

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146775.17 + phase: 0 /pseudo
(140 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
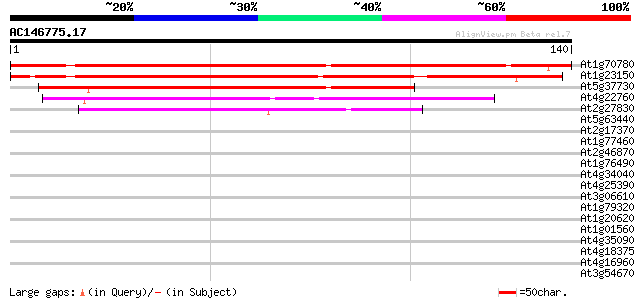
Score E
Sequences producing significant alignments: (bits) Value
At1g70780 unknown protein 147 1e-36
At1g23150 unknown protein 133 3e-32
At5g37730 unknown protein 94 3e-20
At4g22760 unknown protein 54 4e-08
At2g27830 unknown protein 49 9e-07
At5g63440 unknown protein 29 0.71
At2g17370 3-hydroxy-3-methylglutaryl-coenzyme A reductase 2 27 2.7
At1g77460 unknown protein 27 2.7
At2g46870 RAV-like B3 domain DNA binding protein 27 4.6
At1g76490 3-hydroxy-3-methylglutaryl CoA reductase (AA 1-592) 27 4.6
At4g34040 putative protein 26 6.0
At4g25390 receptor kinase-like protein 26 6.0
At3g06610 unknown protein 26 6.0
At1g79320 26 6.0
At1g20620 unknown protein 26 6.0
At1g01560 hypothetical protein 26 6.0
At4g35090 catalase 26 7.8
At4g18375 unknown protein 26 7.8
At4g16960 disease resistance RPP5 like protein 26 7.8
At3g54670 structural maintenance of chromosomes (SMC) - like pro... 26 7.8
>At1g70780 unknown protein
Length = 140
Score = 147 bits (372), Expect = 1e-36
Identities = 82/142 (57%), Positives = 101/142 (70%), Gaps = 6/142 (4%)
Query: 1 MLLSKQKKNQNANNAKRRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLP 60
+L +KQKKNQNA R+LIS+ VLGSAGPIRFV E++LV +VIDT LK YAREGRLP
Sbjct: 2 LLYNKQKKNQNAKG--NRILISVTVLGSAGPIRFVAYEDDLVASVIDTALKGYAREGRLP 59
Query: 61 VLGNDHSAFFLYCPHLGSDAALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTSSL 120
+LG+D + F LYCP +G + ALS WD IGS GARNF+LC+KP+ E + T +
Sbjct: 60 LLGSDFNDFLLYCPMVGPE-ALSTWDAIGSLGARNFMLCRKPEEKKVEESNGRSDSTING 118
Query: 121 PRRGSGSWKSWFN--LNLKVSS 140
R+G GS K+W N NLKVSS
Sbjct: 119 ARKG-GSLKAWINKSFNLKVSS 139
>At1g23150 unknown protein
Length = 141
Score = 133 bits (334), Expect = 3e-32
Identities = 76/143 (53%), Positives = 94/143 (65%), Gaps = 12/143 (8%)
Query: 1 MLLSKQKKNQNANNAKRRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLP 60
MLL K KKNQ R+LIS+ LGSAGPIRFV NE +LV +VIDT LK YAREGRLP
Sbjct: 1 MLLYK-KKNQIVKG--NRILISVTFLGSAGPIRFVANEGDLVASVIDTALKCYAREGRLP 57
Query: 61 VLGNDHSAFFLYCPHLGSDAALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTSSL 120
+LG+D + F YCP +G ALSPW+ IGS G RNF+LCKK E E+ G S+
Sbjct: 58 ILGSDFNDFVFYCPMVG-PGALSPWEAIGSVGVRNFMLCKK---KPEEKKVEEDKGRSNF 113
Query: 121 PRRGS-----GSWKSWFNLNLKV 138
P G+ GS+K+W N +L++
Sbjct: 114 PINGARKGAGGSFKAWINKSLRL 136
>At5g37730 unknown protein
Length = 182
Score = 93.6 bits (231), Expect = 3e-20
Identities = 48/98 (48%), Positives = 66/98 (66%), Gaps = 5/98 (5%)
Query: 8 KNQNANNAKRR----LLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLG 63
+N+N +R LL+S+NVLGS GPIRF+ NE++ V + I+TTLK+YAR+GR+PVLG
Sbjct: 3 RNENVKGVMKRKNKKLLVSVNVLGSVGPIRFLANEDDEVSSAINTTLKAYARQGRIPVLG 62
Query: 64 NDHSAFFLYCPHLGSDAALSPWDKIGSHGARNFVLCKK 101
D F Y + G + L P +KIGS NF++CKK
Sbjct: 63 FDVDNFIFYSINAGFN-TLHPQEKIGSMDVTNFLMCKK 99
>At4g22760 unknown protein
Length = 255
Score = 53.5 bits (127), Expect = 4e-08
Identities = 40/114 (35%), Positives = 56/114 (49%), Gaps = 3/114 (2%)
Query: 9 NQNANNAKR-RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGNDHS 67
NQ N + +++IS+ V GS GP+R +V VE I + Y +EGR P L D S
Sbjct: 113 NQEGNTKEAPKVIISVAVEGSPGPVRAMVKLSCNVEETIKIVVDKYCKEGRTPKLDRD-S 171
Query: 68 AFFLYCPHLGSDAALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTSSLP 121
AF L+ H S L + IG G+R+F + KK A + TS +P
Sbjct: 172 AFELHQSHF-SIQCLEKREIIGELGSRSFYMRKKAPETGGSFAGISPARTSLIP 224
>At2g27830 unknown protein
Length = 190
Score = 48.9 bits (115), Expect = 9e-07
Identities = 26/87 (29%), Positives = 50/87 (56%), Gaps = 2/87 (2%)
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLG-NDHSAFFLYCPHL 76
+LL+++ V GS G ++ +++ E V +ID ++ Y +E R P L ++ S F L+
Sbjct: 71 KLLLNVTVQGSLGAVQIIISPESTVSDLIDAAVRQYVKEARRPFLPESEPSRFDLHYSQF 130
Query: 77 GSDAALSPWDKIGSHGARNFVLCKKPQ 103
++ + +K+ S G+RNF LC + +
Sbjct: 131 SLESIVRD-EKLISLGSRNFFLCGRKE 156
>At5g63440 unknown protein
Length = 232
Score = 29.3 bits (64), Expect = 0.71
Identities = 29/92 (31%), Positives = 37/92 (39%), Gaps = 12/92 (13%)
Query: 49 TLKSYAREGRLPVLGNDHSAFFLYCPHLGS-----DAALSPWDKIGSHGARNFVLCKKPQ 103
T +Y+ E P G D F YC H GS D L K + R+ VL KK
Sbjct: 5 TTHTYSSEDAAPD-GPDSDLFVYYCKHCGSHVLITDTQLQKMPKRKTD--RSNVLDKK-- 59
Query: 104 AATNEAAAEDGSGTSSLPRRGSGSWKSWFNLN 135
T+ A G L +RG G + F +N
Sbjct: 60 --THLARLNVSEGGKVLLKRGEGKMERQFRMN 89
>At2g17370 3-hydroxy-3-methylglutaryl-coenzyme A reductase 2
Length = 562
Score = 27.3 bits (59), Expect = 2.7
Identities = 12/24 (50%), Positives = 18/24 (75%)
Query: 36 VNEEELVEAVIDTTLKSYAREGRL 59
V +EE+V+ VID T+ SY+ E +L
Sbjct: 145 VEDEEIVKLVIDGTIPSYSLETKL 168
>At1g77460 unknown protein
Length = 2110
Score = 27.3 bits (59), Expect = 2.7
Identities = 16/55 (29%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Query: 41 LVEAVIDTTLKSYAREGRLPVLGNDHSAFFLYCPHLGSDAALSPWDKIGSHGARN 95
LVE++ T+ + +E VL + S+ C HL +D ++PW K+ ++ +N
Sbjct: 613 LVESL--TSSREETKEHTASVLADLFSSRQDICGHLATDDIINPWIKLLTNNTQN 665
>At2g46870 RAV-like B3 domain DNA binding protein
Length = 310
Score = 26.6 bits (57), Expect = 4.6
Identities = 17/37 (45%), Positives = 18/37 (47%), Gaps = 2/37 (5%)
Query: 106 TNEAAAEDGSGTSSLPRRGSG--SWKSWFNLNLKVSS 140
TN E S SLPR G G S S+F L L SS
Sbjct: 255 TNSTEEESSSSGGSLPRGGGGGASSSSFFQLRLGSSS 291
>At1g76490 3-hydroxy-3-methylglutaryl CoA reductase (AA 1-592)
Length = 592
Score = 26.6 bits (57), Expect = 4.6
Identities = 11/22 (50%), Positives = 17/22 (77%)
Query: 38 EEELVEAVIDTTLKSYAREGRL 59
+EE+V++VID + SY+ E RL
Sbjct: 172 DEEIVKSVIDGVIPSYSLESRL 193
>At4g34040 putative protein
Length = 666
Score = 26.2 bits (56), Expect = 6.0
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 7/45 (15%)
Query: 82 LSPWDKIGS-------HGARNFVLCKKPQAATNEAAAEDGSGTSS 119
++PW S HGA +L P ++NEAAA GS + S
Sbjct: 440 VAPWSLFPSIESESATHGASLPLLPTGPSVSSNEAAAPSGSSSRS 484
>At4g25390 receptor kinase-like protein
Length = 651
Score = 26.2 bits (56), Expect = 6.0
Identities = 14/31 (45%), Positives = 17/31 (54%)
Query: 100 KKPQAATNEAAAEDGSGTSSLPRRGSGSWKS 130
KK E ++DGS + S RRGSGS S
Sbjct: 442 KKKMTLEAEFCSDDGSSSVSQWRRGSGSGSS 472
>At3g06610 unknown protein
Length = 115
Score = 26.2 bits (56), Expect = 6.0
Identities = 12/52 (23%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query: 2 LLSKQKKNQNANNAKRRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSY 53
+ + ++ + NA + + L S+ + + + F+VNE E+ + V + TL+ +
Sbjct: 49 IAASREADLNAKRLREKELASVKI--NPADVEFIVNELEIEKNVAERTLREH 98
>At1g79320
Length = 368
Score = 26.2 bits (56), Expect = 6.0
Identities = 18/59 (30%), Positives = 27/59 (45%), Gaps = 15/59 (25%)
Query: 15 AKRRLLISINVLGSAGPIRFVVNE---------------EELVEAVIDTTLKSYAREGR 58
AK+ LLI IN +G+ +R VN+ EE ++ +IDT S G+
Sbjct: 2 AKKALLIGINYVGTKAELRGCVNDVRRMRISLVERYGFSEENIKMLIDTDSSSIKPTGK 60
>At1g20620 unknown protein
Length = 492
Score = 26.2 bits (56), Expect = 6.0
Identities = 16/45 (35%), Positives = 24/45 (52%), Gaps = 5/45 (11%)
Query: 32 IRF--VVNEEELVEAVIDT---TLKSYAREGRLPVLGNDHSAFFL 71
+RF VV+E E + D +K Y REG ++GN+ FF+
Sbjct: 101 VRFSTVVHERASPETMRDIRGFAVKFYTREGNFDLVGNNTPVFFI 145
>At1g01560 hypothetical protein
Length = 275
Score = 26.2 bits (56), Expect = 6.0
Identities = 17/50 (34%), Positives = 26/50 (52%), Gaps = 4/50 (8%)
Query: 23 INVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGNDHSAFFLY 72
I+++ P F N+ +V ++DT L R + L +DHS FFLY
Sbjct: 103 IDIIRPPQPDNF--NDVHIVYELMDTDLHHIIRSNQ--PLTDDHSRFFLY 148
>At4g35090 catalase
Length = 492
Score = 25.8 bits (55), Expect = 7.8
Identities = 15/45 (33%), Positives = 24/45 (53%), Gaps = 5/45 (11%)
Query: 32 IRF--VVNEEELVEAVIDT---TLKSYAREGRLPVLGNDHSAFFL 71
+RF V++E E + D +K Y REG ++GN+ FF+
Sbjct: 101 VRFSTVIHERGSPETLRDPRGFAVKFYTREGNFDLVGNNFPVFFI 145
>At4g18375 unknown protein
Length = 606
Score = 25.8 bits (55), Expect = 7.8
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query: 1 MLLSKQKKNQNANNAKRRLLISINVLGSA-GPIRFVVNE 38
+LL + +++A N K +LL+S V+G G V+NE
Sbjct: 381 LLLQEYINDEDAENVKMQLLVSSKVIGCVIGKSGSVINE 419
>At4g16960 disease resistance RPP5 like protein
Length = 1041
Score = 25.8 bits (55), Expect = 7.8
Identities = 11/33 (33%), Positives = 19/33 (57%)
Query: 27 GSAGPIRFVVNEEELVEAVIDTTLKSYAREGRL 59
G+ G RF+ N EE+++ D T+ + +G L
Sbjct: 500 GNPGKRRFLENAEEILDVFTDNTVNENSFQGML 532
>At3g54670 structural maintenance of chromosomes (SMC) - like protein
Length = 1265
Score = 25.8 bits (55), Expect = 7.8
Identities = 16/42 (38%), Positives = 21/42 (49%), Gaps = 3/42 (7%)
Query: 79 DAALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTSSL 120
DAAL D + F+ K QAA + AEDG+G S+
Sbjct: 1187 DAAL---DNLNVAKVAKFIRSKSCQAARDNQDAEDGNGFQSI 1225
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,158,727
Number of Sequences: 26719
Number of extensions: 123597
Number of successful extensions: 221
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 207
Number of HSP's gapped (non-prelim): 21
length of query: 140
length of database: 11,318,596
effective HSP length: 89
effective length of query: 51
effective length of database: 8,940,605
effective search space: 455970855
effective search space used: 455970855
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146775.17


