +++ G+ + I LDWRE++PHPD
Sbjct: 128 LLTMDSPEESKDA-DGFIEKLTIPSVLIDKSFGDDLRQGFQKGKNIVIKLDWRESVPHPD 186
Query: 188 DRVEYELWTNSNDECGPKCDNQINFVKSFKGAAQLLEKKGFTQFTPHYITWYCPKEFLLS 247
RVEYELWTNSNDECG +CD Q++FVK+FKG AQ+LEK G+T FTPHYITW+CP +F+ S
Sbjct: 187 KRVEYELWTNSNDECGARCDEQMDFVKNFKGHAQILEKGGYTAFTPHYITWFCPFQFINS 246
Query: 248 RRCKSQCINHGRYCAPDPEQDFNKGYDGKDVVVQNLRQACFFKVANESGRPWQWWDYVTD 307
CKSQCINHGRYCAPDPE +F +GY+GKDVV++NLRQ C +VANES RPW WWDYVTD
Sbjct: 247 PHCKSQCINHGRYCAPDPEDNFREGYEGKDVVLENLRQLCVHRVANESSRPWVWWDYVTD 306
Query: 308 FSIRCPMKEKKYTEECSDEVIKSLGVDLKKIKDCVGDPLADVENPVLKAEQEAQIGKESR 367
F RC MKEKKY+ +C++ VIKSL + ++KIK C+GDP AD EN VL+ EQ +QIG+ +R
Sbjct: 307 FHSRCSMKEKKYSIDCAESVIKSLNLPIEKIKKCIGDPEADTENQVLRTEQVSQIGRGNR 366
Query: 368 GDVTILPTLVINNRQYRGKLSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGCW 427
GDVTILPTLVINN QYRG+L R AVLKA+CAGF ET+EP+ICL +ETNECLENNGGCW
Sbjct: 367 GDVTILPTLVINNAQYRGRLERTAVLKAICAGFNETSEPAICLNTGLETNECLENNGGCW 426
Query: 428 KEKSSNITACRDTFRGRVCVCPVVNNIKFVGDGYTHCEASGTLSCEFNNGGCWKASHGGR 487
++ +NITAC+DTFRGR+C CPVV +++ GDGYT C G C NNGGCW + G
Sbjct: 427 QDTKANITACQDTFRGRLCECPVVKGVQYKGDGYTSCTPYGPARCTMNNGGCWSDTRNGL 486
Query: 488 LYSACHDDYRKGCECPSGFRGDGVRSCED----------------CFNI---FRILTMLQ 528
+SAC D GC+CP GF+GDG+ +CED C N ++
Sbjct: 487 TFSACSDSVSTGCKCPEGFQGDGL-TCEDINECKERSVCQCSGCRCKNSWGGYKCSCSGD 545
Query: 529 ILMNAKKSRLASVHNANAKIPLGVMSASAIAVCSTHEKMTRVLSGGYAFYKYRIQRYMDT 588
L + + + L + + +AV GY FYKYR + YMD+
Sbjct: 546 RLYINDQDTCIERYGSKTAWWLTFLILAIVAVAGL---------AGYIFYKYRFRSYMDS 596
Query: 589 EIRAIMAQYMPLDNQ 603
EI IM+QYMPL++Q
Sbjct: 597 EIMTIMSQYMPLESQ 611
>At2g34940 putative vacuolar sorting receptor
Length = 618
Score = 650 bits (1678), Expect = 0.0
Identities = 315/618 (50%), Positives = 422/618 (67%), Gaps = 37/618 (5%)
Query: 8 LLLCVLVLFLECCFGRFLVEKNSLRITSPKSLKGSYECAIGNFGVPQYGGTLVGSVVYPN 67
L+L + ++ + RF VEK+SL + + + ++ AI NFG+P+YGG ++GSVVY
Sbjct: 12 LILALTMVVVNGFSSRFFVEKSSLTVLNSWEMGAKHDAAIANFGLPKYGGFMIGSVVYAG 71
Query: 68 VNQKGCKNFTDFSASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGGAAAILVADDREET 127
+ GC +F + +F++ +P +L+DRG C F LK WN Q GAAA+L+AD+ E
Sbjct: 72 QDAYGCNSF---NKTFNTK-SPYPKILLIDRGVCNFALKIWNGQQSGAAAVLLADNIVEP 127
Query: 128 LITMDTPEEGNVVNDDYIEKINIPSALISKSLGDRIKKALSDGEMVHINLDWREALPHPD 187
LITMDTP++ + D+I+K+ IPSALI +S GD +KKAL GE V + +DW E++P+PD
Sbjct: 128 LITMDTPQDED---PDFIDKVKIPSALILRSFGDSLKKALKRGEEVILKMDWSESIPNPD 184
Query: 188 DRVEYELWTNSNDECGPKCDNQINFVKSFKGAAQLLEKKGFTQFTPHYITWYCPKEFLLS 247
+RVEYELW N+NDECG CD QI+F+K+FKG AQ+LEK G+T F PHYI+W CPKE LLS
Sbjct: 185 ERVEYELWANTNDECGVHCDKQIDFIKNFKGMAQILEKGGYTLFRPHYISWVCPKELLLS 244
Query: 248 RRCKSQCINHGRYCAPDPEQDFNKGYDGKDVVVQNLRQACFFKVANESGRPWQWWDYVTD 307
++C++QCIN GRYCA D +Q+F GY+GKDVV +NLRQ C KVA E W WWDYVTD
Sbjct: 245 KQCRTQCINQGRYCALDTKQEFEDGYNGKDVVYENLRQLCVHKVAKEKNTSWVWWDYVTD 304
Query: 308 FSIRCPMKEKKYTEECSDEVIKSLGVDLKKIKDCVGDPLADVENPVLKAEQEAQIGKESR 367
F+IRC MKEKKY+ EC++ +++SLG+ L+KIK C+GDP ADVEN VLKAE+ Q+G+E+R
Sbjct: 305 FNIRCSMKEKKYSRECAETIVESLGLSLEKIKKCIGDPDADVENEVLKAEEAFQLGQENR 364
Query: 368 GDVTILPTLVINNRQYRGKLSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGCW 427
G VTI PTL+INN QYRGKL R AVLKA+C+GF+E TEPSICL D+ETNECL NGGCW
Sbjct: 365 GIVTIFPTLMINNAQYRGKLERTAVLKAICSGFKERTEPSICLNSDIETNECLIENGGCW 424
Query: 428 KEKSSNITACRDTFRGRVCVCPVVNNIKFVGDGYTHCEASGTLSCEFNNGGCWKASHGGR 487
++K SN+TAC+DTFRGRVC CPVV+ +++ GDGYT C+ G C NNG CW + G
Sbjct: 425 QDKRSNVTACKDTFRGRVCECPVVDGVQYKGDGYTSCKPYGPARCSMNNGDCWSETRKGL 484
Query: 488 LYSACHDDYRKGCECPSGFRGDGVRSCED----------------CFNIFR----ILTML 527
+S+C D GC CP GF GDG++ CED C N + +
Sbjct: 485 TFSSCSDSETSGCRCPLGFLGDGLK-CEDIDECKEKSACKCDGCKCKNNWGGYECKCSNN 543
Query: 528 QILMNAKKSRLASVHNANAKIPLGVMSASAIAVCSTHEKMTRVLSGGYAFYKYRIQRYMD 587
I M + + + + ++ ++ +AIA S G Y FYKY +Q YMD
Sbjct: 544 SIYMKEEDTCIERRSGSRSRGLFTIVVLTAIAGISL---------GAYIFYKYHLQSYMD 594
Query: 588 TEIRAIMAQYMPLDNQPL 605
+EI +IM+QY+PLD+Q +
Sbjct: 595 SEIVSIMSQYIPLDSQSI 612
>At2g43070 unknown protein
Length = 543
Score = 51.2 bits (121), Expect = 2e-06
Identities = 29/80 (36%), Positives = 47/80 (58%), Gaps = 7/80 (8%)
Query: 98 RGDCYFTLKAWNAQNGGAAAILVADDREETLITMDTPEEGNVVNDDYIEKINIPSALISK 157
RG+C FT KA +A+ GA+A+LV +D+E+ +E + D ++IP +ISK
Sbjct: 113 RGNCAFTEKAKHAEAAGASALLVINDKEDL-------DEMGCMEKDTSLNVSIPVLMISK 165
Query: 158 SLGDRIKKALSDGEMVHINL 177
S GD + K++ D + V + L
Sbjct: 166 SSGDALNKSMVDNKNVELLL 185
>At1g05820 unknown protein
Length = 441
Score = 51.2 bits (121), Expect = 2e-06
Identities = 31/80 (38%), Positives = 49/80 (60%), Gaps = 7/80 (8%)
Query: 98 RGDCYFTLKAWNAQNGGAAAILVADDREETLITMDTPEEGNVVNDDYIEKINIPSALISK 157
RG+C FT+KA AQ GGAAA+++ +D+EE L M E+ +N ++IP +I+
Sbjct: 109 RGECAFTVKAQVAQAGGAAALVLINDKEE-LDEMVCGEKDTSLN------VSIPILMITT 161
Query: 158 SLGDRIKKALSDGEMVHINL 177
S GD +KK++ + V + L
Sbjct: 162 SSGDALKKSIMQNKKVELLL 181
>At1g63690 unknown protein
Length = 540
Score = 48.5 bits (114), Expect = 1e-05
Identities = 26/84 (30%), Positives = 49/84 (57%), Gaps = 7/84 (8%)
Query: 94 VLVDRGDCYFTLKAWNAQNGGAAAILVADDREETLITMDTPEEGNVVNDDYIEKINIPSA 153
V+V+RG+C FT KA NA+ GA+A+L+ ++++E + P+E ++ I IP+
Sbjct: 108 VIVERGNCRFTAKANNAEAAGASALLIINNQKELYKMVCEPDETDL-------DIQIPAV 160
Query: 154 LISKSLGDRIKKALSDGEMVHINL 177
++ + G ++K L++ V L
Sbjct: 161 MLPQDAGASLQKMLANSSKVSAQL 184
>At1g35625 integral membrane protein, putative
Length = 279
Score = 39.3 bits (90), Expect = 0.007
Identities = 25/82 (30%), Positives = 39/82 (47%), Gaps = 11/82 (13%)
Query: 91 PTFVLVDRGDCYFTLKAWNAQNGGAAAILVADDREETLITMDTPEEGNVVNDDYIEKINI 150
P++VL+ RG C F K NAQ G A +V +DR E L+ + I
Sbjct: 40 PSYVLIVRGGCSFEEKIRNAQEAGYKAAIVYNDRYEELLVRRNS-----------SGVYI 88
Query: 151 PSALISKSLGDRIKKALSDGEM 172
L++++ G+ +K+ S EM
Sbjct: 89 HGVLVTRTSGEVLKEYTSRAEM 110
>At5g66160 ReMembR-H2 protein JR700 (gb|AAF32325.1)
Length = 310
Score = 38.1 bits (87), Expect = 0.015
Identities = 22/75 (29%), Positives = 37/75 (49%), Gaps = 13/75 (17%)
Query: 93 FVLVDRGDCYFTLKAWNAQNGGAAAILVAD--DREETLITMDTPEEGNVVNDDYIEKINI 150
F L+ RG+C F K NAQN G A++V D D E+ ++ P++ I +
Sbjct: 84 FALIIRGECSFEDKLLNAQNSGFQAVIVYDNIDNEDLIVMKVNPQD-----------ITV 132
Query: 151 PSALISKSLGDRIKK 165
+ +S G+ ++K
Sbjct: 133 DAVFVSNVAGEILRK 147
>At1g22670 putative RING zinc finger protein
Length = 422
Score = 36.6 bits (83), Expect = 0.042
Identities = 31/116 (26%), Positives = 45/116 (38%), Gaps = 14/116 (12%)
Query: 58 TLVGSVVYPNVNQKGCKNFTDFSASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGGAAA 117
T+ VVY C+N + P VL+ RG C F K NAQ G A
Sbjct: 49 TVETGVVYVAEPLNACRNLRNKP---EQSPYGTSPLVLIIRGGCSFEYKVRNAQRSGFKA 105
Query: 118 ILVADDREETLIT-MDTPEEGNVVNDDYIEKINIPSALISKSLGDRIKKALSDGEM 172
+V D+ + ++ M +G I I + + K G+ +KK EM
Sbjct: 106 AIVYDNVDRNFLSAMGGDSDG----------IKIQAVFVMKRAGEMLKKYAGSEEM 151
>At1g71980 unknown protein
Length = 448
Score = 35.8 bits (81), Expect = 0.072
Identities = 43/156 (27%), Positives = 64/156 (40%), Gaps = 21/156 (13%)
Query: 13 LVLFLECCFGRFLVEKNSLRITSPKSLKGSYECAIGNFGVPQYGGTLVGSVVYPNVNQKG 72
LVL L C L + + + +L S++ NF P GT VVY
Sbjct: 5 LVLLLYVCTVSCLASSKVILMRNNITL--SFDDIEANFA-PSVKGTGEIGVVYVAEPLDA 61
Query: 73 CKNFTDF--SASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGGAAAILVADDREE-TLI 129
C+N + +S + P FVL+ RG C F K AQ G A ++ D+ + TLI
Sbjct: 62 CQNLMNKPEQSSNETSP-----FVLIVRGGCSFEEKVRKAQRAGFKAAIIYDNEDRGTLI 116
Query: 130 TMDTPEEGNVVNDDYIEKINIPSALISKSLGDRIKK 165
M G I I + ++K G+ +K+
Sbjct: 117 AMAGNSGG----------IRIHAVFVTKETGEVLKE 142
>At1g01650 unknown protein
Length = 491
Score = 34.3 bits (77), Expect = 0.21
Identities = 24/84 (28%), Positives = 41/84 (48%), Gaps = 7/84 (8%)
Query: 94 VLVDRGDCYFTLKAWNAQNGGAAAILVADDREETLITMDTPEEGNVVNDDYIEKINIPSA 153
+LV RG C FT K A+ GA+AIL+ ++ + L M + NV++ I IP
Sbjct: 115 ILVHRGKCSFTTKTKVAEAAGASAILIINNSTD-LFKMVCEKGENVLD------ITIPVV 167
Query: 154 LISKSLGDRIKKALSDGEMVHINL 177
++ G ++ + +V + L
Sbjct: 168 MLPVDAGRSLENIVKSNAIVTLQL 191
>At4g09560 putative protein
Length = 431
Score = 31.2 bits (69), Expect = 1.8
Identities = 17/45 (37%), Positives = 22/45 (48%)
Query: 91 PTFVLVDRGDCYFTLKAWNAQNGGAAAILVADDREETLITMDTPE 135
P +VL+ RG C F K NAQ G A +V D + + T E
Sbjct: 79 PPYVLIIRGGCSFEDKIRNAQKAGYKAAIVYDYEDFGFLVSTTGE 123
>At4g21350 putative protein
Length = 374
Score = 30.8 bits (68), Expect = 2.3
Identities = 13/40 (32%), Positives = 22/40 (54%)
Query: 570 VLSGGYAFYKYRIQRYMDTEIRAIMAQYMPLDNQPLIIPN 609
+L G+ F + IQ+++D+ R +PL P +IPN
Sbjct: 23 ILQSGHTFDRVSIQQWIDSGNRTCPITKLPLSETPYLIPN 62
>At4g04570 receptor-like protein kinase
Length = 654
Score = 30.8 bits (68), Expect = 2.3
Identities = 28/100 (28%), Positives = 43/100 (43%), Gaps = 16/100 (16%)
Query: 191 EYELWTNSNDECGPKCDNQINFVKSFKGAAQLLEKKGFTQFTPHYITWYCPKEFLLSRRC 250
++E N E K + + +K +K EK GFT+F Y+ C + L SR C
Sbjct: 161 QWEEMVNRTLEAATKAEGS-SVLKYYKA-----EKAGFTEFPDVYMLMQCTPD-LSSRDC 213
Query: 251 KSQCINHGRYCAPDPEQDFNKGYDGKDVVVQNLRQACFFK 290
K C D F K Y G+ + +L +C+F+
Sbjct: 214 KQ--------CLGDCVMYFRKDYMGRKGGMASL-PSCYFR 244
>At1g35630 integral membrane protein like protein
Length = 318
Score = 30.4 bits (67), Expect = 3.0
Identities = 23/82 (28%), Positives = 37/82 (45%), Gaps = 11/82 (13%)
Query: 92 TFVLVDRGDCYFTLKAWNAQNGG-AAAILVADDREETLITMDTPEEGNVVNDDYIEKINI 150
++VL+ G C F K AQ G AAI+ D +E L+ M G ++I
Sbjct: 78 SYVLIVLGGCSFEEKVRKAQKAGYKAAIVYNDGYDELLVPMAGNSSG----------VDI 127
Query: 151 PSALISKSLGDRIKKALSDGEM 172
L++++ G+ +K EM
Sbjct: 128 HGLLVTRASGEVLKGYADQDEM 149
>At1g21270 putative protein
Length = 732
Score = 30.4 bits (67), Expect = 3.0
Identities = 13/37 (35%), Positives = 18/37 (48%), Gaps = 1/37 (2%)
Query: 479 CWKASHGGRLYSACHDDYRK-GCECPSGFRGDGVRSC 514
C + H +S C + C CPSG+R D + SC
Sbjct: 282 CISSRHNCSEHSTCENTKGSFNCNCPSGYRKDSLNSC 318
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,370,518
Number of Sequences: 26719
Number of extensions: 715898
Number of successful extensions: 1412
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1361
Number of HSP's gapped (non-prelim): 28
length of query: 617
length of database: 11,318,596
effective HSP length: 105
effective length of query: 512
effective length of database: 8,513,101
effective search space: 4358707712
effective search space used: 4358707712
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146784.6


