

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146760.4 + phase: 0
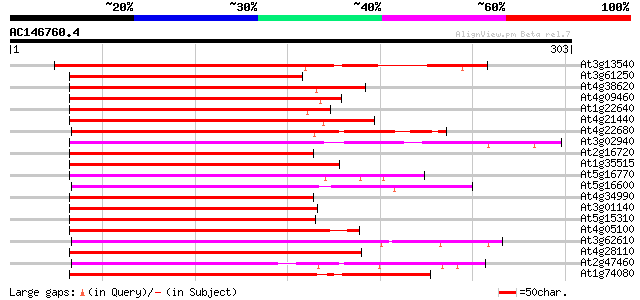
(303 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g13540 myb-related protein 5 263 1e-70
At3g61250 putative transcription factor (MYB17) 208 3e-54
At4g38620 putative transcription factor (MYB4) 204 4e-53
At4g09460 DNA-binding protein 202 2e-52
At1g22640 transcription factor like protein (MYB3) 202 2e-52
At4g21440 myb-related protein M4 201 3e-52
At4g22680 myb-like protein 201 5e-52
At3g02940 MYB family transcription factor like protein 199 2e-51
At2g16720 DNA-binding protein 197 4e-51
At1g35515 putative transcription factor (MYB8) 197 6e-51
At5g16770 putative transcription factor (MYB9) 196 1e-50
At5g16600 transcription factor (gb|AAD53095.1) 196 2e-50
At4g34990 MYB-like protein 195 2e-50
At3g01140 putative Myb-related transcription factor 195 3e-50
At5g15310 myb-related protein - like 194 4e-50
At4g05100 MYB - like protein 194 4e-50
At3g62610 putative transcription factor (MYB11) 194 5e-50
At4g28110 putative transcription factor (MYB41) 193 1e-49
At2g47460 putative MYB family transcription factor 192 1e-49
At1g74080 putative transcription factor 191 4e-49
>At3g13540 myb-related protein 5
Length = 249
Score = 263 bits (671), Expect = 1e-70
Identities = 137/238 (57%), Positives = 158/238 (65%), Gaps = 34/238 (14%)
Query: 25 KKQKTTTTTPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRL 84
KK + TTPCC+K+G+KRGPWTVEEDE+L +IKKEGEGRWR+LPKRAGLLRCGKSCRL
Sbjct: 7 KKPVSKKTTPCCTKMGMKRGPWTVEEDEILVSFIKKEGEGRWRSLPKRAGLLRCGKSCRL 66
Query: 85 RWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKK 144
RWMNYLRPSVKRG I DEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHL KK
Sbjct: 67 RWMNYLRPSVKRGGITSDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLRKK 126
Query: 145 LINQGIDPRTHKPL--NNPSSSSSIIPNTNHQTSTPFFAPSSSDHHHPITITNNEPDPSQ 202
L+ QGIDP+THKPL NN + P SS H T+ + D
Sbjct: 127 LLRQGIDPQTHKPLDANNIHKPEEEVSGGQKYP----LEPISSSHTDDTTVNGGDGD--- 179
Query: 203 TQQAQGCGNLVNDVSAMDNHHVLTNNNRGDYGDDNGNNNYG--GDDVFTSFLDSLIND 258
+ N+ +G ++G ++G DD F+SFL+SLIND
Sbjct: 180 -----------------------SKNSINVFGGEHGYEDFGFCYDDKFSSFLNSLIND 214
>At3g61250 putative transcription factor (MYB17)
Length = 299
Score = 208 bits (530), Expect = 3e-54
Identities = 89/126 (70%), Positives = 105/126 (82%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
TPCC K+GLK+GPWT EEDEVL +IKK G G WRTLPK AGLLRCGKSCRLRW NYLRP
Sbjct: 4 TPCCDKIGLKKGPWTPEEDEVLVAHIKKNGHGSWRTLPKLAGLLRCGKSCRLRWTNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG DEE L+++LH +LGNRW+ IA ++PGRTDNEIKN WNTHL K+L++ G+DP
Sbjct: 64 DIKRGPFTADEEKLVIQLHAILGNRWAAIAAQLPGRTDNEIKNLWNTHLKKRLLSMGLDP 123
Query: 153 RTHKPL 158
RTH+PL
Sbjct: 124 RTHEPL 129
>At4g38620 putative transcription factor (MYB4)
Length = 282
Score = 204 bits (520), Expect = 4e-53
Identities = 95/166 (57%), Positives = 120/166 (72%), Gaps = 6/166 (3%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EEDE L YIK GEG WR+LPK AGLLRCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDERLVAYIKAHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +E++LI++LH LLGN+WSLIAGR+PGRTDNEIKNYWNTH+ +KLIN+GIDP
Sbjct: 64 DLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKLINRGIDP 123
Query: 153 RTHKPLNNPSSS-----SSIIPNTNHQTSTPF-FAPSSSDHHHPIT 192
+H+P+ S+S + + P T++ + F AP H I+
Sbjct: 124 TSHRPIQESSASQDSKPTQLEPVTSNTINISFTSAPKVETFHESIS 169
>At4g09460 DNA-binding protein
Length = 236
Score = 202 bits (513), Expect = 2e-52
Identities = 88/149 (59%), Positives = 112/149 (75%), Gaps = 2/149 (1%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L +YI+ GEG WR+LPK AGLLRCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKAHTNKGAWTKEEDQRLVDYIRNHGEGCWRSLPKSAGLLRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG DE+ +I++LH LLGN+WSLIAGR+PGRTDNEIKNYWNTH+ +KL++ GIDP
Sbjct: 64 DLKRGNFTDDEDQIIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIKRKLLSHGIDP 123
Query: 153 RTHKPLNNPSSSSS--IIPNTNHQTSTPF 179
+TH+ +N + SS ++P N F
Sbjct: 124 QTHRQINESKTVSSQVVVPIQNDAVEYSF 152
>At1g22640 transcription factor like protein (MYB3)
Length = 257
Score = 202 bits (513), Expect = 2e-52
Identities = 87/145 (60%), Positives = 115/145 (79%), Gaps = 4/145 (2%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K + +G WT EED++L +YI+K GEG WR+LP+ AGL RCGKSCRLRWMNYLRP
Sbjct: 4 SPCCEKAHMNKGAWTKEEDQLLVDYIRKHGEGCWRSLPRAAGLQRCGKSCRLRWMNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +E++LI++LH LLGN+WSLIAGR+PGRTDNEIKNYWNTH+ +KL+++GIDP
Sbjct: 64 DLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIKRKLLSRGIDP 123
Query: 153 RTHKPLN----NPSSSSSIIPNTNH 173
+H+ +N +PSS + + T H
Sbjct: 124 NSHRLINESVVSPSSLQNDVVETIH 148
>At4g21440 myb-related protein M4
Length = 350
Score = 201 bits (512), Expect = 3e-52
Identities = 92/171 (53%), Positives = 118/171 (68%), Gaps = 6/171 (3%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K GLK+GPWT EED+ L +YI+K G G WRTLPK AGL RCGKSCRLRW NYLRP
Sbjct: 4 SPCCEKNGLKKGPWTSEEDQKLVDYIQKHGYGNWRTLPKNAGLQRCGKSCRLRWTNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG+ + +EE+ I++LH LGN+WS IA R+PGRTDNEIKN+WNTH+ KKL+ GIDP
Sbjct: 64 DIKRGRFSFEEEETIIQLHSFLGNKWSAIAARLPGRTDNEIKNFWNTHIRKKLLRMGIDP 123
Query: 153 RTHKPLNNPSSSSSII------PNTNHQTSTPFFAPSSSDHHHPITITNNE 197
TH P + SSI+ +++H + ++ HH + N E
Sbjct: 124 VTHSPRLDLLDISSILASSLYNSSSHHMNMSRLMMDTNRRHHQQHPLVNPE 174
>At4g22680 myb-like protein
Length = 266
Score = 201 bits (510), Expect = 5e-52
Identities = 103/206 (50%), Positives = 131/206 (63%), Gaps = 16/206 (7%)
Query: 34 PCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPS 93
PCC K+G+K+GPWTVEED+ L +I G WR LPK AGL RCGKSCRLRW NYLRP
Sbjct: 5 PCCDKLGVKKGPWTVEEDKKLINFILTNGHCCWRALPKLAGLRRCGKSCRLRWTNYLRPD 64
Query: 94 VKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDPR 153
+KRG ++ DEE L++ LH LGN+WS IA R+PGRTDNEIKN+WNTH+ KKL+ GIDP
Sbjct: 65 LKRGLLSHDEEQLVIDLHANLGNKWSKIASRLPGRTDNEIKNHWNTHIKKKLLKMGIDPM 124
Query: 154 THKPLNNPSS---SSSIIPNTNHQTSTPFFAPSSSDHHHPITITNNEPDPSQTQQAQGCG 210
TH+PLN S +S IP+ S +++ + I++T E + S T Q
Sbjct: 125 THQPLNQEPSNIDNSKTIPSNPDDVSVE--PKTTNTKYVEISVTTTEEESSSTVTDQN-- 180
Query: 211 NLVNDVSAMDNHHVLTNNNRGDYGDD 236
S+MDN + L +N Y DD
Sbjct: 181 ------SSMDNENHLIDN---IYDDD 197
>At3g02940 MYB family transcription factor like protein
Length = 321
Score = 199 bits (505), Expect = 2e-51
Identities = 111/277 (40%), Positives = 154/277 (55%), Gaps = 30/277 (10%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC + GLK+GPWT EED+ L +I+K G G WR LPK+AGL RCGKSCRLRW NYLRP
Sbjct: 4 SPCCDESGLKKGPWTPEEDQKLINHIRKHGHGSWRALPKQAGLNRCGKSCRLRWTNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +EE I+ LH LLGN+WS IAG +PGRTDNEIKNYWNTH+ KKLI GIDP
Sbjct: 64 DIKRGNFTAEEEQTIINLHSLLGNKWSSIAGHLPGRTDNEIKNYWNTHIRKKLIQMGIDP 123
Query: 153 RTHKPLNNPSSSSSIIPNTNHQTSTPFFAPSSSDHHHPITITNN-EPDPSQTQQAQGCGN 211
TH+P + + + +P ++++ ++ + + N + D + +AQ +
Sbjct: 124 VTHRPRTDHLNVLAALPQ----------LLAAANFNNLLNLNQNIQLDATSVAKAQLLHS 173
Query: 212 LVNDVSAMDNHHVLTNNNRGDYGD-DNGNNNYGGDDVFTSFLDSLIN--DDAFAAHRSEN 268
++ VL+NNN D + NN G F L ++ N D ++
Sbjct: 174 MI---------QVLSNNNTSSSFDIHHTTNNLFGQSSFLENLPNIENPYDQTQGLSHIDD 224
Query: 269 DPLILSAAPPALWE-------SPPLMMTSTHNFTQNQ 298
PL ++P + PPL+ TS + Q
Sbjct: 225 QPLDSFSSPIRVVAYQHDQNFIPPLISTSPDESKETQ 261
>At2g16720 DNA-binding protein
Length = 269
Score = 197 bits (502), Expect = 4e-51
Identities = 85/132 (64%), Positives = 105/132 (79%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K + +G WT EEDE L YIK GEG WR+LP+ AGLLRCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKEHMNKGAWTKEEDERLVSYIKSHGEGCWRSLPRAAGLLRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG DE++LI++LH LLGN+WSLIA R+PGRTDNEIKNYWNTH+ +KL+++GIDP
Sbjct: 64 DLKRGNFTHDEDELIIKLHSLLGNKWSLIAARLPGRTDNEIKNYWNTHIKRKLLSKGIDP 123
Query: 153 RTHKPLNNPSSS 164
TH+ +N S
Sbjct: 124 ATHRGINEAKIS 135
>At1g35515 putative transcription factor (MYB8)
Length = 212
Score = 197 bits (501), Expect = 6e-51
Identities = 84/146 (57%), Positives = 111/146 (75%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K + +G WT EED+ L +YI+ GEG WR+LPK GLLRCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKAHMNKGAWTKEEDQRLIDYIRNHGEGSWRSLPKSVGLLRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG EE +I++LH L GN+WSLIAG++PGRTDNEIKNYWNTH+ +KL+N+GIDP
Sbjct: 64 DLKRGNFTDGEEQIIVKLHSLFGNKWSLIAGKLPGRTDNEIKNYWNTHIKRKLLNRGIDP 123
Query: 153 RTHKPLNNPSSSSSIIPNTNHQTSTP 178
+TH + P ++S N + +++ P
Sbjct: 124 KTHGSIIEPKTTSFHPRNEDLKSTFP 149
>At5g16770 putative transcription factor (MYB9)
Length = 336
Score = 196 bits (498), Expect = 1e-50
Identities = 98/210 (46%), Positives = 126/210 (59%), Gaps = 18/210 (8%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC + GLK+GPWT EED+ L ++I+K G G WR LPK+AGL RCGKSCRLRW NYLRP
Sbjct: 4 SPCCDENGLKKGPWTQEEDDKLIDHIQKHGHGSWRALPKQAGLNRCGKSCRLRWTNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +EE I+ LH LLGN+WS IAG +PGRTDNEIKNYWNTHL KKL+ GIDP
Sbjct: 64 DIKRGNFTEEEEQTIINLHSLLGNKWSSIAGNLPGRTDNEIKNYWNTHLRKKLLQMGIDP 123
Query: 153 RTHKPLNNPSSSSSIIP--------------NTNHQTSTPFFAPSSSDHH--HPITITNN 196
TH+P + + + +P N N Q A + H ++ NN
Sbjct: 124 VTHRPRTDHLNVLAALPQLIAAANFNSLLNLNQNVQLDATTLAKAQLLHTMIQVLSTNNN 183
Query: 197 EPDP--SQTQQAQGCGNLVNDVSAMDNHHV 224
+P S + NL S ++N ++
Sbjct: 184 TTNPSFSSSTMQNSNTNLFGQASYLENQNL 213
>At5g16600 transcription factor (gb|AAD53095.1)
Length = 327
Score = 196 bits (497), Expect = 2e-50
Identities = 100/225 (44%), Positives = 130/225 (57%), Gaps = 14/225 (6%)
Query: 34 PCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPS 93
PCC KVGLK+GPWT+EED+ L +I G WR LPK +GLLRCGKSCRLRW+NYLRP
Sbjct: 5 PCCDKVGLKKGPWTIEEDKKLINFILTNGHCCWRALPKLSGLLRCGKSCRLRWINYLRPD 64
Query: 94 VKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDPR 153
+KRG ++ EE ++ LH LGNRWS IA +PGRTDNEIKN+WNTH+ KKL GIDP
Sbjct: 65 LKRGLLSEYEEQKVINLHAQLGNRWSKIASHLPGRTDNEIKNHWNTHIKKKLRKMGIDPL 124
Query: 154 THKPLNNPSSSSSIIPNTNHQTSTPFFAPSSSDHHHPITITNNEPDPSQTQQA------- 206
THKPL+ +S Q P + T +E + Q +QA
Sbjct: 125 THKPLSEQEASQQA------QGRKKSLVPHDDKNPKQDQQTKDEQEQHQLEQALEKNNTS 178
Query: 207 -QGCGNLVNDVSAMDNHHVLTNNNRGDYGDDNGNNNYGGDDVFTS 250
G G +++V ++ H +L + + + N +N FTS
Sbjct: 179 VSGDGFCIDEVPLLNPHEILIDISSSHHHHSNDDNVNINTSKFTS 223
>At4g34990 MYB-like protein
Length = 274
Score = 195 bits (496), Expect = 2e-50
Identities = 84/132 (63%), Positives = 104/132 (78%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K +G WT EED+ L YIK GEG WR+LP+ AGL RCGKSCRLRW+NYLRP
Sbjct: 4 SPCCEKDHTNKGAWTKEEDDKLISYIKAHGEGCWRSLPRSAGLQRCGKSCRLRWINYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +E+DLI++LH LLGN+WSLIA R+PGRTDNEIKNYWNTH+ +KL+ +GIDP
Sbjct: 64 DLKRGNFTLEEDDLIIKLHSLLGNKWSLIATRLPGRTDNEIKNYWNTHVKRKLLRKGIDP 123
Query: 153 RTHKPLNNPSSS 164
TH+P+N +S
Sbjct: 124 ATHRPINETKTS 135
>At3g01140 putative Myb-related transcription factor
Length = 345
Score = 195 bits (495), Expect = 3e-50
Identities = 87/134 (64%), Positives = 102/134 (75%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K GLK+GPWT EED+ L YI++ G G WR+LP++AGL RCGKSCRLRW NYLRP
Sbjct: 4 SPCCDKAGLKKGPWTPEEDQKLLAYIEEHGHGSWRSLPEKAGLQRCGKSCRLRWTNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG+ EE I++LH LLGNRWS IA +P RTDNEIKNYWNTHL K+LI GIDP
Sbjct: 64 DIKRGKFTVQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLIKMGIDP 123
Query: 153 RTHKPLNNPSSSSS 166
THK N SSS+
Sbjct: 124 VTHKHKNETLSSST 137
>At5g15310 myb-related protein - like
Length = 326
Score = 194 bits (494), Expect = 4e-50
Identities = 86/133 (64%), Positives = 102/133 (76%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K+GLK+GPWT EED+ L YI++ G G WR+LP++AGL RCGKSCRLRW NYLRP
Sbjct: 4 SPCCDKLGLKKGPWTPEEDQKLLAYIEEHGHGSWRSLPEKAGLHRCGKSCRLRWTNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG+ EE I++LH LLGNRWS IA +P RTDNEIKNYWNTHL K+L+ GIDP
Sbjct: 64 DIKRGKFNLQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLVKMGIDP 123
Query: 153 RTHKPLNNPSSSS 165
THKP N SS
Sbjct: 124 VTHKPKNETPLSS 136
>At4g05100 MYB - like protein
Length = 324
Score = 194 bits (494), Expect = 4e-50
Identities = 90/158 (56%), Positives = 114/158 (71%), Gaps = 10/158 (6%)
Query: 33 TPCCSKV-GLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLR 91
+PCC K GLK+GPWT EED+ L +YI G G WRTLPK AGL RCGKSCRLRW NYLR
Sbjct: 4 SPCCEKKNGLKKGPWTPEEDQKLIDYINIHGYGNWRTLPKNAGLQRCGKSCRLRWTNYLR 63
Query: 92 PSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGID 151
P +KRG+ + +EE+ I++LH ++GN+WS IA R+PGRTDNEIKNYWNTH+ K+L+ GID
Sbjct: 64 PDIKRGRFSFEEEETIIQLHSIMGNKWSAIAARLPGRTDNEIKNYWNTHIRKRLLKMGID 123
Query: 152 PRTHKPLNNPSSSSSIIPNTNHQTSTPFFAPSSSDHHH 189
P TH P + SSI+ ++ + +SS HHH
Sbjct: 124 PVTHTPRLDLLDISSILSSSIY---------NSSHHHH 152
>At3g62610 putative transcription factor (MYB11)
Length = 343
Score = 194 bits (493), Expect = 5e-50
Identities = 107/247 (43%), Positives = 139/247 (55%), Gaps = 15/247 (6%)
Query: 34 PCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPS 93
PCC KVG+K+G WT EED LS+YI+ GEG WR+LPK AGL RCGKSCRLRW+NYLR
Sbjct: 5 PCCEKVGIKKGRWTAEEDRTLSDYIQSNGEGSWRSLPKNAGLKRCGKSCRLRWINYLRSD 64
Query: 94 VKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDPR 153
+KRG I P+EED+I++LH LG RWS IA +PGRTDNEIKNYWN+HLS+KL P
Sbjct: 65 IKRGNITPEEEDVIVKLHSTLGTRWSTIASNLPGRTDNEIKNYWNSHLSRKLHGYFRKPT 124
Query: 154 THKPLNNPSSSSSIIP-NTNHQTSTPFFAPSSSDHHHPITITNNEPD---PSQTQQAQGC 209
+ N P T+ P F + +H P + N+ D P+ T + G
Sbjct: 125 VANTVENAPPPPKRRPGRTSRSAMKPKFILNPKNHKTPNSFKANKSDIVLPTTTIE-NGE 183
Query: 210 GNLVNDVSAMDNHHVLTNNNRG----DYGDDNG-NNNYGGDDVFTSFLDSLIN-----DD 259
G+ + + + + + G YGDD N + GDD D + + DD
Sbjct: 184 GDKEDALMVLSSSSLSGAEEPGLGPCGYGDDGDCNPSINGDDGALCLNDDIFDSCFLLDD 243
Query: 260 AFAAHRS 266
+ A H S
Sbjct: 244 SHAVHVS 250
>At4g28110 putative transcription factor (MYB41)
Length = 282
Score = 193 bits (490), Expect = 1e-49
Identities = 86/158 (54%), Positives = 114/158 (71%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K G+K+GPWT EED+ L +YI+ G G WRTLPK AGL RCGKSCRLRW NYLRP
Sbjct: 4 SPCCDKNGVKKGPWTAEEDQKLIDYIRFHGPGNWRTLPKNAGLHRCGKSCRLRWTNYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG+ + +EE+ I++LH ++GN+WS IA R+PGRTDNEIKN+WNTH+ K+L+ GIDP
Sbjct: 64 DIKRGRFSFEEEETIIQLHSVMGNKWSAIAARLPGRTDNEIKNHWNTHIRKRLVRSGIDP 123
Query: 153 RTHKPLNNPSSSSSIIPNTNHQTSTPFFAPSSSDHHHP 190
TH P + SS++ +Q + A +S +P
Sbjct: 124 VTHSPRLDLLDLSSLLSALFNQPNFSAVATHASSLLNP 161
>At2g47460 putative MYB family transcription factor
Length = 371
Score = 192 bits (489), Expect = 1e-49
Identities = 105/242 (43%), Positives = 142/242 (58%), Gaps = 29/242 (11%)
Query: 34 PCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPS 93
PCC KVG+KRG WT EED++LS YI+ GEG WR+LPK AGL RCGKSCRLRW+NYLR
Sbjct: 5 PCCEKVGIKRGRWTAEEDQILSNYIQSNGEGSWRSLPKNAGLKRCGKSCRLRWINYLRSD 64
Query: 94 VKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDPR 153
+KRG I P+EE+L+++LH LGNRWSLIAG +PGRTDNEIKNYWN+HLS+KL
Sbjct: 65 LKRGNITPEEEELVVKLHSTLGNRWSLIAGHLPGRTDNEIKNYWNSHLSRKL-------- 116
Query: 154 THKPLNNPSSSS--SIIPNTNHQTSTPFFAPSSSDHHHPITITNNEP-----DPSQTQQA 206
H + PS S S + TN ++ P P + + + +P +T++
Sbjct: 117 -HNFIRKPSISQDVSAVIMTNASSAPP--PPQAKRRLGRTSRSAMKPKIHRTKTRKTKKT 173
Query: 207 QGCGNLVNDVSAMDNHHVLTNNNRGD---------YGDDNGNN--NYGGDDVFTSFLDSL 255
DV+ D ++ ++ + YGDD N + GD+ +F D +
Sbjct: 174 SAPPEPNADVAGADKEALMVESSGAEAELGRPCDYYGDDCNKNLMSINGDNGVLTFDDDI 233
Query: 256 IN 257
I+
Sbjct: 234 ID 235
>At1g74080 putative transcription factor
Length = 333
Score = 191 bits (485), Expect = 4e-49
Identities = 95/195 (48%), Positives = 122/195 (61%), Gaps = 9/195 (4%)
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
TPCC GLK+G WT EED+ L Y+++ GEG WRTLP +AGL RCGKSCRLRW NYLRP
Sbjct: 4 TPCCRAEGLKKGAWTQEEDQKLIAYVQRHGEGGWRTLPDKAGLKRCGKSCRLRWANYLRP 63
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG+ + DEED I+ LH + GN+WS IA +IP RTDNEIKN+WNTH+ K L+ +GIDP
Sbjct: 64 DIKRGEFSQDEEDSIINLHAIHGNKWSAIARKIPRRTDNEIKNHWNTHIKKCLVKKGIDP 123
Query: 153 RTHKPLNNPSSSSSIIPNTNHQTSTPFFAPSSSDHHHPITITNNEPDPSQTQQAQGCGNL 212
THK L + + SS +H SS H +N S + A+ +
Sbjct: 124 LTHKSLLDGAGKSS-----DHSA----HPEKSSVHDDKDDQNSNNKKLSGSSSARFLNRV 174
Query: 213 VNDVSAMDNHHVLTN 227
N NH+VL++
Sbjct: 175 ANRFGHRINHNVLSD 189
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.129 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,297,898
Number of Sequences: 26719
Number of extensions: 408154
Number of successful extensions: 1837
Number of sequences better than 10.0: 223
Number of HSP's better than 10.0 without gapping: 170
Number of HSP's successfully gapped in prelim test: 53
Number of HSP's that attempted gapping in prelim test: 1481
Number of HSP's gapped (non-prelim): 298
length of query: 303
length of database: 11,318,596
effective HSP length: 99
effective length of query: 204
effective length of database: 8,673,415
effective search space: 1769376660
effective search space used: 1769376660
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146760.4


