

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146759.1 + phase: 0 /pseudo
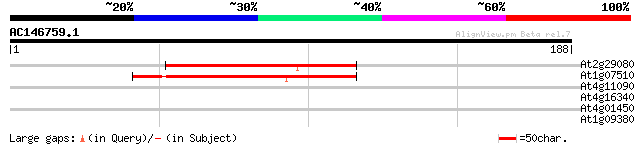
(188 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g29080 AAA-type like ATPase 80 8e-16
At1g07510 unknown protein 75 2e-14
At4g11090 unknown protein 28 3.8
At4g16340 unknown protein 27 8.5
At4g01450 unknown protein 27 8.5
At1g09380 nodulin N21 like protein 27 8.5
>At2g29080 AAA-type like ATPase
Length = 809
Score = 79.7 bits (195), Expect = 8e-16
Identities = 42/83 (50%), Positives = 53/83 (63%), Gaps = 19/83 (22%)
Query: 53 GKNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYVKNNESEV----------------- 95
G+ Q+ISFQEFK K+LEPGLVDHI VSNK +AK+YV++ +
Sbjct: 154 GEQQQISFQEFKNKLLEPGLVDHIDVSNKSVAKVYVRSTPKDQQTTDVVHGNGNGIPAKR 213
Query: 96 --AKYKYYFKIGSVDSFERKLKK 116
+YKYYF IGSVDSFE KL++
Sbjct: 214 TGGQYKYYFNIGSVDSFEEKLEE 236
>At1g07510 unknown protein
Length = 813
Score = 75.1 bits (183), Expect = 2e-14
Identities = 45/94 (47%), Positives = 58/94 (60%), Gaps = 20/94 (21%)
Query: 42 LILSFFYFRPFGKNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYVKNN---------- 91
LILS F + Q+ISFQEFK K+LE GLVDHI VSNKE+AK+YV+++
Sbjct: 148 LILSTFSLGS-REQQQISFQEFKNKLLEAGLVDHIDVSNKEVAKVYVRSSPKSQTTEEVV 206
Query: 92 ---------ESEVAKYKYYFKIGSVDSFERKLKK 116
+ +YKYYF IGSV+SFE KL++
Sbjct: 207 QGPGNGVPAKGRGGQYKYYFNIGSVESFEEKLEE 240
>At4g11090 unknown protein
Length = 432
Score = 27.7 bits (60), Expect = 3.8
Identities = 20/61 (32%), Positives = 29/61 (46%), Gaps = 3/61 (4%)
Query: 19 VAVVGSFQEALKKL---GTLMTLIGGLILSFFYFRPFGKNQEISFQEFKIKVLEPGLVDH 75
V +G E LK L G L+T G + FRPF K++ + Q + PG +DH
Sbjct: 357 VTEMGQESENLKLLDFAGMLLTRPDGHPGPYREFRPFDKDKNATVQNDCLHWCLPGPIDH 416
Query: 76 I 76
+
Sbjct: 417 L 417
>At4g16340 unknown protein
Length = 880
Score = 26.6 bits (57), Expect = 8.5
Identities = 14/38 (36%), Positives = 23/38 (59%), Gaps = 4/38 (10%)
Query: 126 YVLLILLKPTGFRCFSYYLHSCWLSEFSFIIRNVLMEC 163
Y LL +++P C Y L S ++ +FS + ++VL EC
Sbjct: 78 YDLLYIIEP----CQVYELVSLYMDKFSGVCQSVLHEC 111
>At4g01450 unknown protein
Length = 343
Score = 26.6 bits (57), Expect = 8.5
Identities = 11/26 (42%), Positives = 18/26 (68%)
Query: 32 LGTLMTLIGGLILSFFYFRPFGKNQE 57
LGTL++L+GGL+L+ + P + E
Sbjct: 138 LGTLISLVGGLLLTMYQGIPLTNSPE 163
>At1g09380 nodulin N21 like protein
Length = 374
Score = 26.6 bits (57), Expect = 8.5
Identities = 10/24 (41%), Positives = 17/24 (70%)
Query: 30 KKLGTLMTLIGGLILSFFYFRPFG 53
K +GTL+ +IG ++LSF++ G
Sbjct: 134 KVIGTLVCVIGAMVLSFYHGHTIG 157
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.337 0.148 0.450
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,812,293
Number of Sequences: 26719
Number of extensions: 142004
Number of successful extensions: 509
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 503
Number of HSP's gapped (non-prelim): 6
length of query: 188
length of database: 11,318,596
effective HSP length: 93
effective length of query: 95
effective length of database: 8,833,729
effective search space: 839204255
effective search space used: 839204255
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146759.1


