

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146777.12 + phase: 0
(238 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
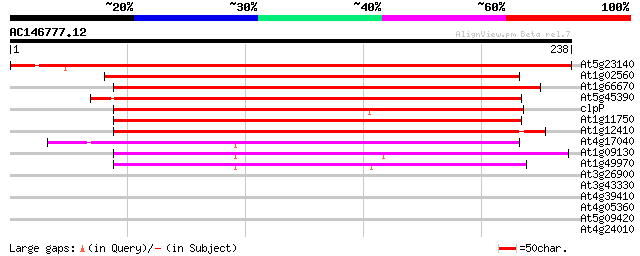
Score E
Sequences producing significant alignments: (bits) Value
At5g23140 ATP-dependent protease proteolytic subunit ClpP-like p... 377 e-105
At1g02560 ATP-dependent clp protease proteolytic subunit (nClpP1) 185 2e-47
At1g66670 nClpP3 protein 162 2e-40
At5g45390 ATP-dependent Clp protease-like protein 157 4e-39
clpP -chloroplast genome- ATP-dependent protease subunit 144 3e-35
At1g11750 putative ATP-dependent Clp protease proteolytic subunit 144 5e-35
At1g12410 ClpP protease complex subunit ClpR2 140 8e-34
At4g17040 ClpP protease complex subunit ClpR4 136 9e-33
At1g09130 ClpP protease complex subunit ClpR3 136 1e-32
At1g49970 ClpP protease complex subunit ClpR1 100 7e-22
At3g26900 shikimate kinase like protein 33 0.13
At3g43330 putative protein 29 1.9
At4g39410 putative WRKY DNA-binding protein 28 3.3
At4g05360 putative protein 28 5.6
At5g09420 putative subunit of TOC complex 27 9.6
At4g24010 putative protein 27 9.6
>At5g23140 ATP-dependent protease proteolytic subunit ClpP-like
protein
Length = 241
Score = 377 bits (969), Expect = e-105
Identities = 193/241 (80%), Positives = 211/241 (87%), Gaps = 4/241 (1%)
Query: 1 MRGLFGITKTLFNGTKSSFSPC---RNRAYSLIPMVIETSSRGERAYDIFSRLLKERIVC 57
MRGL K L + T SS + R+YSLIPMVIE SSRGERAYDIFSRLLKERI+C
Sbjct: 2 MRGLVSGAKML-SSTPSSMATSIATGRRSYSLIPMVIEHSSRGERAYDIFSRLLKERIIC 60
Query: 58 INGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPINTI 117
INGPI+DDT+HVVVAQLL+LESENPSKPI+MYLNSPGG VTAGLAIYDTMQYIRSPI+TI
Sbjct: 61 INGPINDDTSHVVVAQLLYLESENPSKPIHMYLNSPGGHVTAGLAIYDTMQYIRSPISTI 120
Query: 118 CLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAIHTKQIVRMWDALN 177
CLGQAASM SLLL AGAKGQRR+LPNAT+MIHQPSGGYSGQAKDI IHTKQIVR+WDALN
Sbjct: 121 CLGQAASMASLLLAAGAKGQRRSLPNATVMIHQPSGGYSGQAKDITIHTKQIVRVWDALN 180
Query: 178 ELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVADEGKDKGS 237
ELYVKHTGQPLDV+ NMDRD+FMT EEAK FGIIDEVID+RP+ LV DAV +E KDK S
Sbjct: 181 ELYVKHTGQPLDVVANNMDRDHFMTPEEAKAFGIIDEVIDERPLELVKDAVGNESKDKSS 240
Query: 238 N 238
+
Sbjct: 241 S 241
>At1g02560 ATP-dependent clp protease proteolytic subunit (nClpP1)
Length = 298
Score = 185 bits (470), Expect = 2e-47
Identities = 91/176 (51%), Positives = 128/176 (72%)
Query: 41 ERAYDIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAG 100
ER I S+L + RI+ G + DD A+++VAQLL+L++ +P+K I MY+NSPGG+VTAG
Sbjct: 110 ERFQSIISQLFQYRIIRCGGAVDDDMANIIVAQLLYLDAVDPTKDIVMYVNSPGGSVTAG 169
Query: 101 LAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAK 160
+AI+DTM++IR ++T+C+G AASMG+ LL AG KG+R +LPN+ IMIHQP GG G
Sbjct: 170 MAIFDTMRHIRPDVSTVCVGLAASMGAFLLSAGTKGKRYSLPNSRIMIHQPLGGAQGGQT 229
Query: 161 DIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVI 216
DI I +++ LN HTGQ L+ I ++ DRD+FM+A+EAKE+G+ID VI
Sbjct: 230 DIDIQANEMLHHKANLNGYLAYHTGQSLEKINQDTDRDFFMSAKEAKEYGLIDGVI 285
>At1g66670 nClpP3 protein
Length = 309
Score = 162 bits (409), Expect = 2e-40
Identities = 75/181 (41%), Positives = 126/181 (69%)
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D + LL++RIV + + D TA +V++QLL L++E+ + I +++NSPGG++TAG+ IY
Sbjct: 85 DTTNMLLRQRIVFLGSQVDDMTADLVISQLLLLDAEDSERDITLFINSPGGSITAGMGIY 144
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAI 164
D M+ ++ ++T+CLG AASMG+ LL +G+KG+R +PN+ +MIHQP G G+A +++I
Sbjct: 145 DAMKQCKADVSTVCLGLAASMGAFLLASGSKGKRYCMPNSKVMIHQPLGTAGGKATEMSI 204
Query: 165 HTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLV 224
++++ LN+++ + TG+P I+ + DRD F+ EAKE+G+ID VID L+
Sbjct: 205 RIREMMYHKIKLNKIFSRITGKPESEIESDTDRDNFLNPWEAKEYGLIDAVIDDGKPGLI 264
Query: 225 S 225
+
Sbjct: 265 A 265
>At5g45390 ATP-dependent Clp protease-like protein
Length = 292
Score = 157 bits (398), Expect = 4e-39
Identities = 79/183 (43%), Positives = 124/183 (67%), Gaps = 1/183 (0%)
Query: 35 ETSSRGERAYDIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG 94
E++ RG + D+ LL+ERIV + I D A +++QLL L++++P K I +++NSPG
Sbjct: 70 ESAIRGAES-DVMGLLLRERIVFLGSSIDDFVADAIMSQLLLLDAKDPKKDIKLFINSPG 128
Query: 95 GAVTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGG 154
G+++A +AIYD +Q +R+ ++TI LG AAS S++L AG KG+R A+PN IMIHQP GG
Sbjct: 129 GSLSATMAIYDVVQLVRADVSTIALGIAASTASIILGAGTKGKRFAMPNTRIMIHQPLGG 188
Query: 155 YSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDE 214
SGQA D+ I K+++ + + + T + + + K++DRD +M+ EA E+G+ID
Sbjct: 189 ASGQAIDVEIQAKEVMHNKNNVTSIIAGCTSRSFEQVLKDIDRDRYMSPIEAVEYGLIDG 248
Query: 215 VID 217
VID
Sbjct: 249 VID 251
>clpP -chloroplast genome- ATP-dependent protease subunit
Length = 196
Score = 144 bits (364), Expect = 3e-35
Identities = 67/175 (38%), Positives = 118/175 (67%), Gaps = 1/175 (0%)
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
DI++RL +ER+ + + + ++ +++ +++L E +K + +++NSPGG V +G+AIY
Sbjct: 22 DIYNRLYRERLFFLGQEVDTEISNQLISLMIYLSIEKDTKDLYLFINSPGGWVISGMAIY 81
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQP-SGGYSGQAKDIA 163
DTMQ++R + TIC+G AAS+ S +L GA +R A P+A +MIHQP S Y Q +
Sbjct: 82 DTMQFVRPDVQTICMGLAASIASFILVGGAITKRIAFPHARVMIHQPASSFYEAQTGEFI 141
Query: 164 IHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQ 218
+ ++++++ + + +YV+ TG+P+ VI ++M+RD FM+A EA+ GI+D V Q
Sbjct: 142 LEAEELLKLRETITRVYVQRTGKPIWVISEDMERDVFMSATEAQAHGIVDLVAVQ 196
>At1g11750 putative ATP-dependent Clp protease proteolytic subunit
Length = 271
Score = 144 bits (362), Expect = 5e-35
Identities = 71/173 (41%), Positives = 105/173 (60%)
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D+ S L + RI+ I PI+ A V++QL+ L S + I MYLN PGG+ + LAIY
Sbjct: 96 DLSSVLFRNRIIFIGQPINAQVAQRVISQLVTLASIDDKSDILMYLNCPGGSTYSVLAIY 155
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAI 164
D M +I+ + T+ G AAS G+LLL G KG R A+PN +MIHQP G G +D+
Sbjct: 156 DCMSWIKPKVGTVAFGVAASQGALLLAGGEKGMRYAMPNTRVMIHQPQTGCGGHVEDVRR 215
Query: 165 HTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVID 217
+ + ++ +Y TGQPL+ +Q+ +RD F++A EA EFG+ID +++
Sbjct: 216 QVNEAIEARQKIDRMYAAFTGQPLEKVQQYTERDRFLSASEALEFGLIDGLLE 268
>At1g12410 ClpP protease complex subunit ClpR2
Length = 279
Score = 140 bits (352), Expect = 8e-34
Identities = 73/183 (39%), Positives = 116/183 (62%), Gaps = 2/183 (1%)
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
DI++ L +ER++ I I ++ ++ ++A +L+L++ + S+ I MYLN PGG +T LAIY
Sbjct: 89 DIWNALYRERVIFIGQNIDEEFSNQILATMLYLDTLDDSRRIYMYLNGPGGDLTPSLAIY 148
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAI 164
DTM+ ++SP+ T C+G A ++ LL AG KG R A+P + I + P+G GQA DI
Sbjct: 149 DTMKSLKSPVGTHCVGLAYNLAGFLLAAGEKGHRFAMPLSRIALQSPAGAARGQADDIQN 208
Query: 165 HTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLV 224
K++ R+ D L K+TGQP + + K++ R AEEA E+G+ID+++ RP +
Sbjct: 209 EAKELSRIRDYLFNELAKNTGQPAERVFKDLSRVKRFNAEEAIEYGLIDKIV--RPPRIK 266
Query: 225 SDA 227
DA
Sbjct: 267 EDA 269
>At4g17040 ClpP protease complex subunit ClpR4
Length = 305
Score = 136 bits (343), Expect = 9e-33
Identities = 80/208 (38%), Positives = 118/208 (56%), Gaps = 9/208 (4%)
Query: 17 SSFSPCRNRAYSLIPMVIETSSRGERAYDIFSRLLKERIVCINGPISDDTAHVVVAQLLF 76
SS P R +IP + S+ + D+ S L K RIV + + +++A+ L+
Sbjct: 78 SSSKPKRGVVTMVIPFS-KGSAHEQPPPDLASYLFKNRIVYLGMSLVPSVTELILAEFLY 136
Query: 77 LESENPSKPINMYLNSPG--------GAVTAGLAIYDTMQYIRSPINTICLGQAASMGSL 128
L+ E+ KPI +Y+NS G G T AIYD M Y++ PI T+C+G A +L
Sbjct: 137 LQYEDEEKPIYLYINSTGTTKNGEKLGYDTEAFAIYDVMGYVKPPIFTLCVGNAWGEAAL 196
Query: 129 LLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPL 188
LL AGAKG R ALP++TIMI QP + GQA D+ I K+I + + +LY KH G+
Sbjct: 197 LLTAGAKGNRSALPSSTIMIKQPIARFQGQATDVEIARKEIKHIKTEMVKLYSKHIGKSP 256
Query: 189 DVIQKNMDRDYFMTAEEAKEFGIIDEVI 216
+ I+ +M R + + EA E+GIID+V+
Sbjct: 257 EQIEADMKRPKYFSPTEAVEYGIIDKVV 284
>At1g09130 ClpP protease complex subunit ClpR3
Length = 330
Score = 136 bits (342), Expect = 1e-32
Identities = 76/204 (37%), Positives = 117/204 (57%), Gaps = 11/204 (5%)
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG---------G 95
D+ S LL RIV I P+ +VVA+L++L+ +P +PI +Y+NS G G
Sbjct: 121 DLPSMLLDGRIVYIGMPLVPAVTELVVAELMYLQWLDPKEPIYIYINSTGTTRDDGETVG 180
Query: 96 AVTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGY 155
+ G AIYD++ +++ ++T+C+G A LLL AG KG+R +P+A MI QP
Sbjct: 181 MESEGFAIYDSLMQLKNEVHTVCVGAAIGQACLLLSAGTKGKRFMMPHAKAMIQQPRVPS 240
Query: 156 SG--QAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIID 213
SG A D+ I K+++ D L EL KHTG ++ + M R Y+M A +AKEFG+ID
Sbjct: 241 SGLMPASDVLIRAKEVITNRDILVELLSKHTGNSVETVANVMRRPYYMDAPKAKEFGVID 300
Query: 214 EVIDQRPMTLVSDAVADEGKDKGS 237
++ + +++D V E DK +
Sbjct: 301 RILWRGQEKIIADVVPSEEFDKNA 324
>At1g49970 ClpP protease complex subunit ClpR1
Length = 387
Score = 100 bits (249), Expect = 7e-22
Identities = 61/185 (32%), Positives = 100/185 (53%), Gaps = 10/185 (5%)
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG---------G 95
D+ S LL RI + PI ++VAQ ++L+ +NP+KPI +Y+NSPG G
Sbjct: 171 DLPSLLLDARICYLGMPIVPAVTELLVAQFMWLDYDNPTKPIYLYINSPGTQNEKMETVG 230
Query: 96 AVTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPS-GG 154
+ T AI DT+ Y +S + TI G A ++LL G KG R P+++ ++ P
Sbjct: 231 SETEAYAIADTISYCKSDVYTINCGMAFGQAAMLLSLGKKGYRAVQPHSSTKLYLPKVNR 290
Query: 155 YSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDE 214
SG A D+ I K++ + EL K TG+ + I +++ R ++ A+ A ++GI D+
Sbjct: 291 SSGAAIDMWIKAKELDANTEYYIELLAKGTGKSKEQINEDIKRPKYLQAQAAIDYGIADK 350
Query: 215 VIDQR 219
+ D +
Sbjct: 351 IADSQ 355
>At3g26900 shikimate kinase like protein
Length = 280
Score = 33.1 bits (74), Expect = 0.13
Identities = 33/134 (24%), Positives = 60/134 (44%), Gaps = 12/134 (8%)
Query: 55 IVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPI 114
+V IN I +T ++ L + ++ N+ + GG V+A A+ + +
Sbjct: 94 LVGINNSIKTNTGKLLAEALRYYYFDSD----NLITEAAGGNVSAQ-ALKEADEKAFQES 148
Query: 115 NTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPS------GGYSGQAKDIAIHTKQ 168
T L Q +SMG L++CAG G ++L N ++ H S + + D + H++
Sbjct: 149 ETEVLKQLSSMGRLVVCAG-DGAVQSLRNLALLRHGISIWIDVPLDITAKGDDDSFHSEP 207
Query: 169 IVRMWDALNELYVK 182
++D L Y K
Sbjct: 208 SPELFDTLKASYEK 221
>At3g43330 putative protein
Length = 489
Score = 29.3 bits (64), Expect = 1.9
Identities = 12/45 (26%), Positives = 25/45 (54%), Gaps = 1/45 (2%)
Query: 192 QKNMDRDYFMTA-EEAKEFGIIDEVIDQRPMTLVSDAVADEGKDK 235
++N+ D++ ++ EE + +DE ++ M + D + E KDK
Sbjct: 78 EENLQEDFYWSSQEECSSYSSLDEDVNAEDMAQIPDDIIYEAKDK 122
>At4g39410 putative WRKY DNA-binding protein
Length = 304
Score = 28.5 bits (62), Expect = 3.3
Identities = 14/52 (26%), Positives = 27/52 (51%)
Query: 95 GAVTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATI 146
GA+ G++++D Q + +PINT LG S S + + + P++ +
Sbjct: 2 GAINQGISLFDESQTVINPINTNHLGFFFSFPSHSTLSSSSSSSSSSPSSLV 53
>At4g05360 putative protein
Length = 735
Score = 27.7 bits (60), Expect = 5.6
Identities = 21/78 (26%), Positives = 39/78 (49%), Gaps = 9/78 (11%)
Query: 148 IHQPSGGYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAK 207
+ Q SGG+SG+ + +A H K+ + +LN++ T +K R ++ T E
Sbjct: 268 VTQDSGGFSGRTRALAQHLKERYSGYLSLNKILEGKT-------RKIAARMFYETLGEVP 320
Query: 208 EFGIIDEVIDQRPMTLVS 225
E I+ ++I + +L S
Sbjct: 321 E--IVQKIIQEVRQSLKS 336
>At5g09420 putative subunit of TOC complex
Length = 616
Score = 26.9 bits (58), Expect = 9.6
Identities = 21/82 (25%), Positives = 35/82 (42%), Gaps = 12/82 (14%)
Query: 31 PMVIETSSRGERAYDIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYL 90
P +T E+ + + LLK C+ I D+ ++ E+++ PIN +
Sbjct: 94 PQWKKTHEAAEKTAVVVTTLLKNGATCVGKTIMDELGFGIIG-----ENKHYGTPINPLM 148
Query: 91 --NSPGG-----AVTAGLAIYD 105
N PGG AV+ G + D
Sbjct: 149 PDNVPGGCSSGSAVSVGAELVD 170
>At4g24010 putative protein
Length = 727
Score = 26.9 bits (58), Expect = 9.6
Identities = 13/54 (24%), Positives = 25/54 (46%)
Query: 56 VCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQY 109
+C P + VV L + E PS I++Y++ GG+ A+ + ++
Sbjct: 105 ICTADPYKEPPMMVVNTALSVMAYEYPSDKISVYVSDDGGSSLTFFALIEAAKF 158
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,159,006
Number of Sequences: 26719
Number of extensions: 209825
Number of successful extensions: 447
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 429
Number of HSP's gapped (non-prelim): 16
length of query: 238
length of database: 11,318,596
effective HSP length: 96
effective length of query: 142
effective length of database: 8,753,572
effective search space: 1243007224
effective search space used: 1243007224
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146777.12


