

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146776.7 + phase: 0
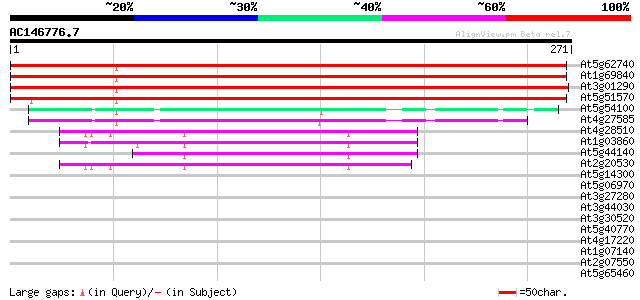
(271 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g62740 unknown protein 459 e-130
At1g69840 unknown protein 431 e-121
At3g01290 unknown protein 407 e-114
At5g51570 unknown protein 299 1e-81
At5g54100 unknown protein 54 1e-07
At4g27585 unknown protein 50 1e-06
At4g28510 prohibitin-like protein 42 3e-04
At1g03860 putative prohibitin 2 42 3e-04
At5g44140 prohibitin 41 6e-04
At2g20530 putative prohibitin 41 8e-04
At5g14300 prohibitin - like protein 37 0.015
At5g06970 unknown protein 35 0.056
At3g27280 prohibitin, putative 33 0.16
At3g44030 putative protein 32 0.36
At3g30520 hypothetical protein 32 0.47
At5g40770 prohibitin (gb|AAC49691.1) 31 0.62
At4g17220 unknown protein 31 0.80
At1g07140 Unknown protein (F10K1.15) 30 1.1
At2g07550 putative retroelement pol polyprotein 30 1.4
At5g65460 putative protein 29 3.1
>At5g62740 unknown protein
Length = 286
Score = 459 bits (1182), Expect = e-130
Identities = 238/282 (84%), Positives = 253/282 (89%), Gaps = 13/282 (4%)
Query: 1 MGNIVCCVQVDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR---------- 50
MGN+ CCVQVDQS VA+KE FGKFE VL PGCH +PW LG ++AG+LSLR
Sbjct: 1 MGNLFCCVQVDQSTVAIKETFGKFEDVLEPGCHFLPWCLGSQVAGYLSLRVQQLDVRCET 60
Query: 51 ---DNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQ 107
DNVFVNVVASIQYRALANKANDA+YKLSNTRGQIQAYVFDVIRASVPKL LDD FEQ
Sbjct: 61 KTKDNVFVNVVASIQYRALANKANDAYYKLSNTRGQIQAYVFDVIRASVPKLLLDDVFEQ 120
Query: 108 KNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEA 167
KN+IAKAVEEE EKAMSAYGYEIVQTLI DIEPD+HVK AMNEINAAARMR+AANEKAEA
Sbjct: 121 KNDIAKAVEEELEKAMSAYGYEIVQTLIVDIEPDEHVKRAMNEINAAARMRLAANEKAEA 180
Query: 168 EKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVPGPSAKDVMDMVLVT 227
EKILQIKRAEGEAESKYLSG+GIARQRQAIVDGLRDSV+GF+ NVPG +AKDVMDMVLVT
Sbjct: 181 EKILQIKRAEGEAESKYLSGLGIARQRQAIVDGLRDSVLGFAVNVPGTTAKDVMDMVLVT 240
Query: 228 QYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGLLQGS 269
QYFDTMKEIGA+SKSSAVFIPHGPGAVRDVASQI DGLLQGS
Sbjct: 241 QYFDTMKEIGASSKSSAVFIPHGPGAVRDVASQIRDGLLQGS 282
>At1g69840 unknown protein
Length = 286
Score = 431 bits (1107), Expect = e-121
Identities = 222/282 (78%), Positives = 244/282 (85%), Gaps = 13/282 (4%)
Query: 1 MGNIVCCVQVDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR---------- 50
MG + C+QVDQS VA+KE FGKF++VL PGCHC+PW LG ++AGHLSLR
Sbjct: 1 MGQALGCIQVDQSNVAIKETFGKFDEVLEPGCHCLPWCLGSQVAGHLSLRVQQLDVRCET 60
Query: 51 ---DNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQ 107
DNVFV VVASIQYRALA A DAFYKLSNTR QIQAYVFDVIRASVPKL+LD TFEQ
Sbjct: 61 KTKDNVFVTVVASIQYRALAESAQDAFYKLSNTRNQIQAYVFDVIRASVPKLDLDSTFEQ 120
Query: 108 KNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEA 167
KN+IAK VE E EKAMS YGYEIVQTLI DIEPD HVK AMNEINAA+RMR AA+EKAEA
Sbjct: 121 KNDIAKTVETELEKAMSHYGYEIVQTLIVDIEPDVHVKRAMNEINAASRMREAASEKAEA 180
Query: 168 EKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVPGPSAKDVMDMVLVT 227
EKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLR+SV+ FSE+VPG S+KDVMDMVLVT
Sbjct: 181 EKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRNSVLAFSESVPGTSSKDVMDMVLVT 240
Query: 228 QYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGLLQGS 269
QYFDT+KEIGA+SKS++VFIPHGPGAVRD+ASQI DGLLQG+
Sbjct: 241 QYFDTLKEIGASSKSNSVFIPHGPGAVRDIASQIRDGLLQGN 282
>At3g01290 unknown protein
Length = 285
Score = 407 bits (1047), Expect = e-114
Identities = 210/283 (74%), Positives = 241/283 (84%), Gaps = 13/283 (4%)
Query: 1 MGNIVCCVQVDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR---------- 50
MGN+ CCV V QS VA+KE FGKF+KVL+PG +PW +G +AG L+LR
Sbjct: 1 MGNLFCCVLVKQSDVAVKERFGKFQKVLNPGLQFVPWVIGDYVAGTLTLRLQQLDVQCET 60
Query: 51 ---DNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQ 107
DNVFV VVASIQYR LA+KA+DAFY+LSN QI+AYVFDVIRA VPKLNLDD FEQ
Sbjct: 61 KTKDNVFVTVVASIQYRVLADKASDAFYRLSNPTTQIKAYVFDVIRACVPKLNLDDVFEQ 120
Query: 108 KNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEA 167
KNEIAK+VEEE +KAM+AYGYEI+QTLI DIEPDQ VK AMNEINAAARMR+AA+EKAEA
Sbjct: 121 KNEIAKSVEEELDKAMTAYGYEILQTLIIDIEPDQQVKRAMNEINAAARMRVAASEKAEA 180
Query: 168 EKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVPGPSAKDVMDMVLVT 227
EKI+QIKRAEGEAESKYLSG+GIARQRQAIVDGLRDSV+GF+ NVPG SAKDV+DMV++T
Sbjct: 181 EKIIQIKRAEGEAESKYLSGLGIARQRQAIVDGLRDSVLGFAGNVPGTSAKDVLDMVMMT 240
Query: 228 QYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGLLQGSH 270
QYFDTM++IGA SKSSAVFIPHGPGAV DVA+QI +GLLQ ++
Sbjct: 241 QYFDTMRDIGATSKSSAVFIPHGPGAVSDVAAQIRNGLLQANN 283
>At5g51570 unknown protein
Length = 292
Score = 299 bits (765), Expect = 1e-81
Identities = 150/284 (52%), Positives = 204/284 (71%), Gaps = 15/284 (5%)
Query: 1 MGNIVCCVQ--VDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR-------- 50
MGN C + ++Q+ V + E +G+FE + PGCH G+ +AG LS R
Sbjct: 1 MGNTYCILGGCIEQASVGVVERWGRFEHIAEPGCHFFNPLAGQWLAGVLSTRIKSLDVKI 60
Query: 51 -----DNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTF 105
DNVFV +V SIQYR + A+DAFY+L N + QIQAYVFDV+RA VP + LD F
Sbjct: 61 ETKTKDNVFVQLVCSIQYRVVKASADDAFYELQNPKEQIQAYVFDVVRALVPMMTLDALF 120
Query: 106 EQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKA 165
EQK E+AK+V EE EK M AYGY I L+ DI PD V+ AMNEINAA R+++A+ K
Sbjct: 121 EQKGEVAKSVLEELEKVMGAYGYSIEHILMVDIIPDPSVRKAMNEINAAQRLQLASVYKG 180
Query: 166 EAEKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVPGPSAKDVMDMVL 225
EAEKILQ+KRAE EAE+KYL G+G+ARQRQAI DGLR++++ FS+ V G SAK+VMD+++
Sbjct: 181 EAEKILQVKRAEAEAEAKYLGGVGVARQRQAITDGLRENILNFSDKVEGTSAKEVMDLIM 240
Query: 226 VTQYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGLLQGS 269
+TQYFDT++++G +SK++ VF+PHGPG VRD++ QI +G+++ +
Sbjct: 241 ITQYFDTIRDLGNSSKNTTVFLPHGPGHVRDISDQIRNGMMEAA 284
>At5g54100 unknown protein
Length = 401
Score = 53.5 bits (127), Expect = 1e-07
Identities = 67/292 (22%), Positives = 118/292 (39%), Gaps = 55/292 (18%)
Query: 10 VDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR--------------DNVFV 55
V + + + E FGKF L G H + F+ RIA SL+ DNV +
Sbjct: 109 VPERKACVIERFGKFHTTLPAGIHFLVPFVD-RIAYVHSLKEEAIPIGNQTAITKDNVSI 167
Query: 56 NVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAV 115
++ + + + K A Y + N + +R+ + K+ LD TFE+++ + + +
Sbjct: 168 HIDGVLYVKIVDPKL--ASYGVENPIYAVMQLAQTTMRSELGKITLDKTFEERDTLNEKI 225
Query: 116 EEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMN----------------------EINA 153
E A +G + ++ I DI P V+ AM IN
Sbjct: 226 VEAINVAAKDWGLQCLRYEIRDIMPPNGVRVAMEMQAEAERKKRAQILESEGERQAHINR 285
Query: 154 AARMRIAANEKAEAEKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVP 213
A + + ++EA + Q+ RA+GEAE+ I + QA GL S+++
Sbjct: 286 ADGKKSSVILESEAAMMDQVNRAQGEAEA-------ILARAQATAKGL----AMVSQSLK 334
Query: 214 GPSAKDVMDMVLVTQYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGL 265
++ + + QY +I A + + + +P V + AS I L
Sbjct: 335 EAGGEEAASLRVAEQYIQAFGKI--AKEGTTMLLPSN---VDNPASMIAQAL 381
>At4g27585 unknown protein
Length = 411
Score = 50.4 bits (119), Expect = 1e-06
Identities = 62/277 (22%), Positives = 113/277 (40%), Gaps = 52/277 (18%)
Query: 10 VDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR--------------DNVFV 55
V + + + E FGK+ L G H + F+ RIA SL+ DNV +
Sbjct: 66 VPERKAFVIERFGKYATTLPSGIHFLIPFVD-RIAYVHSLKEEAIPIPNQTAITKDNVSI 124
Query: 56 NVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAV 115
++ + + + K A Y + + + +R+ + K+ LD TFE+++ + + +
Sbjct: 125 HIDGVLYVKIVDPKL--ASYGVESPIYAVVQLAQTTMRSELGKITLDKTFEERDTLNEKI 182
Query: 116 EEEREKAMSAYGYEIVQTLITDIEPDQHVKTAM----------------------NEINA 153
E A +G + ++ I DI P V+ AM + IN
Sbjct: 183 VEAINVAAKDWGLQCLRYEIRDIMPPHGVRAAMEMQAEAERKKRAQILESEGERQSHINI 242
Query: 154 AARMRIAANEKAEAEKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVP 213
A + + +EA K+ Q+ RA+GEAE+ I + QA GL + S+++
Sbjct: 243 ADGKKSSVILASEAAKMDQVNRAQGEAEA-------ILARAQATAKGL----VLLSQSLK 291
Query: 214 GPSAKDVMDMVLVTQYFDTMKEIGAASKSSAVFIPHG 250
+ + + QY I A + + + +P G
Sbjct: 292 ETGGVEAASLRVAEQYITAFGNI--AKEGTIMLLPSG 326
>At4g28510 prohibitin-like protein
Length = 288
Score = 42.4 bits (98), Expect = 3e-04
Identities = 42/189 (22%), Positives = 79/189 (41%), Gaps = 16/189 (8%)
Query: 25 EKVLHPGCHCM-PWF-----LGKRIAGHL-----SLRDNVFVNVVASIQYRALANKANDA 73
+KV G H M PWF R +L RD V + + R +A++ +
Sbjct: 56 DKVYPEGTHLMIPWFERPVIYDVRARPYLVESTSGSRDLQMVKIGLRVLTRPMADQLPEI 115
Query: 74 FYKLSNTRGQ--IQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGYEIV 131
+ L + + + + + ++A V + N Q+ +++ + + + + + +
Sbjct: 116 YRSLGENYSERVLPSIINETLKAVVAQYNASQLITQREAVSREIRKILTERAANFNVALD 175
Query: 132 QTLITDIEPDQHVKTAMNEINAAARMRIAAN---EKAEAEKILQIKRAEGEAESKYLSGM 188
IT++ + A+ AA+ A EKAE +K + RA+GEA+S L G
Sbjct: 176 DVSITNLTFGKEFTAAIEAKQVAAQEAERAKFIVEKAEQDKRSAVIRAQGEAKSAQLIGQ 235
Query: 189 GIARQRQAI 197
IA + I
Sbjct: 236 AIANNQAFI 244
>At1g03860 putative prohibitin 2
Length = 286
Score = 42.0 bits (97), Expect = 3e-04
Identities = 44/190 (23%), Positives = 77/190 (40%), Gaps = 18/190 (9%)
Query: 25 EKVLHPGCHCM-PWFLGKRIAGHLSLRDNVFVNVVAS-------IQYRALANKANDAFYK 76
EKV G H M PWF + I + R + + S I R L D +
Sbjct: 56 EKVYPEGTHFMVPWF-ERPIIYDVRARPYLVESTTGSHDLQMVKIGLRVLTRPMGDRLPQ 114
Query: 77 LSNTRGQ------IQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGYEI 130
+ T G+ + + + + ++A V + N Q+ +++ + + + S + +
Sbjct: 115 IYRTLGENYSERVLPSIIHETLKAVVAQYNASQLITQREAVSREIRKILTERASNFDIAL 174
Query: 131 VQTLITDIEPDQHVKTAMNEINAAARMRIAAN---EKAEAEKILQIKRAEGEAESKYLSG 187
IT + + A+ AA+ A EKAE ++ + RA+GEA+S L G
Sbjct: 175 DDVSITTLTFGKEFTAAIEAKQVAAQEAERAKFIVEKAEQDRRSAVIRAQGEAKSAQLIG 234
Query: 188 MGIARQRQAI 197
IA + I
Sbjct: 235 QAIANNQAFI 244
>At5g44140 prohibitin
Length = 278
Score = 41.2 bits (95), Expect = 6e-04
Identities = 34/147 (23%), Positives = 64/147 (43%), Gaps = 9/147 (6%)
Query: 60 SIQYRALANKANDAFYKLSNTRGQ------IQAYVFDVIRASVPKLNLDDTFEQKNEIAK 113
+I R L D ++ T GQ + + + + ++A V + N Q+ +++
Sbjct: 98 TIGLRVLTRPMGDRLPEIYRTLGQNYGERVLPSIINETLKAVVAQYNASHLITQREAVSR 157
Query: 114 AVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAAN---EKAEAEKI 170
+ + + + + + IT+++ + A+ + AA+ A EKAE +K
Sbjct: 158 EIRKIVTERAAKFNIALDDVSITNLKFGKEFTEAIEKKQVAAQEAERAKFIVEKAEQDKK 217
Query: 171 LQIKRAEGEAESKYLSGMGIARQRQAI 197
I RA+GEA+S L G IA I
Sbjct: 218 SAIIRAQGEAKSAQLIGQAIANNEAFI 244
>At2g20530 putative prohibitin
Length = 286
Score = 40.8 bits (94), Expect = 8e-04
Identities = 42/186 (22%), Positives = 76/186 (40%), Gaps = 16/186 (8%)
Query: 25 EKVLHPGCHCM-PWF-----LGKRIAGHL-----SLRDNVFVNVVASIQYRALANKANDA 73
+KV G H M PWF R +L RD V + + R +A++ +
Sbjct: 54 DKVYPEGTHLMIPWFERPIIYDVRAKPYLVESTSGSRDLQMVKIGLRVLTRPMADQLPEV 113
Query: 74 FYKLSNTRGQ--IQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGYEIV 131
+ L + + + + + ++A V + N Q+ +++ + + + + +
Sbjct: 114 YRSLGENYRERVLPSIIHETLKAVVAQYNASQLITQRESVSREIRKILTLRAANFHIALD 173
Query: 132 QTLITDIEPDQHVKTAMNEINAAARMRIAAN---EKAEAEKILQIKRAEGEAESKYLSGM 188
IT + + A+ AA+ A EKAE +K + RAEGEA+S L G
Sbjct: 174 DVSITGLTFGKEFTAAIEGKQVAAQEAERAKFIVEKAEQDKRSAVIRAEGEAKSAQLIGQ 233
Query: 189 GIARQR 194
IA +
Sbjct: 234 AIANNQ 239
>At5g14300 prohibitin - like protein
Length = 249
Score = 36.6 bits (83), Expect = 0.015
Identities = 30/130 (23%), Positives = 55/130 (42%), Gaps = 8/130 (6%)
Query: 90 DVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMN 149
DV++A V + N D+ ++ +++ + E K + + IT + + A+
Sbjct: 96 DVVKAVVAQFNADELLTERPQVSALIRETLIKRAKEFNIVLDDVSITGLSYGKEFSLAVE 155
Query: 150 EINAA---ARMRIAANEKAEAEKILQIKRAEGEAE-----SKYLSGMGIARQRQAIVDGL 201
A A KA+ E+ + RAEGE+E SK +G G+ + V+
Sbjct: 156 RKQVAQQEAERSKFVVAKADQERRAAVIRAEGESEAARVISKATAGAGMGLIKLRRVEAA 215
Query: 202 RDSVIGFSEN 211
R+ I S +
Sbjct: 216 REVAITLSNS 225
>At5g06970 unknown protein
Length = 1101
Score = 34.7 bits (78), Expect = 0.056
Identities = 26/97 (26%), Positives = 47/97 (47%), Gaps = 4/97 (4%)
Query: 45 GHLSLRDNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLD-D 103
G L + D V VA I +R L +++ A + S+ R QI++YV I+ + +++L D
Sbjct: 537 GSLVMEDTV---TVAMITWRLLLEESDRAMHSNSSDREQIESYVLSSIKNTFTRMSLAID 593
Query: 104 TFEQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEP 140
++ NE A+ E K + I +++ P
Sbjct: 594 RSDRNNEHHLALLAEETKKLMKKDSTIFMPILSQRHP 630
>At3g27280 prohibitin, putative
Length = 279
Score = 33.1 bits (74), Expect = 0.16
Identities = 27/117 (23%), Positives = 51/117 (43%), Gaps = 12/117 (10%)
Query: 90 DVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMN 149
+V++A V N D ++ +++ V + K + E+ IT + A+
Sbjct: 130 EVLKAVVANFNADQLLTERPQVSALVRDALIKRAREFNIELDDIAITHLSYGAEFSRAV- 188
Query: 150 EINAAARMRIAANE----KAEAEKILQIKRAEGEAESKYL-------SGMGIARQRQ 195
E A+ ++ KA+ E+ + RAEGE+E+ L +GMG+ R+
Sbjct: 189 EAKQVAQQEAERSKFVVMKADQERRAAVIRAEGESEAAQLISDATAKAGMGLIELRR 245
>At3g44030 putative protein
Length = 775
Score = 32.0 bits (71), Expect = 0.36
Identities = 29/103 (28%), Positives = 51/103 (49%), Gaps = 7/103 (6%)
Query: 115 VEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAM---NEI--NAAAR--MRIAANEKAEA 167
V+EE E + + QT + + ++ V +EI N+AAR R N ++ A
Sbjct: 184 VDEEGEDYSDSDSGNMPQTKVPETLEEEEVYRVTIDDDEIFQNSAARRQQRGRPNFQSTA 243
Query: 168 EKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSE 210
+ +R+ G ++ SG +R+RQ+ ++DS+IGF E
Sbjct: 244 RRGSSAQRSGGSSQVSIGSGSRGSRRRQSFETTIQDSIIGFRE 286
>At3g30520 hypothetical protein
Length = 305
Score = 31.6 bits (70), Expect = 0.47
Identities = 28/103 (27%), Positives = 50/103 (48%), Gaps = 7/103 (6%)
Query: 115 VEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAM---NEI--NAAAR--MRIAANEKAEA 167
V+EE E + + QT + + + ++ V +EI N+AAR R N ++ A
Sbjct: 31 VDEEGEDYSDSDSGNMPQTEVPETQEEEEVYRVTIDDDEIFQNSAARRQQRGRPNFQSTA 90
Query: 168 EKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSE 210
+ +R+ G + SG G +R+RQ+ ++DS+ F E
Sbjct: 91 RRGSSAQRSGGTSRVSIGSGSGGSRRRQSFETTIQDSITSFGE 133
>At5g40770 prohibitin (gb|AAC49691.1)
Length = 277
Score = 31.2 bits (69), Expect = 0.62
Identities = 31/139 (22%), Positives = 55/139 (39%), Gaps = 22/139 (15%)
Query: 90 DVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMN 149
+V++A V + N D ++ ++ V E + + IT H+ +
Sbjct: 130 EVLKAVVAQFNADQLLTERPHVSALVRESLITRAKDFNIVLDDVAIT------HLSYGVE 183
Query: 150 EINAAARMRIAANE---------KAEAEKILQIKRAEGEAESKYL-------SGMGIARQ 193
A + ++A E KA+ E+ + RAEGE+E+ L +GMG+
Sbjct: 184 FSRAVEQKQVAQQEAERSKFVVMKADQERRAAVIRAEGESEAAQLISDATAKAGMGLIEL 243
Query: 194 RQAIVDGLRDSVIGFSENV 212
R+ S + S NV
Sbjct: 244 RRIEASREIASTLARSPNV 262
>At4g17220 unknown protein
Length = 513
Score = 30.8 bits (68), Expect = 0.80
Identities = 38/158 (24%), Positives = 70/158 (44%), Gaps = 32/158 (20%)
Query: 47 LSLRDNVFVNVVASIQYRALANK----ANDAFYKLSNTRGQIQAYVFDVIRASVPKLN-- 100
++ +N FV+ +S+Q + + A A + T +++ VF +R +V KL
Sbjct: 1 MTAAENPFVSDTSSLQSQLKEKEKELLAAKAEVEALRTNEELKDRVFKELRENVRKLEEK 60
Query: 101 ---LDDTFEQKNEIAKAVEEEREKAMSA-----------YGYE----------IVQTLIT 136
++ +QK K +EEE+E A++A Y ++ I+ L +
Sbjct: 61 LGATENQVDQKELERKKLEEEKEDALAAQDAAEEALRRVYTHQQDDDSLPLESIIAPLES 120
Query: 137 DIEPDQHVKTAMNEINAAARMRIAANEKA--EAEKILQ 172
I+ +H +A+ E A + E A EAE+IL+
Sbjct: 121 QIKIHKHEISALQEDKKALERLTKSKESALLEAERILR 158
>At1g07140 Unknown protein (F10K1.15)
Length = 228
Score = 30.4 bits (67), Expect = 1.1
Identities = 36/139 (25%), Positives = 64/139 (45%), Gaps = 9/139 (6%)
Query: 47 LSLRDNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFE 106
L + N FV S+Q + N+ + ++ G+++ +F + AS+ TF
Sbjct: 95 LKICANHFVKSGMSVQEH-VGNEKSCVWHARDFADGELKDELFCIRFASIENCK---TFM 150
Query: 107 QK-NEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEK- 164
QK E+A++ EE+ E +A +++ L + E KT + A+ + A EK
Sbjct: 151 QKFKEVAESEEEKEESKDAADTAGLLEKLTVE-ETKTEEKTEAKAVE-TAKTEVKAEEKK 208
Query: 165 -AEAEKILQIKRAEGEAES 182
+EAEK + K+ E S
Sbjct: 209 ESEAEKSGEAKKTEESGPS 227
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 30.0 bits (66), Expect = 1.4
Identities = 16/76 (21%), Positives = 36/76 (47%)
Query: 102 DDTFEQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAA 161
D+ +E+K E +A+EE+++KA SA + ++ I+ + + ++ +
Sbjct: 54 DEDYEEKLEKFEALEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALP 113
Query: 162 NEKAEAEKILQIKRAE 177
N +K+ K +E
Sbjct: 114 NRIYPKQKLYSFKMSE 129
>At5g65460 putative protein
Length = 1264
Score = 28.9 bits (63), Expect = 3.1
Identities = 27/129 (20%), Positives = 58/129 (44%), Gaps = 11/129 (8%)
Query: 98 KLNLDDTFEQKNEIAKAVEEEREKAMSAYGYEI----VQTLITDIEPDQHVKTAMNEINA 153
K+ + F+ +N+IA+ ++ E+E+ + A + +Q+ + D+E + A+
Sbjct: 537 KIEKEQNFQLRNQIAQLLQLEQEQKLQAQQQDSTIQNLQSKVKDLE--SQLSKALKSDMT 594
Query: 154 AARMRIAANEKAEAEKILQ----IKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFS 209
+R + +A AE L K+ E E + + + + + + D L + + S
Sbjct: 595 RSRDPLEPQPRA-AENTLDSSAVTKKLEEELKKRDALIERLHEENEKLFDRLTEKSVASS 653
Query: 210 ENVPGPSAK 218
V PS+K
Sbjct: 654 TQVSSPSSK 662
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,259,494
Number of Sequences: 26719
Number of extensions: 194996
Number of successful extensions: 603
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 580
Number of HSP's gapped (non-prelim): 37
length of query: 271
length of database: 11,318,596
effective HSP length: 98
effective length of query: 173
effective length of database: 8,700,134
effective search space: 1505123182
effective search space used: 1505123182
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146776.7


