

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146777.10 - phase: 1 /pseudo
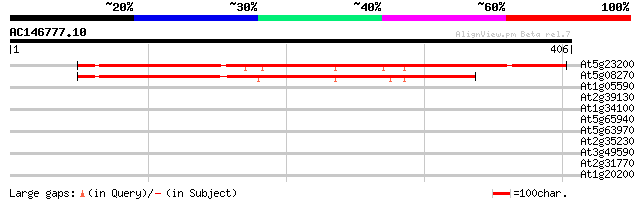
(406 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g23200 unknown protein 372 e-103
At5g08270 hypothetical protein (fragment at BAC end) 295 2e-80
At1g05590 hypothetical protein 33 0.22
At2g39130 unknown protein 30 2.4
At1g34100 putative protein 29 5.3
At5g65940 3-hydroxyisobutyryl-coenzyme A hydrolase 28 6.9
At5g63970 unknown protein 28 6.9
At2g35230 unknown protein 28 6.9
At3g49590 unknown protein 28 9.0
At2g31770 putative ARI-like RING zinc finger protein 28 9.0
At1g20200 proteasome regulatory subunit S3 like protein 28 9.0
>At5g23200 unknown protein
Length = 399
Score = 372 bits (955), Expect = e-103
Identities = 202/392 (51%), Positives = 256/392 (64%), Gaps = 46/392 (11%)
Query: 50 QAVKTLAKSPTFAFARDPRQLQYQIDLNRLFLYTSYNRLGKNASEADAEEIIEIATKASV 109
QAV LAKS TFA ++PRQLQ++ D+N+LF+YTSYNRLG+ A E DAEEIIE+A KA++
Sbjct: 10 QAVGVLAKSTTFA--KNPRQLQFEADVNKLFMYTSYNRLGREAEETDAEEIIEMAGKATL 67
Query: 110 ADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVNDVSFELSQQTKILPQHSDLRSANGKF 169
++QQ VQEN+H Q++ FC+ MD + LP+ ++ S S + P+ S L A G
Sbjct: 68 SEQQKQVQENIHYQVEKFCSLMDGILLPDVRRNESGSQSTSPRP---PRRSGLTFAIGSN 124
Query: 170 ---------VIHQLTILKLKD-------ELGYTLNVKPSQISHKDAGQGLFLDGVVDVGA 213
++ + LKL D ++GYTL KPS I HKDAGQG F+ G DVG
Sbjct: 125 NAFPHADQPLVPETKPLKLNDVSQRLMDQMGYTLETKPSVIPHKDAGQGCFIKGEADVGT 184
Query: 214 VVAFYPGVVYSPAYYHHIPGY--LDEQNPYLITRHDGNVIDAQLWGRGGDKKELWNGR-- 269
V+AFYPGV+YSPA+Y +IPGY +D QN YLITR+DG VI+AQ WG GG+ +E WNG
Sbjct: 185 VLAFYPGVIYSPAFYKYIPGYPKVDSQNSYLITRYDGTVINAQPWGLGGESREAWNGSYT 244
Query: 270 KMVDEKGSQVDNSDD------------------VLERRNPLALAHFANHPSKGMLPNVMI 311
V +N D VLERRNPLA H ANHP+K M PNVMI
Sbjct: 245 PAVKANTKTAENGSDRLWKALSKPLEGSGKGKEVLERRNPLAFGHLANHPAKEMTPNVMI 304
Query: 312 CPYDFPLIENDMRAYIPNVLFGNAAEENTERFGSFWFKSRVPRNNESHVPTTLKTVVLVA 371
CPYDFPL+ D+R YIPN+ FG++ E +RFGSFWFK+ E+ V LKT+VLVA
Sbjct: 305 CPYDFPLMAKDLRPYIPNISFGDSGEIKMKRFGSFWFKTGSKNGLEAPV---LKTLVLVA 361
Query: 372 TRALQDEELLLNYRLGNTKRCPEWYAPVDEEE 403
TR+L +EELLLNYRL N+KR P+WY PV+EEE
Sbjct: 362 TRSLCNEELLLNYRLSNSKRRPDWYTPVNEEE 393
>At5g08270 hypothetical protein (fragment at BAC end)
Length = 325
Score = 295 bits (756), Expect = 2e-80
Identities = 159/318 (50%), Positives = 208/318 (65%), Gaps = 37/318 (11%)
Query: 50 QAVKTLAKSPTFAFARDPRQLQYQIDLNRLFLYTSYNRLGKNASEADAEEIIEIATKASV 109
Q V LAKS TFA ++PRQLQ++ D+N+LF++TSYNRLG++A EADAEEIIE+A KA+
Sbjct: 15 QGVGVLAKSTTFA--KNPRQLQFEADINKLFMFTSYNRLGRDAEEADAEEIIEMAGKATF 72
Query: 110 ADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVNDVSFELSQQTKILPQHSDLRSANGKF 169
++QQ VQEN+H Q++ FC+ M+ + L N K D++ S+ + ++ A K
Sbjct: 73 SEQQKQVQENIHYQLQNFCSSMNEILLTNIDKTKDLNEPGSES-----RRDNIVPAADKP 127
Query: 170 VIHQLTILK-------LKDELGYTLNVKPSQISHKDAGQGLFLDGVVDVGAVVAFYPGVV 222
++ + LK LKD +GYTL +KPS I HKDAGQG F++G DVGAV+AFYPGV+
Sbjct: 128 IVPETKPLKLDEVSHRLKDRVGYTLEIKPSLIPHKDAGQGCFIEGEADVGAVLAFYPGVI 187
Query: 223 YSPAYYHHIPGY--LDEQNPYLITRHDGNVIDAQLWGRGGDKKELWNGRKMVDE---KGS 277
YSPA++ +IPGY +D QN YLIT +DG VI+AQ WGRGG +E+WNG E + +
Sbjct: 188 YSPAFHRYIPGYPNVDAQNSYLITGYDGTVINAQPWGRGGISREVWNGSFTTPEIRTEAT 247
Query: 278 QVDNSDD------------------VLERRNPLALAHFANHPSKGMLPNVMICPYDFPLI 319
+NS D V+E RNPLA HF NHP K M NVMICPYDF L
Sbjct: 248 TTENSSDKVWKMLSRPLEGSGGVEAVIEMRNPLAFGHFINHPGKEMESNVMICPYDFLLS 307
Query: 320 ENDMRAYIPNVLFGNAAE 337
E +MRAYIPNV FGN +
Sbjct: 308 ETEMRAYIPNVAFGNTGD 325
>At1g05590 hypothetical protein
Length = 580
Score = 33.5 bits (75), Expect = 0.22
Identities = 35/123 (28%), Positives = 54/123 (43%), Gaps = 20/123 (16%)
Query: 184 GYTLNVKPSQISHKDAGQGLFL--DGVVD---VGAVVAFYPGVVYSPAYYHHI-PGYLDE 237
GY + V S+ + D G G FL D + D G P + Y + I G L+E
Sbjct: 424 GYRVIVSSSEFYYLDCGHGGFLGNDSIYDQKESGGGSWCAPFKTWQSIYNYDIADGLLNE 483
Query: 238 QNPYLI---------TRHDGNVIDAQLWGRGGDKKE-LWNGRKMVDEKGSQVDNSDDVLE 287
+ L+ + D V+D++LW R E LW+G + DE+G V + ++
Sbjct: 484 EERKLVLGGEVALWSEQADSTVLDSRLWPRASALAESLWSGNR--DERG--VKRCGEAVD 539
Query: 288 RRN 290
R N
Sbjct: 540 RLN 542
>At2g39130 unknown protein
Length = 550
Score = 30.0 bits (66), Expect = 2.4
Identities = 30/116 (25%), Positives = 46/116 (38%), Gaps = 14/116 (12%)
Query: 273 DEKGSQVDNSDDVLERRNPLALAHFANHPSKGMLPNVMICPYDFPLIENDMRAYIPNVLF 332
++ GS D+SDDV + AH P Y + D+ + +P+
Sbjct: 26 EDGGSHSDSSDDVYDENQ----AHIKPSSYTTAWPQ----SYRQSI---DLYSSVPSPGI 74
Query: 333 GNAAEENTERFGSFWFKSRVPRNNESHVPTTLKTVVLVATRALQDEELLLNYRLGN 388
G + RFGS + S + R H P +L TV DE+ L +RL +
Sbjct: 75 GFLGNNSMTRFGSSFLSSGLIRR---HTPESLPTVTKPLLEEQADEQALPKHRLSS 127
>At1g34100 putative protein
Length = 672
Score = 28.9 bits (63), Expect = 5.3
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 11/83 (13%)
Query: 216 AFYPGVVYSPAYYHHIPGYLDEQNPYLITRHDGNVIDAQLWGRGGDKKELWNGR-----K 270
AF G + Y + PG++D +NP + + ++ L+G WN R K
Sbjct: 127 AFLHGKLEETVYMYEPPGFVDNKNPGYVCK-----LNKALYG-FKQAPRAWNARFASYVK 180
Query: 271 MVDEKGSQVDNSDDVLERRNPLA 293
M+ K S+ D S V ++ N +A
Sbjct: 181 MMGFKQSKCDASLFVYKQGNDMA 203
>At5g65940 3-hydroxyisobutyryl-coenzyme A hydrolase
Length = 378
Score = 28.5 bits (62), Expect = 6.9
Identities = 15/45 (33%), Positives = 23/45 (50%)
Query: 261 DKKELWNGRKMVDEKGSQVDNSDDVLERRNPLALAHFANHPSKGM 305
DK W R++ D K S V+ + +ER + L L N P+ G+
Sbjct: 331 DKNPKWEPRRLEDMKDSMVEQYFERVEREDDLKLPPRNNLPALGI 375
>At5g63970 unknown protein
Length = 367
Score = 28.5 bits (62), Expect = 6.9
Identities = 14/52 (26%), Positives = 25/52 (47%)
Query: 146 SFELSQQTKILPQHSDLRSANGKFVIHQLTILKLKDELGYTLNVKPSQISHK 197
+F+ TKI+ +H D F + L + + + +LN KP + SH+
Sbjct: 229 NFQFVNFTKIMSEHKDAAKKEAAFALAALMEIPFQYKATLSLNRKPVRSSHQ 280
>At2g35230 unknown protein
Length = 402
Score = 28.5 bits (62), Expect = 6.9
Identities = 16/55 (29%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query: 288 RRNPLALAHFANHPSKGMLPNVMICPYDFPLIENDMRAYIPNVLFGNAAEENTER 342
R++PL + FA S G P + P + + +ND R+ + L G+ + E+ R
Sbjct: 20 RKSPLHQSTFAASTSNGAAPRLQTQPQVYNISKNDFRSIVQQ-LTGSPSRESLPR 73
>At3g49590 unknown protein
Length = 603
Score = 28.1 bits (61), Expect = 9.0
Identities = 13/29 (44%), Positives = 18/29 (61%)
Query: 49 FQAVKTLAKSPTFAFARDPRQLQYQIDLN 77
F V + + SP FAF+R P +L Q DL+
Sbjct: 449 FSGVLSSSDSPRFAFSRSPSRLSSQDDLD 477
>At2g31770 putative ARI-like RING zinc finger protein
Length = 543
Score = 28.1 bits (61), Expect = 9.0
Identities = 15/49 (30%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Query: 355 NNESHVPTTLKTVVLVATRALQDEELLLNYRLGNTKRCPEWYAPVDEEE 403
+ ++H P TV + QDE N+ L N+K CPE P+++ +
Sbjct: 251 SEDAHSPVDCDTVSKWIFKN-QDESENKNWMLANSKPCPECKRPIEKND 298
>At1g20200 proteasome regulatory subunit S3 like protein
Length = 488
Score = 28.1 bits (61), Expect = 9.0
Identities = 16/61 (26%), Positives = 35/61 (57%), Gaps = 3/61 (4%)
Query: 84 SYNRLGKNASEADAEEIIEIATKASVADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVN 143
+++RL + D E +E+ T +S Q +++ A+++ +C F+ +FL ++KK N
Sbjct: 80 AHSRLSSFVPKGD-EHDMEVDTASSAT--QAAPSKHLPAELEIYCYFIVLLFLIDQKKYN 136
Query: 144 D 144
+
Sbjct: 137 E 137
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,341,671
Number of Sequences: 26719
Number of extensions: 411058
Number of successful extensions: 773
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 760
Number of HSP's gapped (non-prelim): 11
length of query: 406
length of database: 11,318,596
effective HSP length: 102
effective length of query: 304
effective length of database: 8,593,258
effective search space: 2612350432
effective search space used: 2612350432
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146777.10


