

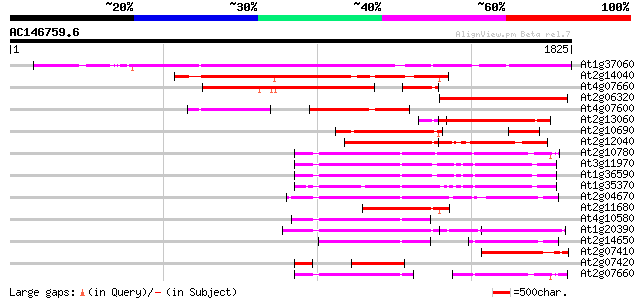
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146759.6 - phase: 0
(1825 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g37060 Athila retroelment ORF 1, putative 1390 0.0
At2g14040 putative retroelement pol polyprotein 741 0.0
At4g07660 putative athila transposon protein 562 e-160
At2g06320 putative retroelement pol polyprotein 526 e-149
At4g07600 453 e-127
At2g13060 pseudogene 451 e-126
At2g10690 putative retroelement pol polyprotein 433 e-121
At2g12040 T10J7.2 395 e-109
At2g10780 pseudogene 362 1e-99
At3g11970 hypothetical protein 362 1e-99
At1g36590 hypothetical protein 356 8e-98
At1g35370 hypothetical protein 351 3e-96
At2g04670 putative retroelement pol polyprotein 350 3e-96
At2g11680 putative retroelement pol polyprotein 338 2e-92
At4g10580 putative reverse-transcriptase -like protein 294 3e-79
At1g20390 hypothetical protein 275 2e-73
At2g14650 putative retroelement pol polyprotein 273 5e-73
At2g07410 putative retroelement pol polyprotein 259 8e-69
At2g07420 putative retroelement pol polyprotein 250 5e-66
At2g07660 putative retroelement pol polyprotein 246 9e-65
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 1390 bits (3599), Expect = 0.0
Identities = 790/1812 (43%), Positives = 1086/1812 (59%), Gaps = 167/1812 (9%)
Query: 79 NHRGRLTHVFRPANPVAFEIKASVQNGLKERQFDGTGTISPHEHLSHFAETCEFCVPPAN 138
NH R V P FEIK+ + ++ +F G P +HL F C
Sbjct: 22 NHNQRNGIVPPPVQNNNFEIKSGLIAMVQSNKFHGLPMEDPLDHLDEFDRLCSL-TKINR 80
Query: 139 VNEDQKKLRLFPFTLIGKAKDWLLTLPNGTIQTWEELELKFLEKYFPMSKYLDKKQEIAS 198
V+ED KLRLFPF+L KA W +LP G+I +W + + FL K+F S+ + +I+
Sbjct: 81 VSEDGFKLRLFPFSLGDKAHQWEKSLPQGSITSWNDCKKAFLAKFFSNSRTARLRNDISG 140
Query: 199 FRQGEGESLYDAWERFNLLLKRCPGHEFSEKQYLQFFTEGLTHSNRMFLDASAGGSLRVK 258
F Q E+ Y+AWERF +CP HE M LD ++ G+ K
Sbjct: 141 FTQTNNETFYEAWERFKGYQTQCPHHE-------------------MLLDTASNGNFLNK 181
Query: 259 TDHEVQILIENMASNEYRAETKKKERGVFGVSDTTSILANQA-AMNKQIE--ILTKEVHA 315
+ ++EN+A ++ + +R + SD+ + AMN +++ +L ++ H
Sbjct: 182 DVEDGWEVVENLAQSDGNYN-EDYDRSIRTSSDSDEKHRREMKAMNDKLDKLLLMQQKHI 240
Query: 316 YQLSNKQQAAAIRCDLCGEGHPNGECVPEGASEEANYVNYQRQNPYSMN------KHPNL 369
+ L + + +GE + EE +YV Q Q Y+ HPNL
Sbjct: 241 HFLGDDETLQV----------QDGETLQ---LEEVSYV--QNQGGYNKGFNNFKQNHPNL 285
Query: 370 SYSNNNTLN---PLFPNPQQQQQPRKPPGI-----------EETMMNFMK-MTQSNFEEM 414
SY + N N ++P+ QQQ QP+ P + ++ N+ + + F +
Sbjct: 286 SYRSTNVANRQDQVYPS-QQQNQPK--PFVPYNQGQGYVPKQQYQGNYQQQLPPPGFTQQ 342
Query: 415 KKSQDLERKNNEASRKMLETQLGQ------LAKQLAE-QNKGGFSGNTQENPKNETCNVI 467
++ L +++ + + GQ L+K++AE NK S N + I
Sbjct: 343 QQQPALTTPDSDLKNMLQQILQGQATGAMDLSKRMAEIHNKVDCSYNDINIKVEALTSKI 402
Query: 468 ELRSKKVLAPLVPKVPNKVDESVVEVDENIE------DEEVVEKESDQGVVENEKKKKTE 521
+ + PK +S+ E +E+ KES E+ + E
Sbjct: 403 RYIEGQTGSTAAPKFTGPSGKSMSNSKEYAHAITLRSGKELPTKESPNQNTEDSLDQDGE 462
Query: 522 -----GEKSERLIDEDSI------LRKSKSQILKDG-----DEPQIIPSYVKLPYPHLAK 565
G +E+ I+E + L + S +++ E IP K P P +
Sbjct: 463 DFCQNGNSAEKAIEEPILHQPTRPLAPAASPLVEKPAAAKTKENVFIPPPYKPPLPFPGR 522
Query: 566 KKKKEEGQFKKFMQL-FSQLQVNIPFGDALDQMPVYAKFMKEMLTGRRKPKDDENISLSE 624
KK ++K ++ L+V +P D L +P K++K+M+T R K + LS
Sbjct: 523 FKKVMIQKYKALLEKQLKNLEVTMPLVDCLALIPDSNKYVKDMITERIKEVQGM-VVLSH 581
Query: 625 NCSAILQRKL-PPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLGGGEPK 683
CSAI+Q+K+ P KL DPG+FT+PC++GP+ + LCDLGAS++LMPL + KKLG + K
Sbjct: 582 ECSAIIQQKIIPKKLGDPGSFTLPCALGPLAFSKCLCDLGASVSLMPLPVAKKLGFNKYK 641
Query: 684 PTRMTLTLADRSISYPFGVLEDVLVKVNDLIFPADFVILDMAEDEDMPLILGRPFLATGR 743
P ++L LADRS+ G+LED+ V + + P DFV+L+M E+ PLILGRPFLA R
Sbjct: 642 PCNISLILADRSVRISHGLLEDLPVMIGVVEVPTDFVVLEMDEEPKDPLILGRPFLARAR 701
Query: 744 ALIDVEMGQLMLRFHNE-QVVFNIFEAMKHRAENPKCYRIDVIDEIVEDNSREHKPTHPL 802
A+IDV+ G++ L + ++ F+I MK + I+ +D + + E T L
Sbjct: 702 AIIDVKKGKIDLNLGRDLKMTFDITNTMKKPTIEGNIFWIEEMDMLADKMLEELGETDHL 761
Query: 803 EETIVNSIENCEL-VEDLEVQECVKQLE--ANEQLLKPTKLEE-MSNEGKLGAKSLSNEE 858
+ + + +L +E L + + + + T L+ S+E L + S +
Sbjct: 762 QSALTKDSKEGDLHLETLGGEYILDHITRPTAHSVYSTTSLDHNSSSEANLVSDDWSEIK 821
Query: 859 EKIPELKELPSHLKYVFLSKDVSKPAIISSTLTPLEEEKLMRVLRENEGALGWKISDLKG 918
+LK LP L+YVFL + + P I++ LT + L+ L + A+G+ + D+KG
Sbjct: 822 APKVDLKPLPKGLRYVFLGLNSTYPVIVNDELTADQVNLLITELMKYRKAIGYSLDDIKG 881
Query: 919 ISPAYCMHRIHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVH 978
ISP C HRIH+E E S ++PQRRLNP +KEVVKKE+LKLLDAG+IYPISDS+WVSPVH
Sbjct: 882 ISPTLCTHRIHLENESYSSIEPQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVH 941
Query: 979 VVPKKGGMTVVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQ 1038
VPKKGGMTVV N K+ELIPTRT+TG RMCI+YR+LN A+RK+HFPLPF+D M+ERLA
Sbjct: 942 CVPKKGGMTVVKNSKDELIPTRTITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANH 1001
Query: 1039 AFYCFLDGYSGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSD 1098
+YCFLD YSG+ QI + P DQ KT FTCP+G FAY+RMPFGLC APATFQRCM SIFSD
Sbjct: 1002 PYYCFLDSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSD 1061
Query: 1099 MIEKNIEVFMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKI 1158
+IE+ +EVFMDDFSV+G SF CL +L VLKRC ETNL+LNWEKCHFMV EGIVLG KI
Sbjct: 1062 LIEEMVEVFMDDFSVYGSSFSSCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKI 1121
Query: 1159 SSKGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETE 1218
S +GIEVD+AKI+V+ +L PP VK +RSFLGHAGFYR FIKDFSK+A+PL LL KETE
Sbjct: 1122 SEEGIEVDKAKIDVMMQLQPPKTVKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETE 1181
Query: 1219 FDFDVECLNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIY 1278
F FD ECL AF LIK L+TAPI+ APNWD FE++
Sbjct: 1182 FAFDDECLTAFKLIKEALITAPIVQAPNWDFPFEII------------------------ 1217
Query: 1279 YASKVLNESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPR 1338
++++QV Y+TTEKELLAV+FA EKFRSYL+GSKV ++TDHAAL+++ K D+KPR
Sbjct: 1218 ----TMDDAQVRYATTEKELLAVVFAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKPR 1273
Query: 1339 LLRWVLLLQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAI---- 1394
LLRW+LLLQEFD+EI DKKG+EN VADHLSR+ + E I P+EQL AI
Sbjct: 1274 LLRWILLLQEFDMEIVDKKGIENGVADHLSRMR----IEDEVLIDDSMPEEQLMAIQQLN 1329
Query: 1395 -RERPWFADMANFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAG 1453
++ PW+AD N+ P ++ +++KKFFKD NH+ WD+PYL+ + D + R CV+
Sbjct: 1330 EKKLPWYADHVNYLVSGEEPPNLSSYEKKKFFKDINHFYWDEPYLYTLCKDKIYRTCVSE 1389
Query: 1454 EEIKNIVWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISK 1513
+EI+ I+ HCH AYGGH + +T +K+LQ+GFWWP++FKD +F+ +CD+
Sbjct: 1390 DEIEGILLHCHGFAYGGHFATFKTMSKILQAGFWWPSMFKDAQEFISKCDS--------- 1440
Query: 1514 RNEMPLTGIIEVEPFDCWGIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVV 1573
E FD WGIDFMGPFP S +ILV +DYV+KWVEAI ND++ V+
Sbjct: 1441 -----------FENFDVWGIDFMGPFPSSYGNKYILVAIDYVSKWVEAIASHTNDARVVL 1489
Query: 1574 NFLRKNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNR 1633
+ IF RFG PR++ISDGGKHF N E +LKK+ +KHK TSGQVE+SNR
Sbjct: 1490 KLFKTIIFPRFGVPRIVISDGGKHFINKGFENLLKKHGVKHK--------TSGQVEISNR 1541
Query: 1634 QLKQILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAY 1693
++K ILEKTV S+RKDWS KL+D LWAYR AFKT +G +P+ L++GK+CHLPVELE+KA
Sbjct: 1542 EIKAILEKTVGSTRKDWSAKLNDTLWAYRTAFKTPIGTTPFNLLYGKSCHLPVELEYKAM 1601
Query: 1694 WAIKALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQL 1753
WA+K LNFD A +KRL++LN+L ++RL AYE++ IYKERTK +HDK +V R+F VG
Sbjct: 1602 WAVKLLNFDIKTAEEKRLIQLNDLNKIRLEAYESSKIYKERTKSFHDKKIVSRDFKVGDQ 1661
Query: 1754 VLLFNSRLKLFPGKLKSKWSGPFMIESISPYGAVELSKPGEPGTFKVNAQRIKPYLGGEL 1813
VLLFNSRL+LFPGKLKS+WSGPF + ++ PYGA+ L+ G+ G F VN QR+K Y+ +
Sbjct: 1662 VLLFNSRLRLFPGKLKSRWSGPFSVTAVRPYGAITLA--GKNGDFTVNGQRLKKYMIDQF 1719
Query: 1814 PTNKGGVVLNDP 1825
V L +P
Sbjct: 1720 IPEGTSVPLEEP 1731
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 741 bits (1912), Expect = 0.0
Identities = 417/922 (45%), Positives = 559/922 (60%), Gaps = 124/922 (13%)
Query: 537 KSKSQILKDGDEPQIIPSYV-KLPYPHLAKKKKKEEGQFKKFMQLFSQLQVNIPFGDALD 595
K + Q L++ + ++P YV KLP+P +K ++E+ ++ F ++ QLQ
Sbjct: 2 KLRKQTLEEKAKKTVLPPYVPKLPFPGRQRKIQREK-EYSLFDEIMRQLQ---------- 50
Query: 596 QMPVYAKFMKEMLTGRRKPKDDENISLSENCSAILQRKLPPKLKDPGAFTIPCSIGPVDI 655
S CSAILQ +P K +DPG+F +P IG
Sbjct: 51 -------------------------RYSPECSAILQNVIPVKREDPGSFVLPSRIGEYTF 85
Query: 656 GRALCDLGASINLMPLSMMKKLGGGEPKPTRMTLTLADRSISYPFGVLEDVLVKVNDLIF 715
R LCDLGA ++LMP S+ K+LG PT+M+L L DRSIS+P GV EDV V+V +
Sbjct: 86 DRCLCDLGAGVSLMPFSVAKRLGDTNFTPTKMSLVLGDRSISFPVGVAEDVQVRVGNFYI 145
Query: 716 PADFVILDMAEDEDMPLILGRPFLATGRALIDVEMGQLMLRFHNEQVVFNIFEAMKHRAE 775
P DFVI+++ E+ LILGRPFL ALIDV ++ LR + FN+ M
Sbjct: 146 PTDFVIIELDEEPRHRLILGRPFLNIVAALIDVRKSKINLRIGDIVQEFNMERIMSKPTT 205
Query: 776 NPKCYRIDVIDEIVEDNSREHKPTHPLEETIVNSIENCELVEDLEVQECVKQLEANEQLL 835
+ + +D++DE+V + E PL+ + + + + + L+++ +
Sbjct: 206 ECQTFWVDIMDELVNELLAELNTEDPLQTVLTKEESEFGYLGEATTR-FARILDSSSPMT 264
Query: 836 KPTKLEEMSNEGKLGAKSLSNEEE-------KIP--ELKELPSHLKYVFLSKDVSKPAII 886
K E+ + A +S+ ++ P ELK LP+ L+ FL + + II
Sbjct: 265 KVVAFAELGDNEVEKALVVSSPKDCDDWSELNAPKMELKPLPARLRVAFLGPNSTYLVII 324
Query: 887 SSTLTPLEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEYKSVVQPQRRLNP 946
+ L +E L+ LR+ ALG+ + D+ GISP CMH IH+E E + V+ QRRLN
Sbjct: 325 NPELNNVESALLLCELRKYRKALGYSLDDITGISPTLCMHMIHLEGESITSVEHQRRLNS 384
Query: 947 TMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPTRTVTGRR 1006
+++VVKKE++KLLDAG+IYPISDS+WVSPVHVVPKKGG+ V+ NEKNELIPTRTVTG R
Sbjct: 385 NLRDVVKKEIMKLLDAGIIYPISDSTWVSPVHVVPKKGGVIVIKNEKNELIPTRTVTGHR 444
Query: 1007 MCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFT 1066
MCIDYR+LN+ATRKD+FPL F+DQM+ERL+ Q +YCFLDGY G+ QI + P+DQEKT FT
Sbjct: 445 MCIDYRKLNSATRKDNFPLSFIDQMLERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTFT 504
Query: 1067 CPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLN 1126
CP+G FAYRRMPFGLC APATFQ CM IFSDMIE +EVF+DDF
Sbjct: 505 CPYGTFAYRRMPFGLCNAPATFQHCMKYIFSDMIEDFMEVFIDDF--------------- 549
Query: 1127 AVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGVR 1186
VL+RC + +L+LNWEK HFMV +GIVLGHKIS KG+EVD+AKIE++
Sbjct: 550 LVLQRCEDKHLVLNWEKSHFMVRDGIVLGHKISEKGVEVDRAKIEIMR------------ 597
Query: 1187 SFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIAPN 1246
FLGHAGFYRRFIKDFSKIA+P LL KE + +FD CL AF+++K LV+A I+ P
Sbjct: 598 -FLGHAGFYRRFIKDFSKIARPCTQLLCKEQKSEFDDTCLQAFNIVKESLVSALIVQPPE 656
Query: 1247 WDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFALE 1306
W+L FE++CDASDY VGAVLGQRK+K HAIYYAS+ L+
Sbjct: 657 WELPFEVICDASDYVVGAVLGQRKDKKLHAIYYASRTLD--------------------- 695
Query: 1307 KFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENVVADH 1366
G++VIV T HAAL+YLL+K D+KPRLLRW+LLLQ+FDLEI+DKKG+EN VAD+
Sbjct: 696 -------GAQVIVHTVHAALRYLLSKKDAKPRLLRWILLLQDFDLEIKDKKGIENEVADN 748
Query: 1367 LSRLE-NNEVTKKE---GAIMAEFPD---EQLFAIR--------------ERPWFADMAN 1405
LSRL EV ++ G +A D +++ +R PW+AD AN
Sbjct: 749 LSRLRVQEEVLMRDILPGENLASIEDCYMDEVGRLRVSTLELMTLHTGESNLPWYADFAN 808
Query: 1406 FKAGNIIPDDMEQHQRKKFFKD 1427
+ + + P D + +KK K+
Sbjct: 809 YLSCEVPPPDFTGYFKKKLLKE 830
>At4g07660 putative athila transposon protein
Length = 724
Score = 562 bits (1449), Expect = e-160
Identities = 298/596 (50%), Positives = 395/596 (66%), Gaps = 38/596 (6%)
Query: 628 AILQRKL-PPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLGGGEPKPTR 686
AI Q+K+ P KL DPG+FT+PCS+GP+ R LCDLGA ++LMPLS+ K+LG + K
Sbjct: 3 AITQKKIVPKKLIDPGSFTLPCSLGPLAFKRCLCDLGALVSLMPLSVAKRLGFTQYKSCN 62
Query: 687 MTLTLADRSISYPFGVLEDVLVKVNDLIFPADFVILDMAEDEDMPLILGRPFLATGRALI 746
++L LADRS+ P + E++ +++ + P DFV+L+M E+ PLILGRPFLAT A+
Sbjct: 63 ISLILADRSVRIPHSLFENLPIRIGAVDIPTDFVVLEMDEEPKDPLILGRPFLATAGAMN 122
Query: 747 DVEMGQLMLRFHNE-QVVFNIFEAMKHRAENPKCYRIDVIDEIVEDNSREHKPTHPLEET 805
DV+ G++ L ++ F++ +AMK + + I+ ID++ ++ E L
Sbjct: 123 DVKKGKIDLNLGKYCRMTFDVKDAMKKPTIKGQLFWIEEIDQLADELLEERAEEDHLYSA 182
Query: 806 IVNS-------IENCELVEDLEVQECVKQLEANEQLLKP-TKLEEMSNEGKL-------- 849
+ +E + L+ + +++ E E+L P T++ MS EG
Sbjct: 183 LTKRGEDGFLHLETLGYQKLLDSHKAMEESEPFEELNGPETEVMVMSEEGSTQVQPAHSR 242
Query: 850 ------GAKSLSNEEEKI--------------PELKELPSHLKYVFLSKDVSKPAIISST 889
++SN E I +LK LP L+YVF + + P II+
Sbjct: 243 TYSTNNSTSTISNSGELIIPTSDDWSELKAPKVDLKSLPKGLRYVFFGPNSTYPVIINVE 302
Query: 890 LTPLEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEYKSVVQPQRRLNPTMK 949
L E L+ LR+ + A+G+ +SD+K ISP+ C HRIH+E E S ++PQRRLN +K
Sbjct: 303 LNDNEVNLLLSELRKYKRAIGYSLSDIKRISPSLCNHRIHLENESYSSIEPQRRLNLNLK 362
Query: 950 EVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPTRTVTGRRMCI 1009
EVVKKE+LKLLDAG+IYPISDS+WV PVH VPKKGGMTVV NEK+ELIPTRT+TG R+CI
Sbjct: 363 EVVKKEILKLLDAGVIYPISDSTWVFPVHCVPKKGGMTVVKNEKDELIPTRTITGHRVCI 422
Query: 1010 DYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFTCPF 1069
DYR+LN A+RKDHFPLPF +QM+E LA + CFLDGYSG+ QI + P DQEKT FTCP+
Sbjct: 423 DYRKLNAASRKDHFPLPFTNQMLEGLANHLYNCFLDGYSGFFQIPIHPNDQEKTTFTCPY 482
Query: 1070 GVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLNAVL 1129
G FAY+RMPFGLC AP TFQRCM SIFSD+IEK +EVFMDDFSV+G SF CL +L VL
Sbjct: 483 GTFAYKRMPFGLCNAPTTFQRCMTSIFSDLIEKMVEVFMDDFSVYGPSFSSCLLNLGRVL 542
Query: 1130 KRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGV 1185
+ ETNL+LNWEKC+FMV EGIVLGHKIS KGIEVD+ KI+V+ +L PP VK +
Sbjct: 543 TKWEETNLVLNWEKCYFMVKEGIVLGHKISEKGIEVDKEKIKVMMQLQPPKTVKDI 598
Score = 135 bits (340), Expect = 2e-31
Identities = 69/118 (58%), Positives = 88/118 (74%), Gaps = 4/118 (3%)
Query: 1277 IYYASKVLNESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSK 1336
IYYAS+ L+E+Q Y+TTEKELL V+FA +KFRSYL+ SKV V+TDHAAL+++ K D+K
Sbjct: 598 IYYASRTLDEAQGRYATTEKELLVVVFAFKKFRSYLVESKVTVYTDHAALRHMYAKKDTK 657
Query: 1337 PRLLRWVLLLQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAI 1394
PRLLR +LLLQEFD+EI +KKG+EN ADHLSR+ E I P+EQL +
Sbjct: 658 PRLLRGILLLQEFDMEIVEKKGIENGAADHLSRMRIEEPI----LIDDSMPEEQLMVV 711
>At2g06320 putative retroelement pol polyprotein
Length = 466
Score = 526 bits (1356), Expect = e-149
Identities = 243/416 (58%), Positives = 315/416 (75%), Gaps = 2/416 (0%)
Query: 1398 PWFADMANFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIK 1457
PW+AD N+ A I P ++ ++RK FF+D +HY WD+PYL+ + D + R V+ +E++
Sbjct: 38 PWYADHVNYLACGIEPPNLTSYERKNFFRDIHHYYWDEPYLYTLCKDKIYRSYVSEDEVE 97
Query: 1458 NIVWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEM 1517
I+ HCH SAYGGH + +T +K+LQ+GFWWPT+FKD +FV +CD+CQR G+IS+RNEM
Sbjct: 98 GILLHCHDSAYGGHFATFKTVSKILQAGFWWPTMFKDAQEFVSKCDSCQRKGNISRRNEM 157
Query: 1518 PLTGIIEVEPFDCWGIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVVNFLR 1577
P I+EV+ FD WGIDFMGPFP S +ILV VDYV+KWVEAI ND+K V+ +
Sbjct: 158 PQNPILEVDIFDVWGIDFMGPFPSSYGNKYILVVVDYVSKWVEAIASPTNDAKVVLKLFK 217
Query: 1578 KNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQ 1637
IF RFG V+ISDGGKHF N E +LKK+ +KHKVATPYHPQTSGQVE+SNR++K
Sbjct: 218 TIIFPRFGVSWVVISDGGKHFINKVFENLLKKHGVKHKVATPYHPQTSGQVEISNREIKT 277
Query: 1638 ILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAYWAIK 1697
ILEKTV +RKDWS KLDDALWAY+ AFKT +G +P+ L+ K+CHL VELE+KA WA+K
Sbjct: 278 ILEKTVGITRKDWSTKLDDALWAYKTAFKTPIGTTPFNLLCVKSCHLHVELEYKAMWAVK 337
Query: 1698 ALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQLVLLF 1757
LNFD A +KRL++L+EL+E+RL AYE++ IYKERTK +HDK ++ ++F VG VLLF
Sbjct: 338 LLNFDIKTAEEKRLIQLSELDEIRLEAYESSKIYKERTKLFHDKKIITKDFQVGDQVLLF 397
Query: 1758 NSRLKLFPGKLKSKWSGPFMIESISPYGAVELSKPGEPGTFKVNAQRIKPYLGGEL 1813
NSRLK+FPGKLKS+WSGPF I + PYGAV L+ G+ G F VN QR+K YL ++
Sbjct: 398 NSRLKIFPGKLKSRWSGPFCITEVRPYGAVTLA--GKSGVFTVNGQRLKKYLANQI 451
>At4g07600
Length = 630
Score = 453 bits (1165), Expect = e-127
Identities = 217/325 (66%), Positives = 250/325 (76%), Gaps = 18/325 (5%)
Query: 974 VSPVHVVPKKGGMTVVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIE 1033
VSPV +PKKGG+TV+ NEK+ELIP RT+TG RMCIDYR LN A+R DHFPLPF DQM+E
Sbjct: 324 VSPVQCIPKKGGITVIKNEKDELIPIRTITGLRMCIDYRNLNAASRNDHFPLPFTDQMLE 383
Query: 1034 RLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCML 1093
RLA +YCFLDGYSG+ QI + P D EKT FTCP+G FAY RMPFGLC APATFQRCM
Sbjct: 384 RLANHPYYCFLDGYSGFFQIPIHPNDHEKTTFTCPYGTFAYERMPFGLCNAPATFQRCMT 443
Query: 1094 SIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIV 1153
SIFSD+IE+ +EVFMDDFSV+G SF CL +L VL RC ETNL+LNWEKCHFMV EGI+
Sbjct: 444 SIFSDIIEEMVEVFMDDFSVYGPSFSSCLLNLGRVLTRCEETNLVLNWEKCHFMVKEGIM 503
Query: 1154 LGHKISSKGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLL 1213
LGHKIS KGIEVD+ FLGHAGFYRRFIKDFSKIA+PL LL
Sbjct: 504 LGHKISEKGIEVDKG------------------CFLGHAGFYRRFIKDFSKIARPLTRLL 545
Query: 1214 VKETEFDFDVECLNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKF 1273
KETEF+FD +C +F IK LV+AP++ A NWD FE+MCDA DYAVGAVLGQ+ +K
Sbjct: 546 CKETEFEFDEDCPKSFHTIKEALVSAPVVRATNWDYPFEIMCDALDYAVGAVLGQKIDKK 605
Query: 1274 FHAIYYASKVLNESQVNYSTTEKEL 1298
H IYYAS+ L+E+Q Y+TTEKEL
Sbjct: 606 LHVIYYASRTLDEAQGRYATTEKEL 630
Score = 149 bits (377), Expect = 1e-35
Identities = 98/281 (34%), Positives = 165/281 (57%), Gaps = 11/281 (3%)
Query: 577 FMQLFSQLQVNIPFGDALDQMPVYAKFMKEMLTGRRKPKDDENISLSENCSAILQRKL-P 635
F + ++++ IP DAL + KF+K+++ R + + LS CSAI+Q+K+ P
Sbjct: 2 FAKNIKEVELRIPLVDALALILDTHKFLKDLIVERIQEVQGM-VVLSHECSAIIQKKIVP 60
Query: 636 PKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLGGGEPKPTRMTLTLADRS 695
KL DPG+FT+PC +G V R LCDLGA ++ MPLS+ K+LG + K ++L LADRS
Sbjct: 61 KKLSDPGSFTLPCFLGTVAFNRCLCDLGALVSPMPLSIAKRLGFTQYKSCNISLILADRS 120
Query: 696 ISYPFGVLEDVLVKVNDLIFPADFVILDMAEDEDMPLILGRPFLATGRALIDVEMGQLML 755
+ G+L+++ +++ DFVIL+M E+ PLIL RPFLAT A+IDV+ G++ L
Sbjct: 121 VRISHGLLKNLPIRIGAAEISTDFVILEMDEEPKDPLILRRPFLATAGAMIDVKKGKIDL 180
Query: 756 RFHNE-QVVFNIFEAMKHRAENPKCYRID----VIDEIVEDNSREHKPTHPLEETIVNSI 810
+ ++ F+I +AMK + + I+ + DE++E+ ++E L +T +
Sbjct: 181 NLGKDFRMTFDIKDAMKKPNIEGQLFWIEEMDQLADELLEELAQEDHLNSALTKTGQDGF 240
Query: 811 ENCELVEDLEVQECVKQLEAN---EQLLKP-TKLEEMSNEG 847
+ +++ ++ + K +E + E+L P TK+ M+ EG
Sbjct: 241 LHLKVLGYQKLLDSHKAMEESKPFEELNGPATKVMVMNEEG 281
>At2g13060 pseudogene
Length = 930
Score = 451 bits (1161), Expect = e-126
Identities = 205/363 (56%), Positives = 271/363 (74%), Gaps = 6/363 (1%)
Query: 1395 RERPWFADMANFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGE 1454
++ PW+AD+ N+ A ++ PD+ + +K+F ++ Y WD+PYL+K S DG+ RRC+A
Sbjct: 134 KDYPWYADIVNYLAADVEPDNFTDYNKKRFLREIRRYQWDEPYLYKHSYDGIYRRCIAAT 193
Query: 1455 EIKNIVWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKR 1514
E+ +I+ HCHSS+YGGH + +T +KVLQ+ FWWPT+F+D F+ +CD CQR G ISKR
Sbjct: 194 EVPSILSHCHSSSYGGHFATFKTVSKVLQADFWWPTMFRDTQKFISQCDPCQRRGKISKR 253
Query: 1515 NEMPLTGIIEVEPFDCWGIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVVN 1574
NEMP +EVE FD WGIDFMGPFPPS+ L+ILV VDYV+KWVEAI + NDS V+
Sbjct: 254 NEMPPNFRLEVEVFDRWGIDFMGPFPPSNKNLYILVDVDYVSKWVEAIASLKNDSAVVMK 313
Query: 1575 FLRKNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQ 1634
+ IF RFG PR++ISDG KHF N LE +L +Y ++H+VATPYHPQTSGQVEVSNRQ
Sbjct: 314 LFKSIIFPRFGVPRIVISDGDKHFINKILEKLLLQYGVQHRVATPYHPQTSGQVEVSNRQ 373
Query: 1635 LKQILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAYW 1694
+K+ILEKTV ++K+WS KL DALWAY+ AFKT LG +P+ L++GKACHLPVELEHKA W
Sbjct: 374 IKEILEKTVGKAKKEWSYKLYDALWAYKTAFKTPLGTTPFHLLYGKACHLPVELEHKAAW 433
Query: 1695 AIKALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQLV 1754
A+K +NFD AG+ +EL+E+R+ AY+N+ +YKERTK YHDK ++ R F +
Sbjct: 434 AVKMMNFDIKSAGE------SELDEIRIHAYDNSKLYKERTKAYHDKKILTRTFEPNDQI 487
Query: 1755 LLF 1757
L F
Sbjct: 488 LRF 490
Score = 70.1 bits (170), Expect = 1e-11
Identities = 39/92 (42%), Positives = 55/92 (59%), Gaps = 19/92 (20%)
Query: 1330 LTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDE 1389
+ K D+KPRLL W+LLLQEFD+E+RDKKGVEN VADHLSR+ ++ + I P+E
Sbjct: 1 MQKKDAKPRLLIWILLLQEFDIEVRDKKGVENGVADHLSRIRIDD----DVPINDFLPEE 56
Query: 1390 QLFAIRERPWFADMANFKAGNIIPDDMEQHQR 1421
++ I + + DD+ +H R
Sbjct: 57 NIYMI---------------DTVEDDVYKHDR 73
>At2g10690 putative retroelement pol polyprotein
Length = 622
Score = 433 bits (1114), Expect = e-121
Identities = 223/368 (60%), Positives = 267/368 (71%), Gaps = 29/368 (7%)
Query: 1060 QEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFD 1119
++KT FTCP G FAY+RM FGLC AP TFQR M SIFSD IE+ +EVFMDDFS F
Sbjct: 163 KKKTTFTCPNGTFAYKRMSFGLCNAPGTFQRSMTSIFSDFIEEIMEVFMDDFS----GFS 218
Query: 1120 QCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPP 1179
CL +L VL+RC ETNL+LNWEKCHFMV EGIVLGHKIS KGIEVD+AKI+V+ +L PP
Sbjct: 219 SCLLNLCRVLERCEETNLVLNWEKCHFMVHEGIVLGHKISGKGIEVDKAKIDVMIQLQPP 278
Query: 1180 MNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLVTA 1239
VK +RSFLGHA FYRRFIKDFSKIA+PL LL KETEF+FD +CL AF LIK LV+A
Sbjct: 279 KTVKDIRSFLGHAEFYRRFIKDFSKIARPLTRLLCKETEFNFDEDCLKAFHLIKETLVSA 338
Query: 1240 PIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKELL 1299
PI APNWD FE+MCDA DYAVGAVL Q+ + H IYYAS+ ++E+Q Y+T EKELL
Sbjct: 339 PIFQAPNWDHPFEIMCDAFDYAVGAVLDQKIDDKLHVIYYASRTMDEAQTRYATIEKELL 398
Query: 1300 AVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKKGV 1359
AV+FA EK RSYL+G KV V+ DHAAL+++ K ++K RLLRW+LLLQEFD+EI DKKGV
Sbjct: 399 AVVFAFEKIRSYLVGFKVKVYIDHAALRHIYAKKETKSRLLRWILLLQEFDMEIIDKKGV 458
Query: 1360 ENVVADHLSRLENNEVTKKEGAIMAEFPDEQLF-------------------AIRER--P 1398
EN VADHLSR+ + + + P+EQL A+ E P
Sbjct: 459 ENGVADHLSRMRIEDSVPIDDTM----PEEQLMFYDLVNKSFDTKDMPEEADAVEEEKLP 514
Query: 1399 WFADMANF 1406
W+AD+ N+
Sbjct: 515 WYADLVNY 522
Score = 127 bits (320), Expect = 5e-29
Identities = 59/98 (60%), Positives = 78/98 (79%)
Query: 1624 TSGQVEVSNRQLKQILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACH 1683
TSGQVEVSNRQ+K IL K V S++DWS KLD+ LWAYR A+KT +G +P+Q+++GK+CH
Sbjct: 524 TSGQVEVSNRQMKDILTKVVGVSKRDWSTKLDETLWAYRTAYKTPIGRTPFQMLYGKSCH 583
Query: 1684 LPVELEHKAYWAIKALNFDQTLAGKKRLLKLNELEEMR 1721
LPVE+E+KA WA K LN + A +KR + L+ELEE+R
Sbjct: 584 LPVEVEYKAIWATKLLNLEIKGAQEKRPIDLHELEEIR 621
Score = 35.8 bits (81), Expect = 0.24
Identities = 24/70 (34%), Positives = 35/70 (49%), Gaps = 2/70 (2%)
Query: 842 EMSNEGKLGAKSLSNEEEKIP--ELKELPSHLKYVFLSKDVSKPAIISSTLTPLEEEKLM 899
EM + G S E K P +LK LP L+Y FL + + P II+ L+ + +L+
Sbjct: 81 EMLDHGGENISSDDLSELKAPKLDLKPLPKGLRYAFLGPNDTYPVIINDGLSDEQVNQLL 140
Query: 900 RVLRENEGAL 909
LR+ G L
Sbjct: 141 NELRKYRGQL 150
>At2g12040 T10J7.2
Length = 976
Score = 395 bits (1014), Expect = e-109
Identities = 196/306 (64%), Positives = 242/306 (79%), Gaps = 4/306 (1%)
Query: 1089 QRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMV 1148
+R M+SIF+DMIE +EVFMDDFSV+G F+ CL +L VL RC E +L+LNWEKCHF V
Sbjct: 79 ERGMMSIFTDMIEDIMEVFMDDFSVYGSLFEDCLENLYKVLARCEEKHLVLNWEKCHFRV 138
Query: 1149 TEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKP 1208
+GIVLGH+IS GIE D+AKIEV+ L N+K VRSFLGHAGFYRRFIKDFSKIA+P
Sbjct: 139 QDGIVLGHRISEYGIEADRAKIEVMTSLQALDNLKAVRSFLGHAGFYRRFIKDFSKIARP 198
Query: 1209 LCNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQ 1268
L +LL KE +F+F EC +AF IK L++API+ P+WDL FE+MCDASD+AVGAVLGQ
Sbjct: 199 LTSLLCKEVKFEFTQECHDAFQQIKQALISAPIVQPPDWDLPFEVMCDASDFAVGAVLGQ 258
Query: 1269 RKNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKY 1328
RK+K HAIYYAS+ L++++ NY+TTEKE LAV+FA EKFRSYL+G KVIV TDHAALKY
Sbjct: 259 RKDKKLHAIYYASRTLDDAKRNYATTEKEFLAVVFAFEKFRSYLVGLKVIVHTDHAALKY 318
Query: 1329 LLTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPD 1388
L+ D+KPRLLRW+LLLQEFD+E+RDKKGVE VADHLSR+ ++ + I P+
Sbjct: 319 LMQNKDAKPRLLRWILLLQEFDIEVRDKKGVEKCVADHLSRIRIDD----DVPINDFLPE 374
Query: 1389 EQLFAI 1394
E ++ I
Sbjct: 375 ENIYMI 380
Score = 311 bits (798), Expect = 2e-84
Identities = 164/354 (46%), Positives = 216/354 (60%), Gaps = 40/354 (11%)
Query: 1395 RERPWFADMANFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGE 1454
++ PW+AD+ + ++ D+ + +K+F ++ Y WD PYL ++ GE
Sbjct: 456 KDYPWYADIVIYLGADVELDNFIDYNKKRFLREIRRYYWDKPYL----PISIVLMEFIGE 511
Query: 1455 EIKNIVWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKR 1514
HC + S R SG C + D CQR G IS
Sbjct: 512 ------MHC-CNRDSIQSSSSRLLVAYNVSG---------CSEIHIASDPCQRRGKISNC 555
Query: 1515 NEMPLTGIIEVEPFDCWGIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVVN 1574
NEMP I+EV+ FDCWGI+FMGP PPS+ L+ILV VDYV+KWVEAI NDS V+
Sbjct: 556 NEMPQKFILEVKVFDCWGINFMGPIPPSNKNLYILVVVDYVSKWVEAIASPKNDSAVVMK 615
Query: 1575 FLRKNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQ 1634
+ IF FG PR++ISDGGKHF N L +L +Y ++H+VATPYHPQTSGQVEVSNRQ
Sbjct: 616 LFKCIIFPHFGVPRIVISDGGKHFINKILTKLLLQYGVQHRVATPYHPQTSGQVEVSNRQ 675
Query: 1635 LKQILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAYW 1694
+K+ILEKTV AFKT LG +P+ L++GKACHLPVELEHKA W
Sbjct: 676 IKEILEKTV--------------------AFKTPLGTTPFHLLYGKACHLPVELEHKAAW 715
Query: 1695 AIKALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREF 1748
+K LNFD AG++RL++LNEL+E+++ AY+N+ +YKERTK YHDK ++ R F
Sbjct: 716 TVKMLNFDIKSAGERRLIQLNELDEIQIHAYDNSKLYKERTKAYHDKKILTRTF 769
Score = 37.7 bits (86), Expect = 0.062
Identities = 18/33 (54%), Positives = 24/33 (72%)
Query: 925 MHRIHMEAEYKSVVQPQRRLNPTMKEVVKKEVL 957
MHRIH+E E KS V+ QRRLN K+ ++K +L
Sbjct: 1 MHRIHLEDESKSSVEHQRRLNLIQKKRLRKRLL 33
>At2g10780 pseudogene
Length = 1611
Score = 362 bits (929), Expect = 1e-99
Identities = 268/889 (30%), Positives = 424/889 (47%), Gaps = 83/889 (9%)
Query: 928 IHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMT 987
I +E + + R+ P +KK++ +LLD G I P S S W +PV V KK G
Sbjct: 651 IELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRP-SSSPWGAPVLFVKKKDGSF 709
Query: 988 VVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGY 1047
R+CIDYR LN T K+ +PLP +D+++++L G ++ +D
Sbjct: 710 ------------------RLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLA 751
Query: 1048 SGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVF 1107
SGY+QI + P D KTAF + F + MPFGL APA F + M +F D +++ + +F
Sbjct: 752 SGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIF 811
Query: 1108 MDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQ 1167
++D V+ KS++ HL AVL+R E L KC F LGH IS +G+ VD
Sbjct: 812 INDILVYSKSWEAHQEHLRAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDP 871
Query: 1168 AKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLN 1227
KI I++ P P N +RSFLG AG+YRRF+ F+ +A+PL L K+T F++ EC
Sbjct: 872 EKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEK 931
Query: 1228 AFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNES 1287
+F +K L AP+++ P + + DAS +G VL Q+ + I YAS+ L +
Sbjct: 932 SFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGS----VIAYASRQLRKH 987
Query: 1288 QVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQ 1347
+ NY T + E+ AV+F L+ +RSYL G+KV ++TDH +LKY+ T+ + R RW+ L+
Sbjct: 988 EKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQRRWMELVA 1047
Query: 1348 EFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAIRERPWFADMANFK 1407
+++L+I G N VAD LSR + ++ + + + ++
Sbjct: 1048 DYNLDIAYHPGKANQVADALSRRRSEVEAERSQVDLV----NMMGTLHVNALSKEVEPLG 1103
Query: 1408 AGNIIPDDMEQHQRKKFFKDANHYLW--DDPYLFKVSTDGLI----RRCVAGEEI--KNI 1459
G D+ R +D W ++ ++ S +G I R CV + + I
Sbjct: 1104 LGAADQADLLSRIRLAQERDEEIKGWAQNNKTEYQTSNNGTIVVNGRVCVPNDRALKEEI 1163
Query: 1460 VWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEMPL 1519
+ H S + H G + L+ + W + KD +V +C CQ + +++P
Sbjct: 1164 LREAHQSKF-SIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQ---LVKAEHQVP- 1218
Query: 1520 TGIIEVEPFDCW-----GIDFMGPFPPSSSYLH--ILVCVDYVTKWVEAIPCVAND-SKT 1571
+G+++ P W +DF+ P H + V VD +TK + D ++
Sbjct: 1219 SGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEI 1278
Query: 1572 VVNFLRKNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVS 1631
+ I G P ++SD F + F + K + ++T YHPQT Q E +
Sbjct: 1279 IAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQSERT 1338
Query: 1632 NRQLKQILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHK 1691
+ L+ +L V +W + L +AY +F+ +G+SPY+ ++G+AC P+
Sbjct: 1339 IQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRACRTPL----- 1393
Query: 1692 AYWAIKALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKE---RTKRYHDKGLVRREF 1748
W T G++RL ++E + KE R K Y +K EF
Sbjct: 1394 -CW---------TPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANKRRKELEF 1443
Query: 1749 YVGQLVLL----------FNSRLKLFPGKLKSKWSGPFMIESISPYGAV 1787
VG LV L F SR KL P ++ GP+ + I GAV
Sbjct: 1444 QVGDLVYLKAMTYKGAGRFTSRKKLSP-----RYVGPYKV--IERVGAV 1485
Score = 34.7 bits (78), Expect = 0.52
Identities = 21/73 (28%), Positives = 36/73 (48%), Gaps = 3/73 (4%)
Query: 681 EPKPTRMTLTLADRSISYPFGVLEDVLVKVNDLIFPADFVILDMAEDEDMPLILGRPFLA 740
EP + A YP G++ + V VN + PAD +I+ + + + +ILG +L
Sbjct: 499 EPNTDYGMVRAAGGQAMYPTGLVRGISVVVNGVNMPADLIIVPLKKHD---VILGMDWLG 555
Query: 741 TGRALIDVEMGQL 753
+A ID G++
Sbjct: 556 KYKAHIDCHRGRI 568
>At3g11970 hypothetical protein
Length = 1499
Score = 362 bits (928), Expect = 1e-99
Identities = 277/869 (31%), Positives = 413/869 (46%), Gaps = 77/869 (8%)
Query: 926 HRIHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGG 985
H+I + V Q R + K + K V LL G + S S + SPV +V KK G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQA-SSSPYASPVVLVKKKDG 649
Query: 986 MTVVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLD 1045
R+C+DYR LN T KD FP+P ++ +++ L G + +D
Sbjct: 650 TW------------------RLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKID 691
Query: 1046 GYSGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIE 1105
+GY+Q+ + P+D +KTAF G F Y MPFGL APATFQ M IF + K +
Sbjct: 692 LRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVL 751
Query: 1106 VFMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEV 1165
VF DD V+ S ++ HL V + L KC F V + LGH IS++GIE
Sbjct: 752 VFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIET 811
Query: 1166 DQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVEC 1225
D AKI+ +++ P P +K +R FLG AG+YRRF++ F IA PL + L K F++
Sbjct: 812 DPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPL-HALTKTDAFEWTAVA 870
Query: 1226 LNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLN 1285
AF +K L AP++ P +D F + DA +GAVL Q H + Y S+ L
Sbjct: 871 QQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEG----HPLAYISRQLK 926
Query: 1286 ESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLL 1345
Q++ S EKELLAVIFA+ K+R YL+ S I+ TD +LKYLL + + P +W+
Sbjct: 927 GKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPK 986
Query: 1346 LQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAIRERPWFADM-A 1404
L EFD EI+ ++G ENVVAD LSR+E +EV + E D+ A
Sbjct: 987 LLEFDYEIQYRQGKENVVADALSRVEGSEVL------------HMAMTVVECDLLKDIQA 1034
Query: 1405 NFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKN-IVWHC 1463
+ + + D + QR K ++ W L + S + A + IKN I+
Sbjct: 1035 GYANDSQLQDIITALQRDPDSK--KYFSWSQNILRRKSKIVV----PANDNIKNTILLWL 1088
Query: 1464 HSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEMPLTGII 1523
H S GG HSG + ++ F+W + KD ++R C CQ+ K + G++
Sbjct: 1089 HGSGVGG-HSGRDVTHQRVKGLFYWKGMIKDIQAYIRSCGTCQQ----CKSDPAASPGLL 1143
Query: 1524 EVEPF--DCW---GIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCV-ANDSKTVVNFLR 1577
+ P W +DF+ P S I+V VD ++K I + TV +
Sbjct: 1144 QPLPIPDTIWSEVSMDFIEGLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAHAYL 1203
Query: 1578 KNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQ 1637
N+F G P ++SD F + F + K+ + YHPQ+ GQ EV NR L+
Sbjct: 1204 DNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLET 1263
Query: 1638 ILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKA--CHLPVELEHKAYWA 1695
L + WS+ L A + Y + + ++P+++V+G+ HLP L ++ A
Sbjct: 1264 YLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPY-LPGESKVA 1322
Query: 1696 IKALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQLVL 1755
+ A + L E E+M L + + + R K++ D+ REF +G V
Sbjct: 1323 VVARS-------------LQEREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVY 1369
Query: 1756 L------FNSRLKLFPGKLKSKWSGPFMI 1778
+ S + KL K+ GP+ I
Sbjct: 1370 VKLQPYRQQSVVMRANQKLSPKYFGPYKI 1398
>At1g36590 hypothetical protein
Length = 1499
Score = 356 bits (913), Expect = 8e-98
Identities = 276/869 (31%), Positives = 411/869 (46%), Gaps = 77/869 (8%)
Query: 926 HRIHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGG 985
H+I + V Q R + K + K V LL G + S S + SPV +V KK G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQA-SSSPYASPVVLVKKKDG 649
Query: 986 MTVVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLD 1045
R+C+DYR LN T KD FP+P ++ +++ L G + +D
Sbjct: 650 TW------------------RLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKID 691
Query: 1046 GYSGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIE 1105
+GY+Q+ + P+D +KTAF G F Y MPFGL APATFQ M IF + K +
Sbjct: 692 LRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVL 751
Query: 1106 VFMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEV 1165
VF DD V+ S ++ HL V + L KC F V + LGH IS++GIE
Sbjct: 752 VFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIET 811
Query: 1166 DQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVEC 1225
D AKI+ +++ P P +K +R FLG AG+YRRF++ F IA PL + L K F++
Sbjct: 812 DPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPL-HALTKTDAFEWTAVA 870
Query: 1226 LNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLN 1285
AF +K L AP++ P +D F + DA +GAVL Q H + Y S+ L
Sbjct: 871 QQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEG----HPLAYISRQLK 926
Query: 1286 ESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLL 1345
Q++ S EKELLAVIFA+ K+R YL+ S I+ TD +LKYLL + + P +W+
Sbjct: 927 GKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPK 986
Query: 1346 LQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAIRERPWFADM-A 1404
L EFD EI+ ++G ENVVAD LSR+E +EV + E D+ A
Sbjct: 987 LLEFDYEIQYRQGKENVVADALSRVEGSEVL------------HMAMTVVECDLLKDIQA 1034
Query: 1405 NFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKN-IVWHC 1463
+ + + D + QR K ++ W L + S + A + IKN I+
Sbjct: 1035 GYANDSQLQDIITALQRDPDSK--KYFSWSQNILRRKSKIVV----PANDNIKNTILLWL 1088
Query: 1464 HSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEMPLTGII 1523
H S GG HSG + ++ F+ + KD ++R C CQ+ K + G++
Sbjct: 1089 HGSGVGG-HSGRDVTHQRVKGLFYSKGMIKDIQAYIRSCGTCQQ----CKSDPAASPGLL 1143
Query: 1524 EVEPF--DCW---GIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCV-ANDSKTVVNFLR 1577
+ P W +DF+ P S I+V VD ++K I + TV
Sbjct: 1144 QPLPIPDTIWSEVSMDFIEGLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAQAYL 1203
Query: 1578 KNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQ 1637
N+F G P ++SD F + F + K+ + YHPQ+ GQ EV NR L+
Sbjct: 1204 DNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLET 1263
Query: 1638 ILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKA--CHLPVELEHKAYWA 1695
L + WS+ L A + Y + + ++P+++V+G+ HLP L ++ A
Sbjct: 1264 YLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPY-LPGESKVA 1322
Query: 1696 IKALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQLVL 1755
+ A + L E E+M L + + + R K++ D+ REF +G V
Sbjct: 1323 VVARS-------------LQEREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVY 1369
Query: 1756 L------FNSRLKLFPGKLKSKWSGPFMI 1778
+ S + KL K+ GP+ I
Sbjct: 1370 VKLQPYRQQSVVMRANQKLSPKYFGPYKI 1398
>At1g35370 hypothetical protein
Length = 1447
Score = 351 bits (900), Expect = 3e-96
Identities = 270/871 (30%), Positives = 416/871 (46%), Gaps = 89/871 (10%)
Query: 926 HRIHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGG 985
H+I + V Q R K+ + K V ++ +G I +S S + SPV +V KK G
Sbjct: 570 HKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQ-VSSSPFASPVVLVKKKDG 628
Query: 986 MTVVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLD 1045
R+C+DY LN T KD F +P ++ +++ L G + +D
Sbjct: 629 TW------------------RLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKID 670
Query: 1046 GYSGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIE 1105
+GY+Q+ + P+D +KTAF G F Y M FGL APATFQ M S+F D + K +
Sbjct: 671 LRAGYHQVRMDPDDIQKTAFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVL 730
Query: 1106 VFMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEV 1165
VF DD ++ S ++ HL V + L K H LGH IS++ IE
Sbjct: 731 VFFDDILIYSSSIEEHKEHLRLVFEVMRLHKLFAKGSKEH--------LGHFISAREIET 782
Query: 1166 DQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVEC 1225
D AKI+ +++ P P VK VR FLG AG+YRRF+++F IA PL + L K F + +E
Sbjct: 783 DPAKIQAVKEWPTPTTVKQVRGFLGFAGYYRRFVRNFGVIAGPL-HALTKTDGFCWSLEA 841
Query: 1226 LNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLN 1285
+AF +K L AP++ P +D F + DA + AVL Q+ H + Y S+ L
Sbjct: 842 QSAFDTLKAVLCNAPVLALPVFDKQFMVETDACGQGIRAVLMQKG----HPLAYISRQLK 897
Query: 1286 ESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLL 1345
Q++ S EKELLA IFA+ K+R YL+ S I+ TD +LKYLL + + P +W+
Sbjct: 898 GKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLLEQRLNTPVQQQWLPK 957
Query: 1346 LQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIM-AEFPDEQLFAIRERPWFADMA 1404
L EFD EI+ ++G EN+VAD LSR+E +EV +I+ +F E A D
Sbjct: 958 LLEFDYEIQYRQGKENLVADALSRVEGSEVLHMALSIVECDFLKEIQVAYESDGVLKD-- 1015
Query: 1405 NFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIK---NIVW 1461
I ++QH K HY W L + S + V +++ ++
Sbjct: 1016 -------IISALQQHPDAK-----KHYSWSQDILRRKS------KIVVPNDVEITNKLLQ 1057
Query: 1462 HCHSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTGSISKRNEMPLTG 1521
H S GG SG + + ++S F+W + KD F+R C CQ+ K + G
Sbjct: 1058 WLHCSGMGG-RSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQ----CKSDNAAYPG 1112
Query: 1522 IIEVEPF--DCW---GIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCV-ANDSKTVVNF 1575
+++ P W +DF+ P S I+V VD ++K + + TV
Sbjct: 1113 LLQPLPIPDKIWCDVSMDFIEGLPNSGGKSVIMVVVDRLSKAAHFVALAHPYSALTVAQA 1172
Query: 1576 LRKNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQL 1635
N++ G P ++SD F ++F + K ++ ++++ YHPQ+ GQ EV NR L
Sbjct: 1173 FLDNVYKHHGCPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCL 1232
Query: 1636 KQILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKA--CHLPVELEHKAY 1693
+ L + W++ L A + Y + + ++P++LV+G+A HLP L K+
Sbjct: 1233 ENYLRCMCHARPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLPY-LPGKSK 1291
Query: 1694 WAIKALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQL 1753
A+ A + L E E M L + + + R K++ D+ R F +G
Sbjct: 1292 VAVVARS-------------LQERENMLLFLKFHLMRAQHRMKQFADQHRTERTFDIGDF 1338
Query: 1754 VLL------FNSRLKLFPGKLKSKWSGPFMI 1778
V + S + KL K+ GP+ I
Sbjct: 1339 VYVKLQPYRQQSVVLRVNQKLSPKYFGPYKI 1369
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 350 bits (899), Expect = 3e-96
Identities = 265/903 (29%), Positives = 418/903 (45%), Gaps = 96/903 (10%)
Query: 899 MRVLRENEGALGWKISDLKGISPAYC-MHRIHMEAEYKSVVQPQRRLNPTMKEVVKKEVL 957
+RV++E E L+G+ P+ I +E + + R+ P +KK++
Sbjct: 483 IRVIQEFEDVF----QSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMTELKKQLE 538
Query: 958 KLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPTRTVTGRRMCIDYRRLNTA 1017
LL G I P S S W +PV V KK G R+CIDYR LN
Sbjct: 539 DLLGKGFIRP-STSPWGAPVLFVKKKDGSF------------------RLCIDYRGLNWV 579
Query: 1018 TRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFTCPFGVFAYRRM 1077
T K+ +PLP +D+++++L G + +D SGY+QI +A D KTAF +G F + M
Sbjct: 580 TVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVM 639
Query: 1078 PFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLNAVLKRCTETNL 1137
PF L APA F R M S+F + +++ + +F+DD V+ KS ++ HL V+++ E L
Sbjct: 640 PFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLREQKL 699
Query: 1138 ILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRR 1197
KC F E LGH +S++G+ VD KIE I P P N +RSFL G+YRR
Sbjct: 700 FAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLRLTGYYRR 759
Query: 1198 FIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIAPNWDLHFELMCDA 1257
F+K F+ +A+P+ L K+ F + EC F +K L + P++ P + + DA
Sbjct: 760 FVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDA 819
Query: 1258 SDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFALEKFRSYLIGSKV 1317
S +G VL QR I YAS+ L + + NY T + E+ VIFAL+ +RSYL G KV
Sbjct: 820 SRVGLGCVLMQRGK----VIAYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYGGKV 875
Query: 1318 IVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENVVADHLSRLENNEVT- 1376
VFTDH +LKY+ + + R +RW+ L+ ++DLEI G NVVAD LSR
Sbjct: 876 QVFTDHKSLKYIFNQPELNLRQMRWMELVADYDLEIAYHPGKANVVADALSRKRVGAAPG 935
Query: 1377 KKEGAIMAEFPDEQLFAIRERPW---FADMANFKAGNIIPDDMEQHQRKKFFKDANHYLW 1433
+ A+++E +L + P D A+ + + ++ + + Y
Sbjct: 936 QSVEALVSEIGALRLCVVAREPLGLEAVDRADLLTRARLAQEKDEGLIAASKAEGSEY-- 993
Query: 1434 DDPYLFKVSTDGLI----RRCVAGEE--IKNIVWHCHSSAYGGHHSGERTAAKVLQSGFW 1487
+ + +G I R CV +E + I+ H+S + H G + L+ +
Sbjct: 994 ------QFAANGTIFVYGRVCVPKDEELRREILSEAHASMF-SIHPGATKMYRDLKRYYQ 1046
Query: 1488 WPTLFKDCHDFVRRCDNCQRTGSISKRNEMPLTGIIEVEPFDCWGIDFMGPFPPSSSYLH 1547
W + +D ++V CD CQ + +++P
Sbjct: 1047 WVGMKRDVANWVAECDVCQ---LVKAEHQVPDA--------------------------- 1076
Query: 1548 ILVCVDYVTKWVEAIPCVANDSKTVV-NFLRKNIFTRFGTPRVLISDGGKHFCNNFLETV 1606
I V +D +TK + D V+ I G P ++SD F F
Sbjct: 1077 IWVIMDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAFWRAF 1136
Query: 1607 LKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSRKLDDALWAYRIAFK 1666
K K +++T YHPQT GQ E + + L+ +L V W+ L +AY +++
Sbjct: 1137 QAKMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQ 1196
Query: 1667 THLGLSPYQLVFGKACHLPVELEHKAYWAIKALNFDQTLAGKKRLLKLNELEEMRLGAYE 1726
+G++P++ ++G+ C P+ +I ++ Q + R+LKLN E
Sbjct: 1197 ASIGMAPFEALYGRPCWTPLRWTQVEERSIYGADYVQETTERIRVLKLNMKEA------- 1249
Query: 1727 NAVIYKERTKRYHDKGLVRREFYVGQLVLLFNSRLK-----LFPGKLKSKWSGPF-MIES 1780
+ R + Y DK EF VG V L + L+ + KL ++ GPF ++E
Sbjct: 1250 -----QARQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSILETKLSPRYMGPFRIVER 1304
Query: 1781 ISP 1783
+ P
Sbjct: 1305 VGP 1307
>At2g11680 putative retroelement pol polyprotein
Length = 301
Score = 338 bits (866), Expect = 2e-92
Identities = 172/305 (56%), Positives = 218/305 (71%), Gaps = 24/305 (7%)
Query: 1147 MVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIA 1206
MV EGIVLGHKIS KG++V++AK++V+ +L PP K +RSFLGHAGFYRRFIKDFSK+A
Sbjct: 1 MVREGIVLGHKISKKGVQVNKAKVDVMMQLQPPKTGKDIRSFLGHAGFYRRFIKDFSKLA 60
Query: 1207 KPLCNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVL 1266
+PL LL KE EF F+ ECL AF K LV+API+ APN D FE+MCD SDYAVGAVL
Sbjct: 61 RPLTRLLCKEAEFIFEEECLTAFKSTKEALVSAPIVQAPNRDYPFEIMCDDSDYAVGAVL 120
Query: 1267 GQRKNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAAL 1326
GQ+ ++ H IYYAS ++++QV Y+TTEKELLAV+FA EKFRSYL+GSKV V+TDHAAL
Sbjct: 121 GQKIDRKLHVIYYASGTMDDTQVRYATTEKELLAVVFAFEKFRSYLVGSKVTVYTDHAAL 180
Query: 1327 KYLLTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENVVADHLSRLENNEVTKKEGAIMAEF 1386
+++ K D+KPRLLRW+LLLQEFD+EI DKKG+EN VADHLSR+ + E I
Sbjct: 181 RHIYAKKDTKPRLLRWILLLQEFDMEIVDKKGIENGVADHLSRMR----IEDEVPIDDSM 236
Query: 1387 PDEQLFAIRE--------------------RPWFADMANFKAGNIIPDDMEQHQRKKFFK 1426
P+EQL AI++ W+AD + G P ++ +++KKFFK
Sbjct: 237 PEEQLMAIQQLNESAQINKSLDQVCAVEDKLMWYADHVKYLFGGEEPPNLSSYEKKKFFK 296
Query: 1427 DANHY 1431
D NH+
Sbjct: 297 DINHF 301
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 294 bits (753), Expect = 3e-79
Identities = 174/455 (38%), Positives = 249/455 (54%), Gaps = 24/455 (5%)
Query: 916 LKGISPAYC-MHRIHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWV 974
L+G+ P+ I +E + + R+ P +KK++ LL G I P S S W
Sbjct: 470 LQGLPPSQSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRP-STSPWG 528
Query: 975 SPVHVVPKKGGMTVVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIER 1034
+PV V KK G R+CIDYR LN T K+ +PLP +D+++++
Sbjct: 529 APVLFVKKKDGSF------------------RLCIDYRELNRVTVKNRYPLPRIDELLDQ 570
Query: 1035 LAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLS 1094
L G + +D SGY+QI +A D KTAF +G F + MPFGL APA F R M S
Sbjct: 571 LRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNS 630
Query: 1095 IFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVL 1154
+F + +++ + +F+DD V+ KS ++ HL V+++ E L KC F E L
Sbjct: 631 VFQEFLDEFVIIFIDDILVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFL 690
Query: 1155 GHKISSKGIEVDQAKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLV 1214
GH +S++G+ VD KIE I P P N +RSFLG AG+YRRF+K F+ +A+P+ L
Sbjct: 691 GHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTG 750
Query: 1215 KETEFDFDVECLNAFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFF 1274
K+ F + EC F +K L + P++ P + + DAS +G VL Q
Sbjct: 751 KDVPFVWSQECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQHGK--- 807
Query: 1275 HAIYYASKVLNESQVNYSTTEKELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGD 1334
I YAS+ L + + NY T + E+ AVIFAL+ +RSYL G KV VFTDH +LKY+ T+ +
Sbjct: 808 -VIAYASRQLMKHEGNYPTHDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPE 866
Query: 1335 SKPRLLRWVLLLQEFDLEIRDKKGVENVVADHLSR 1369
R RW+ L+ ++DLEI G NVV D LSR
Sbjct: 867 LNLRQRRWMELVADYDLEIAYHPGKANVVVDALSR 901
>At1g20390 hypothetical protein
Length = 1791
Score = 275 bits (702), Expect = 2e-73
Identities = 197/659 (29%), Positives = 304/659 (45%), Gaps = 40/659 (6%)
Query: 886 ISSTLTPLEEEKLMRVLRENEGALGWKISDLKGISPAYCMHRIHMEAEYKSVVQPQRRLN 945
+ + ++P +L+ +L+ N W I D+KGI PA H ++++ +K V Q +R+L
Sbjct: 727 VGAEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNVDPTFKPVKQKRRKLG 786
Query: 946 PTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPTRTVTGR 1005
P V +EV KLL AG I + W++ VV KK G
Sbjct: 787 PERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKW------------------ 828
Query: 1006 RMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAF 1065
R+C+DY LN A KD +PLP +D+++E +G F+D +SGYNQI + +DQEKT+F
Sbjct: 829 RVCVDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSF 888
Query: 1066 TCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHL 1125
G + Y+ M FGL A AT+QR + + +D I + +EV++DD V + + HL
Sbjct: 889 VTDRGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHL 948
Query: 1126 NAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGV 1185
+ + LN KC F VT G LG+ ++ +GIE + +I I +LP P N + V
Sbjct: 949 SKCFDVLNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREV 1008
Query: 1186 RSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIAP 1245
+ G RFI + P NLL + +FD+D + AF +K+ L T PI++ P
Sbjct: 1009 QRLTGRIAALNRFISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKP 1068
Query: 1246 NWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFAL 1305
L SD+AV +VL + I+Y SK L E++ Y EK LAV+ +
Sbjct: 1069 EVGETLYLYIAVSDHAVSSVLVREDRGEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSA 1128
Query: 1306 EKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKKGVEN-VVA 1364
K R Y + V TD L+ L R+ +W + L E+D++ R + +++ V+A
Sbjct: 1129 RKLRPYFQSHTIAVLTDQ-PLRVALHSPSQSGRMTKWAVELSEYDIDFRPRPAMKSQVLA 1187
Query: 1365 DHLSRLENNEVTKKEGAIMAEFPDE-QLFAIRERPWFADMANFKAGNIIPDDMEQHQRKK 1423
D L L + E A+ +E L+ + + + +EQ R +
Sbjct: 1188 DFLIEL---PLQSAERAVSGNRGEEWSLYVDGSSSARGSGIGIRLVSPTAEVLEQSFRLR 1244
Query: 1424 FFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKNIVWHCHSSAYGGHHSGERTAAKVLQ 1483
F N ++V GL R AG +I I S G SGE A
Sbjct: 1245 FVATNN------VAEYEVLIAGL--RLAAGMQITTIHAFTDSQLIAGQLSGEYEAKNEKM 1296
Query: 1484 SGFW--WPTLFKDCHDF----VRRCDN--CQRTGSISKRNEMPLTGIIEVEPFDCWGID 1534
+ + KD +F + R DN +++ ++ L II VE D ID
Sbjct: 1297 DAYLKIVQLMTKDFENFKLSKIPRGDNAPADALAALALTSDSDLRRIIPVESIDKPSID 1355
Score = 195 bits (495), Expect = 2e-49
Identities = 123/416 (29%), Positives = 202/416 (47%), Gaps = 12/416 (2%)
Query: 1399 WFADMANFKAGNIIPDDMEQHQRKKFFKDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKN 1458
W ++ ++ + +P D +R + K A + L + +L KVS G + C+ G EI
Sbjct: 1380 WRVEIRDYLSDGTLPSDKWTARRLRI-KAAKYTLMKE-HLLKVSAFGAMLNCLHGTEINE 1437
Query: 1459 IVWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQRTG-SISKRNEM 1517
I+ H A G H G A K+ + GF+WPT+ DC F +C+ CQR +I + E+
Sbjct: 1438 IMKETHEGAAGNHSGGRALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQPTEL 1497
Query: 1518 PLTGIIEVEPFDCWGIDFMGPFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVVNFLR 1577
G+ PF W +D +GP P S ILV DY TKWVEA + V NF+
Sbjct: 1498 LRAGVAPY-PFMRWAMDIVGPMPASRQKRFILVMTDYFTKWVEAESYATIRANDVQNFVW 1556
Query: 1578 KNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQ 1637
K I R G P +I+D G F + E + I+ +TP +PQ +GQ E +N+ +
Sbjct: 1557 KFIICRHGLPYEIITDNGSQFISLSFENFCASWKIRLNKSTPRYPQGNGQAEATNKTILS 1616
Query: 1638 ILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAYWAIK 1697
L+K + + W+ +LD LW+YR ++ +P+ +G P E+ + +
Sbjct: 1617 GLKKRLDEKKGAWADELDGVLWSYRTTPRSATDQTPFAHAYGMEAMAPAEVGYSSLRRSM 1676
Query: 1698 ALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQLVL-- 1755
+ + L + L +L++LEE+R A Y+ +++++ + R F VG LVL
Sbjct: 1677 MVK-NPELNDRMMLDRLDDLEEIRNAALCRIQNYQLAAAKHYNQKVHNRHFDVGDLVLRK 1735
Query: 1756 LFNSRLKLFPGKLKSKWSGPFMIESI---SPYGAVELSKPGEPGTFKVNAQRIKPY 1808
+F + ++ GKL + W G + + I Y + +S P T+ N+ +K Y
Sbjct: 1736 VFENTAEINAGKLGANWEGSYQVSKIVRPGDYELLTMSGTAVPRTW--NSMHLKRY 1789
Score = 40.4 bits (93), Expect = 0.010
Identities = 23/79 (29%), Positives = 39/79 (49%)
Query: 140 NEDQKKLRLFPFTLIGKAKDWLLTLPNGTIQTWEELELKFLEKYFPMSKYLDKKQEIASF 199
N D + ++F L G A +W L +I ++ +L FL+ Y P+ + ++ S
Sbjct: 203 NFDAGRCQIFIEHLTGPAHNWFSRLKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWSI 262
Query: 200 RQGEGESLYDAWERFNLLL 218
QG ESL +RF L++
Sbjct: 263 SQGAKESLRSFVDRFKLVV 281
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 273 bits (699), Expect = 5e-73
Identities = 148/363 (40%), Positives = 214/363 (58%), Gaps = 4/363 (1%)
Query: 1006 RMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAF 1065
R+CIDYR LN T K+ +PLP +D+++++L G + +D SGY+ I +A D KTAF
Sbjct: 517 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAF 576
Query: 1066 TCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHL 1125
+G F + MPFGL APA F R M S+F +++++ + +F+DD V+ KS ++ HL
Sbjct: 577 RTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHEVHL 636
Query: 1126 NAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKLPPPMNVKGV 1185
V+++ E L KC F E LGH +S++G+ VD KIE I P N +
Sbjct: 637 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTPTNATEI 696
Query: 1186 RSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIAP 1245
RSFLG AG+YRRF+K F+ +A+P+ L K+ F + EC F +K L + P++ P
Sbjct: 697 RSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALP 756
Query: 1246 NWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFAL 1305
+ + DAS +G VL QR I YAS+ L + + NY T + E+ AVIFAL
Sbjct: 757 EHGEPYMVYTDASGVGLGCVLMQRGK----VIAYASRQLRKHEGNYPTHDLEMAAVIFAL 812
Query: 1306 EKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENVVAD 1365
+ +RSYL G V VFTDH +LKY+ T+ + R +W+ L+ ++DLEI G NVVAD
Sbjct: 813 KIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQRQWMELVADYDLEIAYHPGKANVVAD 872
Query: 1366 HLS 1368
LS
Sbjct: 873 ALS 875
Score = 90.1 bits (222), Expect = 1e-17
Identities = 78/303 (25%), Positives = 133/303 (43%), Gaps = 28/303 (9%)
Query: 1493 KDCHDFVRRCDNCQRTGSISKRNEMPLTGIIEVEPFDCWG-----IDFMGPFPPSSSYLH 1547
+D ++V CD CQ + +++P G+++ P W IDF+ P S +
Sbjct: 941 EDVANWVAECDVCQL---VKAEHQVP-GGMLQSLPIPEWKWDFITIDFVVGLPVSRTKDA 996
Query: 1548 ILVCVDYVTKWVEAIPCVANDSKTVV-NFLRKNIFTRFGTPRVLISDGGKHFCNNFLETV 1606
I V VD +TK + D V+ I G P ++SD F + F
Sbjct: 997 IWVIVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAF 1056
Query: 1607 LKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSRKLDDALWAYRIAFK 1666
+ K +++T YHPQT GQ E + + L+ +L V W+ L +AY ++
Sbjct: 1057 QAEMGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYP 1116
Query: 1667 THLGLSPYQLVFGKACHLPVELEHKAYWAIKALNFDQTLAGKKRLLKLNELEEMRLGAYE 1726
+G++P++ ++ + C P+ L +I ++ Q + R+LKLN E
Sbjct: 1117 ASIGMAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTERIRVLKLNMKEA------- 1169
Query: 1727 NAVIYKERTKRYHDKGLVRREFYVGQLVLLFNSRLK-----LFPGKLKSKWSGPF-MIES 1780
++R + Y DK EF VG V L + L+ + KL ++ GPF ++E
Sbjct: 1170 -----QDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVER 1224
Query: 1781 ISP 1783
+ P
Sbjct: 1225 VGP 1227
>At2g07410 putative retroelement pol polyprotein
Length = 411
Score = 259 bits (663), Expect = 8e-69
Identities = 143/287 (49%), Positives = 182/287 (62%), Gaps = 38/287 (13%)
Query: 1536 MGPFPPSSSYLHILVCVDYVTKWVEAIPCVANDSKTVVNFLRKNIFTRFGTPRVLISDGG 1595
MG FP S +ILV VDYV+KWVEAI ND+K V+ + IFTRFG V+ISDG
Sbjct: 1 MGSFPSSYGNKYILVAVDYVSKWVEAIVSPTNDAKVVLKLFKTIIFTRFGVHMVVISDGR 60
Query: 1596 KHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSRKLD 1655
KHF N E +LKK+ +KHKVAT YHPQTSGQVE+SNR++K ILEKTV +RKD S KLD
Sbjct: 61 KHFINKAFENLLKKHGVKHKVATSYHPQTSGQVEISNREIKAILEKTVGVTRKDLSAKLD 120
Query: 1656 DALWAYRIAFKTH----LGLSPYQLVFGKACHLPVELEHKAYWAIKALNFDQTLAGKKRL 1711
DALWAY+ AFKT +G +P+ L++ K+ HLP+ELE KA WA+K +NFD A ++RL
Sbjct: 121 DALWAYKTAFKTAFKTPIGTTPFNLLYAKSGHLPIELEFKAMWAVKLMNFDIKTAEEERL 180
Query: 1712 LKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRREFYVGQLVLLFNSRLKLFPGKLKSK 1771
++LN+L+E+RL AYE++ YK + K LKS+
Sbjct: 181 IQLNDLDEIRLEAYESSKNYKGKRK-------------------------------LKSR 209
Query: 1772 WSGPFMIESISPYGAVELSKPGEPGTFKVNAQRIKPYLGGE-LPTNK 1817
GPF + I PYGAV L+ + G F VN QR+K YL + LP NK
Sbjct: 210 LFGPFCVTEIRPYGAVTLAV--KSGDFTVNGQRLKKYLEDQILPKNK 254
>At2g07420 putative retroelement pol polyprotein
Length = 1664
Score = 250 bits (639), Expect = 5e-66
Identities = 119/172 (69%), Positives = 136/172 (78%)
Query: 1111 FSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKI 1170
F + SF CL +L VL RC ETNL+LNWEKCHFMV EGIVLGHKIS KGIEVD+ KI
Sbjct: 1483 FLGYSPSFSSCLLNLGRVLTRCKETNLVLNWEKCHFMVKEGIVLGHKISKKGIEVDKGKI 1542
Query: 1171 EVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFS 1230
+VI +L PP VK +RSFLGHAGFYRRFIKDFSKIA+PL LL KETEF+FD +CL +F
Sbjct: 1543 KVIMQLQPPKTVKDIRSFLGHAGFYRRFIKDFSKIARPLTRLLCKETEFEFDEDCLKSFH 1602
Query: 1231 LIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASK 1282
IK LV+AP++ APNWD FE+MCDASDYAVGA+LGQR K H IYYAS+
Sbjct: 1603 TIKEALVSAPVVRAPNWDYPFEIMCDASDYAVGAILGQRIGKKLHVIYYASR 1654
Score = 80.1 bits (196), Expect = 1e-14
Identities = 37/57 (64%), Positives = 47/57 (81%)
Query: 927 RIHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKK 983
RIH+ S ++PQRRLNP +KEVVKKE+LKL+DAG+ YPIS+S+W+S VH VPKK
Sbjct: 1405 RIHVGNVSYSSIEPQRRLNPNLKEVVKKEILKLVDAGVKYPISESTWISLVHCVPKK 1461
Score = 37.0 bits (84), Expect = 0.11
Identities = 32/127 (25%), Positives = 52/127 (40%), Gaps = 29/127 (22%)
Query: 1539 FPPSSSYLHILVCVDYVTKWVEAIPCVANDS-KTVVNFLRKN-----------------I 1580
FP SS+ C D + V PC++ ++ K V F+ +
Sbjct: 483 FPKSSTIYE--KCFDLIHSDVWTSPCLSRENHKYFVTFIDEKSKFTWFTLLPSKDRVLEA 540
Query: 1581 FTRFGTP---------RVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVS 1631
FT F T ++L SD + ++ + L K+ I H+ + PY PQ +G E
Sbjct: 541 FTNFQTYVTNHYDAKIKILRSDNRGEYTSHAFKQHLNKHGIIHQTSCPYTPQQNGVAERK 600
Query: 1632 NRQLKQI 1638
NR L ++
Sbjct: 601 NRHLMEV 607
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 246 bits (628), Expect = 9e-65
Identities = 145/386 (37%), Positives = 213/386 (54%), Gaps = 23/386 (5%)
Query: 928 IHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMT 987
I +E + + R+ P +KK++ +LL G I P S S W +PV V KK G
Sbjct: 146 IELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRP-SSSPWGAPVLFVKKKDGSF 204
Query: 988 VVVNEKNELIPTRTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGY 1047
R+CIDYR LN T K+ +PLP +D+++++L G ++ +D
Sbjct: 205 ------------------RLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLA 246
Query: 1048 SGYNQIAVAPEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVF 1107
SGY+QI + P D KTAF +G F + MPFGL APA F + M +F D +++ + +F
Sbjct: 247 SGYHQIPIEPTDVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIF 306
Query: 1108 MDDFSVFGKSFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQ 1167
+DD V KS++ HL AVL+R E L K F LGH IS +G+ VD
Sbjct: 307 IDDILVHSKSWEAHQEHLRAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDP 366
Query: 1168 AKIEVIEKLPPPMNVKGVRSFLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLN 1227
KI I++ P P N +RSFLG AG+YRRF+ F+ +A+PL L K+T F++ EC
Sbjct: 367 EKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEK 426
Query: 1228 AFSLIKNKLVTAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNES 1287
+F +K L+ AP+++ P + + DAS +G VL Q+ + I YAS+ L +
Sbjct: 427 SFLELKAMLINAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGS----VIAYASRQLRKH 482
Query: 1288 QVNYSTTEKELLAVIFALEKFRSYLI 1313
+ NY T + E+ AV+FAL+ +RSYLI
Sbjct: 483 EKNYPTHDLEMAAVVFALKIWRSYLI 508
Score = 96.3 bits (238), Expect = 1e-19
Identities = 102/405 (25%), Positives = 169/405 (41%), Gaps = 58/405 (14%)
Query: 1439 FKVSTDGLI----RRCVAGEEI--KNIVWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLF 1492
++ S +G I R CV + + I+ H S + H G + L+ + W +
Sbjct: 529 YQTSNNGTIVVNGRVCVPNDRALKEEILREAHQSKFS-IHPGSNKMYRDLKRYYHWVGMR 587
Query: 1493 KDCHDFVRRCDNCQRTGSISKRNEMPLTGIIEVEPFDCWG-----IDFMGPFPPSSSYLH 1547
KD +V +C CQ + +++P +G+++ P W +DF+ P H
Sbjct: 588 KDVARWVAKCPTCQL---VKAEHQVP-SGLLQNLPISEWKWDHITMDFVTRLPTGIKSKH 643
Query: 1548 --ILVCVDYVTKWVEAIPCVANDSKTVV-NFLRKNIFTRFGTPRVLISDGGKHFCNNFLE 1604
+ V VD +TK + D ++ I G P ++SD F + F
Sbjct: 644 NAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIMRLHGIPVSIVSDRDTRFTSKFWN 703
Query: 1605 TVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSRKLDDALWAYRIA 1664
K + ++T YHPQT GQ E + + L+ +L V +W + L +AY +
Sbjct: 704 AFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLIEFAYNNS 763
Query: 1665 FKTHLGLSPYQLVFGKACHLPVELEHKAYWAIKALNFDQTLAGKKRLLKLNELEEMRLGA 1724
F+ +G+SPY+ ++G+AC P+ W T G++RL ++E
Sbjct: 764 FQASIGMSPYEALYGRACRTPL------CW---------TPVGERRLFGPTIVDETTERM 808
Query: 1725 YENAVIYKE---RTKRYHDKGLVRREFYVGQLVLL----------FNSRLKLFPGKLKSK 1771
+ KE R K Y +K EF VG LV L F SR KL P +
Sbjct: 809 KFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSP-----R 863
Query: 1772 WSGPFMIESISPYGAV--ELSKPGEPGTFK--VNAQRIKPYLGGE 1812
+ GP+ + I GAV +L P + F + +++ YL +
Sbjct: 864 YVGPYKV--IERVGAVAYKLDLPPKLNVFHNVFHVSQLRKYLSDQ 906
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,240,979
Number of Sequences: 26719
Number of extensions: 2020620
Number of successful extensions: 8978
Number of sequences better than 10.0: 318
Number of HSP's better than 10.0 without gapping: 169
Number of HSP's successfully gapped in prelim test: 151
Number of HSP's that attempted gapping in prelim test: 7785
Number of HSP's gapped (non-prelim): 893
length of query: 1825
length of database: 11,318,596
effective HSP length: 114
effective length of query: 1711
effective length of database: 8,272,630
effective search space: 14154469930
effective search space used: 14154469930
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146759.6


