

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146775.16 - phase: 0
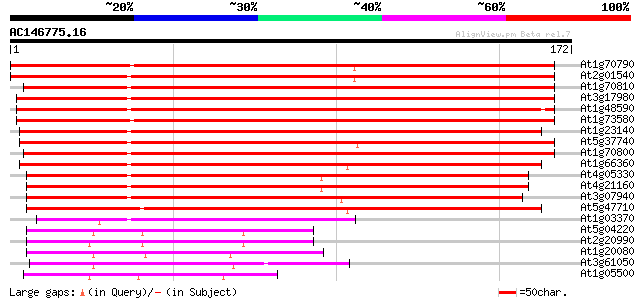
(172 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g70790 unknown protein 230 2e-61
At2g01540 unknown protein (At2g01540) 214 1e-56
At1g70810 unknown protein 207 2e-54
At3g17980 unknown protein 203 3e-53
At1g48590 unknown protein 201 2e-52
At1g73580 hypothetical protein 196 7e-51
At1g23140 unknown protein 191 1e-49
At5g37740 unknown protein 184 2e-47
At1g70800 unknown protein 184 2e-47
At1g66360 unknown protein 183 3e-47
At4g05330 unknown protein 130 5e-31
At4g21160 unknown protein 128 1e-30
At3g07940 putative GTPase activating protein 127 2e-30
At5g47710 unknown protein 123 6e-29
At1g03370 unknown protein 63 7e-11
At5g04220 calcium lipid binding protein - like 52 2e-07
At2g20990 unknown protein 50 8e-07
At1g20080 unknown protein 49 2e-06
At3g61050 CaLB protein 44 4e-05
At1g05500 Ca2+-dependent lipid-binding protein, putative 44 4e-05
>At1g70790 unknown protein
Length = 185
Score = 230 bits (587), Expect = 2e-61
Identities = 110/169 (65%), Positives = 137/169 (80%), Gaps = 3/169 (1%)
Query: 1 MDNNVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTL 60
M++ LG+L++ +KRGINLAIRD+ +SDPYVV+ + + QKLKTRV+ NNCNP WNE+LTL
Sbjct: 1 MEDKPLGILRVHVKRGINLAIRDATTSDPYVVITLAN-QKLKTRVINNNCNPVWNEQLTL 59
Query: 61 SIRDVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLD--TLPNGCAIKRVQAN 118
SI+DV PI LTVFDKD F DDKMGDAEID +P+ + +M+LD LPNGCAIKR++
Sbjct: 60 SIKDVNDPIRLTVFDKDRFSGDDKMGDAEIDFRPFLEAHQMELDFQKLPNGCAIKRIRPG 119
Query: 119 RTNCLAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
RTNCLAEESS W NGK++QEMILRL+NVE GE+ + +EW D PGCKGL
Sbjct: 120 RTNCLAEESSITWSNGKIMQEMILRLKNVECGEVELMLEWTDGPGCKGL 168
>At2g01540 unknown protein (At2g01540)
Length = 180
Score = 214 bits (546), Expect = 1e-56
Identities = 102/169 (60%), Positives = 131/169 (77%), Gaps = 3/169 (1%)
Query: 1 MDNNVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTL 60
MD LGLL + +KRGINLAIRD SSDPY+V+N+ +Q LKTRVVK NCNP WNEE+T+
Sbjct: 1 MDQKPLGLLTIHVKRGINLAIRDHRSSDPYIVLNVA-DQTLKTRVVKKNCNPVWNEEMTV 59
Query: 61 SIRDVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLD--TLPNGCAIKRVQAN 118
+I+D VPI LTVFD D F DDKMGDA ID++PY + +KM ++ LPNGCAIKRVQ +
Sbjct: 60 AIKDPNVPIRLTVFDWDKFTGDDKMGDANIDIQPYLEALKMGMELLRLPNGCAIKRVQPS 119
Query: 119 RTNCLAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
R NCL++ESS +W NGK+ Q++ILRL NVE GE+ + +EW + GC+G+
Sbjct: 120 RHNCLSDESSIVWNNGKITQDLILRLNNVECGEIEIMLEWHEGAGCRGI 168
>At1g70810 unknown protein
Length = 165
Score = 207 bits (528), Expect = 2e-54
Identities = 100/163 (61%), Positives = 129/163 (78%), Gaps = 1/163 (0%)
Query: 5 VLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRD 64
++GLL++R+KRGINLA RD+ SSDP+VV+ +G QKLKTRVV+NNCNPEWNEELTL++R
Sbjct: 4 LVGLLRIRVKRGINLAQRDTLSSDPFVVITMG-SQKLKTRVVENNCNPEWNEELTLALRH 62
Query: 65 VRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRTNCLA 124
P+ L V+DKDTF DKMGDA+ID+KP+ + KM L LP+G IKRV NR NCLA
Sbjct: 63 PDEPVNLIVYDKDTFTSHDKMGDAKIDIKPFLEVHKMGLQELPDGTEIKRVVPNRENCLA 122
Query: 125 EESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
E SS + NGK++Q MIL LRNVE GE+ +++EW+D+PG +GL
Sbjct: 123 EASSIVSNNGKIVQNMILLLRNVECGEVEIQLEWIDIPGSRGL 165
>At3g17980 unknown protein
Length = 165
Score = 203 bits (517), Expect = 3e-53
Identities = 92/165 (55%), Positives = 132/165 (79%), Gaps = 1/165 (0%)
Query: 3 NNVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSI 62
+++LGLL++RIKRG+NLA+RD +SSDPYVVV +G +QKLKTRV+ + NPEWNE+LTLS+
Sbjct: 2 DDLLGLLRIRIKRGVNLAVRDISSSDPYVVVKMG-KQKLKTRVINKDVNPEWNEDLTLSV 60
Query: 63 RDVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRTNC 122
D + + LTV+D D F DDKMGDAE ++KPY + ++M+LD LP+G + V+ +R NC
Sbjct: 61 TDSNLTVLLTVYDHDMFSKDDKMGDAEFEIKPYIEALRMQLDGLPSGTIVTTVKPSRRNC 120
Query: 123 LAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
LAEES W +GK++Q+++LRLR+VE GE+ +++W+D+PG KGL
Sbjct: 121 LAEESRVTWVDGKLVQDLVLRLRHVECGEVEAQLQWIDLPGSKGL 165
>At1g48590 unknown protein
Length = 185
Score = 201 bits (510), Expect = 2e-52
Identities = 95/165 (57%), Positives = 129/165 (77%), Gaps = 2/165 (1%)
Query: 3 NNVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSI 62
+++LGLL++RIKRG+NLA+RD NSSDPYVVV + +QKLKTRV+ N NPEWNE+LTLS+
Sbjct: 23 DSLLGLLRIRIKRGVNLAVRDLNSSDPYVVVKMA-KQKLKTRVIYKNVNPEWNEDLTLSV 81
Query: 63 RDVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRTNC 122
D + + LTV+D DTF DDKMGDAE +KP+ +KM L LP+G + VQ +R NC
Sbjct: 82 SDPNLTVLLTVYDYDTFTKDDKMGDAEFGIKPFVNALKMHLHDLPSGTIVTTVQPSRDNC 141
Query: 123 LAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
LAEES IW +GK++Q+++LRLR+VE GE+ +++W+D+PG KGL
Sbjct: 142 LAEESRVIWSDGKLVQDIVLRLRHVECGEVEAQLQWIDLPG-KGL 185
>At1g73580 hypothetical protein
Length = 168
Score = 196 bits (497), Expect = 7e-51
Identities = 89/165 (53%), Positives = 127/165 (76%), Gaps = 1/165 (0%)
Query: 3 NNVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSI 62
+N+LG+L++R++RG+NLA+RD +SSDPYVV+ +G QKLKT+VVK N NP+W E+L+ ++
Sbjct: 5 DNLLGILRVRVQRGVNLAVRDVSSSDPYVVLKLGR-QKLKTKVVKQNVNPQWQEDLSFTV 63
Query: 63 RDVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRTNC 122
D +P+ L V+D D F DDKMGDAEIDLKPY + ++M+L LP+G I + +R NC
Sbjct: 64 TDPNLPLTLIVYDHDFFSKDDKMGDAEIDLKPYIEALRMELSGLPDGTIISTIGPSRGNC 123
Query: 123 LAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
LAEES W N +++Q + LRLRNVE GE+ +E++W+D+PG KGL
Sbjct: 124 LAEESYIRWINDRIVQHICLRLRNVERGEVEIELQWIDLPGSKGL 168
>At1g23140 unknown protein
Length = 165
Score = 191 bits (486), Expect = 1e-49
Identities = 93/160 (58%), Positives = 123/160 (76%), Gaps = 1/160 (0%)
Query: 4 NVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIR 63
N++GLL++R+KRGINL RDSN+SDP+VVV +G QKLKTR V+N+CNPEW++ELTL I
Sbjct: 3 NLVGLLRIRVKRGINLVSRDSNTSDPFVVVTMG-SQKLKTRGVENSCNPEWDDELTLGIN 61
Query: 64 DVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRTNCL 123
D + L V+DKDTF D MGDAEID+KP+ + + L NG I+RV+ + NCL
Sbjct: 62 DPNQHVTLEVYDKDTFTSHDPMGDAEIDIKPFFEVQGTDIQELTNGTEIRRVKPSGDNCL 121
Query: 124 AEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPG 163
AEES I+ NGK+LQ+MIL+LRNVESGE+ ++IEW++V G
Sbjct: 122 AEESRIIFSNGKILQDMILQLRNVESGEVEIQIEWINVTG 161
>At5g37740 unknown protein
Length = 168
Score = 184 bits (468), Expect = 2e-47
Identities = 86/167 (51%), Positives = 125/167 (74%), Gaps = 4/167 (2%)
Query: 4 NVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIR 63
N++GLL++ +KRG+NLAIRD +SSDPY+VV+ G +QKLKTRVVK++ NPEWN++LTLS+
Sbjct: 3 NLVGLLRIHVKRGVNLAIRDISSSDPYIVVHCG-KQKLKTRVVKHSVNPEWNDDLTLSVT 61
Query: 64 DVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDT---LPNGCAIKRVQANRT 120
D +PI LTV+D D DDKMG+AE + P+ + +K LPNG IK+++ +R
Sbjct: 62 DPNLPIKLTVYDYDLLSADDKMGEAEFHIGPFIEAIKFAHQLGPGLPNGTIIKKIEPSRK 121
Query: 121 NCLAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
NCL+E S + GK++Q M LRL++VE GE+ +++EW+DVPG +G+
Sbjct: 122 NCLSESSHIVLNQGKIVQNMFLRLQHVECGEVELQLEWIDVPGSRGI 168
>At1g70800 unknown protein
Length = 174
Score = 184 bits (468), Expect = 2e-47
Identities = 88/163 (53%), Positives = 123/163 (74%), Gaps = 1/163 (0%)
Query: 5 VLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRD 64
++GL+++ +KRGI+LA RD+ SSDP+VV+ +G QKLK+ VKNNCNPEWNEELTL+I D
Sbjct: 13 LVGLVRILVKRGIDLARRDALSSDPFVVITMG-PQKLKSFTVKNNCNPEWNEELTLAIED 71
Query: 65 VRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRTNCLA 124
P+ L V+DKDTF DDKMGDA+ID+KP+ K+ L LP+G +KR+ R NCL+
Sbjct: 72 PNEPVKLMVYDKDTFTADDKMGDAQIDMKPFLDVHKLGLKELPHGKELKRIVPTRDNCLS 131
Query: 125 EESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
E+S + NGK++Q+MIL L+NVE G++ +++EW+ PG GL
Sbjct: 132 EDSIIVSDNGKIVQDMILLLKNVECGKVEIQLEWLKNPGGSGL 174
>At1g66360 unknown protein
Length = 174
Score = 183 bits (465), Expect = 3e-47
Identities = 85/162 (52%), Positives = 122/162 (74%), Gaps = 3/162 (1%)
Query: 4 NVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIR 63
N+LGLL+L + RG+NLAIRDS SSDPYV+V +G +QKL+TRV+K N N EWNE+LTLS+
Sbjct: 3 NMLGLLRLHVIRGVNLAIRDSQSSDPYVIVRMG-KQKLRTRVMKKNLNTEWNEDLTLSVT 61
Query: 64 DVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMK--LDTLPNGCAIKRVQANRTN 121
D +P+ + V+D+D F DDKMGDA + P+ + ++++ L LP G I ++QA+R N
Sbjct: 62 DPTLPVKIMVYDRDRFSRDDKMGDAIFHIDPFLEAIRIQNQLGGLPEGTVIMKIQASRQN 121
Query: 122 CLAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPG 163
CL+EES +W GK++Q M L+L+NVE GE+ +++EW+DV G
Sbjct: 122 CLSEESKIVWHKGKIVQNMFLKLQNVERGEIELQLEWIDVSG 163
>At4g05330 unknown protein
Length = 336
Score = 130 bits (326), Expect = 5e-31
Identities = 72/155 (46%), Positives = 104/155 (66%), Gaps = 2/155 (1%)
Query: 6 LGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
+GLLK+ IK+G NLAIRD SSDPYVV+N+G +QKL+T V+ +N NP WN+EL LS+ +
Sbjct: 179 IGLLKVTIKKGTNLAIRDMMSSDPYVVLNLG-KQKLQTTVMNSNLNPVWNQELMLSVPES 237
Query: 66 RVPICLTVFDKDTFFVDDKMGDAEIDLKP-YTQCVKMKLDTLPNGCAIKRVQANRTNCLA 124
P+ L V+D DTF DD MG+A+ID++P T + + I + + N L
Sbjct: 238 YGPVKLQVYDYDTFSADDIMGEADIDIQPLITSAMAFGDPEMFGDMQIGKWLKSHDNPLI 297
Query: 125 EESSCIWKNGKVLQEMILRLRNVESGELVVEIEWV 159
++S +GKV QE+ ++L+NVESGEL +E+EW+
Sbjct: 298 DDSIINIVDGKVKQEVQIKLQNVESGELELEMEWL 332
>At4g21160 unknown protein
Length = 337
Score = 128 bits (322), Expect = 1e-30
Identities = 73/155 (47%), Positives = 101/155 (65%), Gaps = 2/155 (1%)
Query: 6 LGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
+GLLK+ IK+G N+AIRD SSDPYVV+ +G +QK ++ VVK+N NP WNEEL LS+
Sbjct: 180 IGLLKVTIKKGTNMAIRDMMSSDPYVVLTLG-QQKAQSTVVKSNLNPVWNEELMLSVPHN 238
Query: 66 RVPICLTVFDKDTFFVDDKMGDAEIDLKP-YTQCVKMKLDTLPNGCAIKRVQANRTNCLA 124
+ L VFD DTF DD MG+AEID++P T + + I + + N L
Sbjct: 239 YGSVKLQVFDYDTFSADDIMGEAEIDIQPLITSAMAFGDPEMFGDMQIGKWLKSHDNALI 298
Query: 125 EESSCIWKNGKVLQEMILRLRNVESGELVVEIEWV 159
E+S +GKV QE+ ++L+NVESGEL +E+EW+
Sbjct: 299 EDSIINIADGKVKQEVQIKLQNVESGELELEMEWL 333
>At3g07940 putative GTPase activating protein
Length = 385
Score = 127 bits (320), Expect = 2e-30
Identities = 65/153 (42%), Positives = 101/153 (65%), Gaps = 2/153 (1%)
Query: 6 LGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
+GL+K+ + +G NLA+RD +SDPYV++ +G +Q +KTRV+KNN NP WNE L LSI +
Sbjct: 228 VGLIKVNVVKGTNLAVRDVMTSDPYVILALG-QQSVKTRVIKNNLNPVWNETLMLSIPEP 286
Query: 66 RVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVK-MKLDTLPNGCAIKRVQANRTNCLA 124
P+ + V+DKDTF DD MG+AEID++P K + ++ + A++ N L
Sbjct: 287 MPPLKVLVYDKDTFSTDDFMGEAEIDIQPLVSAAKAYETSSIKEPMQLGSWVASKENTLV 346
Query: 125 EESSCIWKNGKVLQEMILRLRNVESGELVVEIE 157
+ + ++GKV Q++ LRL+NVE G L +++E
Sbjct: 347 SDGIILLEDGKVKQDISLRLQNVERGVLEIQLE 379
>At5g47710 unknown protein
Length = 166
Score = 123 bits (308), Expect = 6e-29
Identities = 65/160 (40%), Positives = 100/160 (61%), Gaps = 3/160 (1%)
Query: 6 LGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
LGLL++ + +G L IRD SSDPYV+V +G+E KT+V+ N NP WNEEL +++D
Sbjct: 5 LGLLQVTVIQGKKLVIRDFKSSDPYVIVKLGNESA-KTKVINNCLNPVWNEELNFTLKDP 63
Query: 66 RVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMK--LDTLPNGCAIKRVQANRTNCL 123
+ L VFDKD F DDKMG A + L+P +++ + +++V + NC+
Sbjct: 64 AAVLALEVFDKDRFKADDKMGHASLSLQPLISVARLRHVVRVSSGETTLRKVLPDPENCV 123
Query: 124 AEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPG 163
+ ES+ +G+V+Q + LRL VESGE+ ++I+ +D PG
Sbjct: 124 SRESTISCIDGEVVQSVWLRLCAVESGEIELKIKLIDPPG 163
>At1g03370 unknown protein
Length = 1859
Score = 63.2 bits (152), Expect = 7e-11
Identities = 32/99 (32%), Positives = 55/99 (55%), Gaps = 2/99 (2%)
Query: 9 LKLRIKRGINLAIRDSNS-SDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRV 67
L++R+ NL D N SDPYV + +G +Q+ +T+VVK N NP+W E+ + + D+
Sbjct: 829 LQVRVVEARNLPAMDLNGFSDPYVRLQLG-KQRSRTKVVKKNLNPKWTEDFSFGVDDLND 887
Query: 68 PICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTL 106
+ ++V D+D +F DD +G + + L T+
Sbjct: 888 ELVVSVLDEDKYFNDDFVGQVRVSVSLVFDAENQSLGTV 926
Score = 32.3 bits (72), Expect = 0.13
Identities = 28/88 (31%), Positives = 44/88 (49%), Gaps = 6/88 (6%)
Query: 8 LLKLRIKRGINLAIRD-SNSSDPYVVVNIGHEQKLKTRVVK-NNCNPEWNEELTL-SIRD 64
LL + + G++LA D S DPY+V K +T +K NP+WNE ++ D
Sbjct: 1362 LLTVALIEGVDLAAVDPSGHCDPYIVFT--SNGKTRTSSIKFQKSNPQWNEIFEFDAMAD 1419
Query: 65 VRVPICLTVFDKDTFFVDD-KMGDAEID 91
+ + VFD D F + +G AE++
Sbjct: 1420 PPSVLNVEVFDFDGPFDEAVSLGHAEVN 1447
>At5g04220 calcium lipid binding protein - like
Length = 540
Score = 52.0 bits (123), Expect = 2e-07
Identities = 35/92 (38%), Positives = 50/92 (54%), Gaps = 4/92 (4%)
Query: 6 LGLLKLRIKRGINLAIRDS-NSSDPYVVVNIGHEQ--KLKTRVVKNNCNPEWNEELTLSI 62
+GLL + I R NL +D +SDPYV +++ E+ KT + K N NPEWNE L +
Sbjct: 260 VGLLHVSILRARNLLKKDLLGTSDPYVKLSLTGEKLPAKKTTIKKRNLNPEWNEHFKLIV 319
Query: 63 RDVRVPIC-LTVFDKDTFFVDDKMGDAEIDLK 93
+D + L VFD D D++G I L+
Sbjct: 320 KDPNSQVLQLEVFDWDKVGGHDRLGMQMIPLQ 351
Score = 37.7 bits (86), Expect = 0.003
Identities = 27/91 (29%), Positives = 50/91 (54%), Gaps = 7/91 (7%)
Query: 7 GLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRD-- 64
GLL + ++ ++ + +S+ PY VV E+K KT+++K +P WNEE ++ +
Sbjct: 420 GLLSVAVQSAKDVEGKKKHSN-PYAVVLFRGEKK-KTKMLKKTRDPRWNEEFQFTLEEPP 477
Query: 65 VRVPICLTVFDKDT---FFVDDKMGDAEIDL 92
V+ I + V K T F +++G +I+L
Sbjct: 478 VKESIRVEVMSKGTGFHFRSKEELGHVDINL 508
>At2g20990 unknown protein
Length = 541
Score = 49.7 bits (117), Expect = 8e-07
Identities = 28/92 (30%), Positives = 50/92 (53%), Gaps = 4/92 (4%)
Query: 6 LGLLKLRIKRGINLAIRD-SNSSDPYVVVNIGHEQ--KLKTRVVKNNCNPEWNEELTLSI 62
+G++ +++ R + L +D +DP+V + + ++ KT V N NPEWNEE S+
Sbjct: 259 VGIVHVKVVRAVGLRKKDLMGGADPFVKIKLSEDKIPSKKTTVKHKNLNPEWNEEFKFSV 318
Query: 63 RDVRVPIC-LTVFDKDTFFVDDKMGDAEIDLK 93
RD + + +V+D + +KMG + LK
Sbjct: 319 RDPQTQVLEFSVYDWEQVGNPEKMGMNVLALK 350
Score = 34.3 bits (77), Expect = 0.034
Identities = 15/44 (34%), Positives = 26/44 (59%), Gaps = 1/44 (2%)
Query: 21 IRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRD 64
+ + ++PYV + E++ KT+ VK N +P WNEE T + +
Sbjct: 432 VEGKHHTNPYVRIYFKGEER-KTKHVKKNRDPRWNEEFTFMLEE 474
>At1g20080 unknown protein
Length = 535
Score = 48.5 bits (114), Expect = 2e-06
Identities = 33/95 (34%), Positives = 51/95 (52%), Gaps = 4/95 (4%)
Query: 6 LGLLKLRIKRGINLAIRDS-NSSDPYVVVNIGHEQK--LKTRVVKNNCNPEWNEELTLSI 62
+GLL +++ + I L +D SDPYV + + ++ KT V +N NPEWNEE L +
Sbjct: 257 VGLLSVKVIKAIKLKKKDLLGGSDPYVKLTLSGDKVPGKKTVVKHSNLNPEWNEEFDLVV 316
Query: 63 RDVR-VPICLTVFDKDTFFVDDKMGDAEIDLKPYT 96
++ + L V+D + DK+G I LK T
Sbjct: 317 KEPESQELQLIVYDWEQVGKHDKIGMNVIQLKDLT 351
>At3g61050 CaLB protein
Length = 510
Score = 43.9 bits (102), Expect = 4e-05
Identities = 29/100 (29%), Positives = 49/100 (49%), Gaps = 3/100 (3%)
Query: 7 GLLKLRIKRGINLAIRDS-NSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
G L + + + NL ++ SDPY + I K KT+ ++NN NP W++ L D
Sbjct: 263 GKLIVTVVKATNLKNKELIGKSDPYATIYIRPVFKYKTKAIENNLNPVWDQTFELIAEDK 322
Query: 66 RV-PICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLD 104
+ + VFDKD D+++G ++ L V +L+
Sbjct: 323 ETQSLTVEVFDKDV-GQDERLGLVKLPLSSLEAGVTKELE 361
>At1g05500 Ca2+-dependent lipid-binding protein, putative
Length = 560
Score = 43.9 bits (102), Expect = 4e-05
Identities = 28/81 (34%), Positives = 44/81 (53%), Gaps = 3/81 (3%)
Query: 5 VLGLLKLRIKRGINLAIRD-SNSSDPYVVVNIGHE-QKLKTRVVKNNCNPEWNEELTLSI 62
V G+L + + + I+D +DPYVV+++ K KTRVV ++ NP WN+ +
Sbjct: 433 VRGVLSVTVISAEEIPIQDLMGKADPYVVLSMKKSGAKSKTRVVNDSLNPVWNQTFDFVV 492
Query: 63 RD-VRVPICLTVFDKDTFFVD 82
D + + L V+D DTF D
Sbjct: 493 EDGLHDMLVLEVWDHDTFGKD 513
Score = 37.7 bits (86), Expect = 0.003
Identities = 22/91 (24%), Positives = 47/91 (51%), Gaps = 4/91 (4%)
Query: 6 LGLLKLRIKRGINLAIRDS-NSSDPYVVVNIG--HEQKLKTRVVKNNCNPEWNEELTLSI 62
+G+L++++ + NL +D SDP+ + I E+ +++ + N+ NP WNE +
Sbjct: 261 VGMLEVKLVQAKNLTNKDLVGKSDPFAKMFIRPLREKTKRSKTINNDLNPIWNEHFEFVV 320
Query: 63 RDVRVP-ICLTVFDKDTFFVDDKMGDAEIDL 92
D + + ++D + + +G A+I L
Sbjct: 321 EDASTQHLVVRIYDDEGVQASELIGCAQIRL 351
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,002,837
Number of Sequences: 26719
Number of extensions: 163426
Number of successful extensions: 481
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 360
Number of HSP's gapped (non-prelim): 116
length of query: 172
length of database: 11,318,596
effective HSP length: 92
effective length of query: 80
effective length of database: 8,860,448
effective search space: 708835840
effective search space used: 708835840
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146775.16


