

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146758.2 + phase: 0
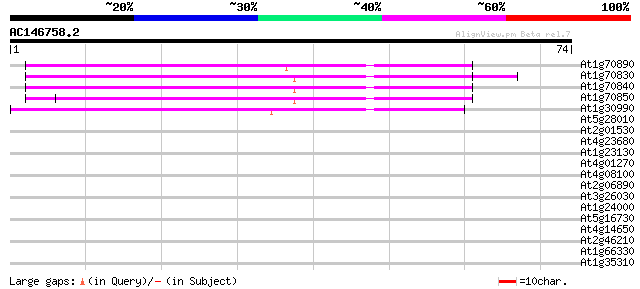
(74 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g70890 unknown protein 46 4e-06
At1g70830 unknown protein 45 7e-06
At1g70840 unknown protein (At1g70840) 44 2e-05
At1g70850 putative protein 43 3e-05
At1g30990 F17F8.9 39 6e-04
At5g28010 major latex protein homolog - like 37 0.001
At2g01530 unknown protein 32 0.044
At4g23680 putative major latex protein 32 0.075
At1g23130 unknown protein 32 0.075
At4g01270 putative RING zinc finger protein 28 0.83
At4g08100 putative polyprotein 28 1.1
At2g06890 putative retroelement integrase 28 1.1
At3g26030 protein phosphatase 2A regulatory subunit isoform B' d... 27 1.8
At1g24000 hypothetical protein 27 1.8
At5g16730 putative protein 25 5.4
At4g14650 hypothetical protein 25 7.0
At2g46210 putative fatty acid desaturase/cytochrome b5 fusion pr... 25 9.2
At1g66330 unknown protein 25 9.2
At1g35310 unknown protein 25 9.2
>At1g70890 unknown protein
Length = 158
Score = 45.8 bits (107), Expect = 4e-06
Identities = 24/60 (40%), Positives = 33/60 (55%), Gaps = 2/60 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNIC-ESVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL TE+ IKAS+ KF +F H V + +H +LH+GD W S+ W +V
Sbjct: 7 LVGKLETEVEIKASAKKFHHMFTERPHHVSKATPDKIHGCELHEGD-WGKVGSIVIWKYV 65
>At1g70830 unknown protein
Length = 335
Score = 45.1 bits (105), Expect = 7e-06
Identities = 24/60 (40%), Positives = 33/60 (55%), Gaps = 2/60 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL T++ IKAS+DKF +FA H V ++ LH+GD W S+ W +V
Sbjct: 22 LVGKLETDVEIKASADKFHHMFAGKPHHVSKASPGNIQGCDLHEGD-WGTVGSIVFWNYV 80
Score = 44.7 bits (104), Expect = 9e-06
Identities = 24/66 (36%), Positives = 35/66 (52%), Gaps = 2/66 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL T++ IKAS++KF +FA H V ++ LH+GD W S+ W +V
Sbjct: 184 LVGKLETDVEIKASAEKFHHMFAGKPHHVSKASPGNIQGCDLHEGD-WGQVGSIVFWNYV 242
Query: 62 RVLSSK 67
+K
Sbjct: 243 HDREAK 248
>At1g70840 unknown protein (At1g70840)
Length = 171
Score = 43.5 bits (101), Expect = 2e-05
Identities = 24/60 (40%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL T+I IKAS+ KF +FA H V + +LH+GD W S+ W +V
Sbjct: 20 LCGKLETDIEIKASAGKFHHMFAGRPHHVSKATPGKIQGCELHEGD-WGKVGSIVFWNYV 78
>At1g70850 putative protein
Length = 316
Score = 42.7 bits (99), Expect = 3e-05
Identities = 23/60 (38%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL TE+ IKAS+ +F +FA H V ++ LH+GD W S+ W +V
Sbjct: 9 LVGKLETEVEIKASAGQFHHMFAGKPHHVSKASPGNIQSCDLHEGD-WGTVGSIVFWNYV 67
Score = 39.7 bits (91), Expect = 3e-04
Identities = 21/56 (37%), Positives = 30/56 (53%), Gaps = 2/56 (3%)
Query: 7 LSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
L TE+ IKAS++KF +FA H V ++ LH+GD W S+ W +V
Sbjct: 169 LETEVEIKASAEKFHHMFAGKPHHVSKATPGNIQSCDLHEGD-WGTVGSIVFWNYV 223
>At1g30990 F17F8.9
Length = 152
Score = 38.5 bits (88), Expect = 6e-04
Identities = 18/61 (29%), Positives = 34/61 (55%), Gaps = 2/61 (3%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQN-ICESVHETKLHQGDEWHHSDSVKHWT 59
M ++G T++ + S++K +KL++S H + + I + LH+GD W S+K W
Sbjct: 1 MAMSGTYMTDVPLNGSAEKHYKLWSSETHRIPDTIGHLIQGVILHEGD-WDSHGSIKTWK 59
Query: 60 H 60
+
Sbjct: 60 Y 60
>At5g28010 major latex protein homolog - like
Length = 166
Score = 37.4 bits (85), Expect = 0.001
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 2/60 (3%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNIC-ESVHETKLHQGDEWHHSDSVKHWTHV 61
L GKL E+ IKA + F+ ++A H V +V LH G EW S+ +W +V
Sbjct: 15 LLGKLEVEVEIKAPAAIFYHIYAGRPHHVAKATPRNVQSCDLHDG-EWGTVGSIVYWNYV 73
>At2g01530 unknown protein
Length = 151
Score = 32.3 bits (72), Expect = 0.044
Identities = 17/61 (27%), Positives = 30/61 (48%), Gaps = 2/61 (3%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQN-ICESVHETKLHQGDEWHHSDSVKHWT 59
M +G TE+ +K S+DK +K + H + I + +H G EW +++K W
Sbjct: 1 MATSGTYVTEVPLKGSADKHYKRWRDENHLFPDAIGHHIQGVTVHDG-EWDSHEAIKIWN 59
Query: 60 H 60
+
Sbjct: 60 Y 60
>At4g23680 putative major latex protein
Length = 151
Score = 31.6 bits (70), Expect = 0.075
Identities = 15/61 (24%), Positives = 32/61 (51%), Gaps = 2/61 (3%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQN-ICESVHETKLHQGDEWHHSDSVKHWT 59
M +G TE+ +K S++K++K + + H + I + +H+G+ H S++ W
Sbjct: 1 MATSGTYVTEVPLKGSAEKYYKRWKNENHVFPDAIGHHIQNVTVHEGEHDSHG-SIRSWN 59
Query: 60 H 60
+
Sbjct: 60 Y 60
>At1g23130 unknown protein
Length = 160
Score = 31.6 bits (70), Expect = 0.075
Identities = 18/58 (31%), Positives = 28/58 (48%), Gaps = 1/58 (1%)
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTH 60
L G+L +I IKAS+ KF + + +V + LH+G E+ SV W +
Sbjct: 10 LQGELEVDIEIKASAKKFHHMLSGRPQDVAKATPDIQGCALHEG-EFGKVGSVIFWNY 66
>At4g01270 putative RING zinc finger protein
Length = 506
Score = 28.1 bits (61), Expect = 0.83
Identities = 13/30 (43%), Positives = 18/30 (59%)
Query: 42 KLHQGDEWHHSDSVKHWTHVRVLSSKQRLL 71
KLHQ +E D VK W ++ +S+ Q LL
Sbjct: 129 KLHQCNEQLKEDKVKRWEALQEISTTQHLL 158
>At4g08100 putative polyprotein
Length = 1054
Score = 27.7 bits (60), Expect = 1.1
Identities = 11/32 (34%), Positives = 18/32 (55%)
Query: 18 DKFFKLFASNIHEVQNICESVHETKLHQGDEW 49
D KL S+I ++ H+T++ +GDEW
Sbjct: 547 DTLDKLHGSSIFSKIDLKSGYHQTRMKEGDEW 578
>At2g06890 putative retroelement integrase
Length = 1215
Score = 27.7 bits (60), Expect = 1.1
Identities = 11/40 (27%), Positives = 21/40 (52%)
Query: 18 DKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKH 57
D +L S+I ++ H+ ++++GDEW + KH
Sbjct: 494 DMLDELHGSSIFSKIDLKSGYHQIRMNEGDEWKTAFKTKH 533
>At3g26030 protein phosphatase 2A regulatory subunit isoform B'
delta
Length = 477
Score = 26.9 bits (58), Expect = 1.8
Identities = 15/52 (28%), Positives = 23/52 (43%)
Query: 4 AGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSV 55
A K TE +K SD LF +H+ Q + +TK G+ W + +
Sbjct: 420 AVKNLTENVLKVLSDTNPDLFEECLHKFQEDQQKAEDTKKKNGETWRQLEEI 471
>At1g24000 hypothetical protein
Length = 122
Score = 26.9 bits (58), Expect = 1.8
Identities = 11/27 (40%), Positives = 15/27 (54%)
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASN 27
M L G LS + +K +DKFF F +
Sbjct: 1 MTLKGALSVKFDVKCPADKFFSAFVED 27
>At5g16730 putative protein
Length = 853
Score = 25.4 bits (54), Expect = 5.4
Identities = 10/32 (31%), Positives = 19/32 (59%)
Query: 13 IKASSDKFFKLFASNIHEVQNICESVHETKLH 44
IKA+++K+ + HE+ + +V +TK H
Sbjct: 506 IKATNEKYENMLDEARHEIDVLVSAVEQTKKH 537
>At4g14650 hypothetical protein
Length = 644
Score = 25.0 bits (53), Expect = 7.0
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query: 11 IGIKASSDKFFKLFASNI--HEVQNICESVHETKLHQGD 47
I I+ + D+FF N E +++C+ V ET+ H D
Sbjct: 193 IPIETTEDQFFLAGEGNDGDEEKESLCDGVEETEPHSKD 231
>At2g46210 putative fatty acid desaturase/cytochrome b5 fusion
protein
Length = 449
Score = 24.6 bits (52), Expect = 9.2
Identities = 8/19 (42%), Positives = 11/19 (57%)
Query: 44 HQGDEWHHSDSVKHWTHVR 62
H G WHH + + + HVR
Sbjct: 65 HPGTAWHHLEKLHNGYHVR 83
>At1g66330 unknown protein
Length = 417
Score = 24.6 bits (52), Expect = 9.2
Identities = 11/36 (30%), Positives = 19/36 (52%)
Query: 18 DKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSD 53
D+ K A +H + I +++ E+KL + H SD
Sbjct: 164 DRDVKRKAERLHHIATIFKNIAESKLKNAADKHWSD 199
>At1g35310 unknown protein
Length = 151
Score = 24.6 bits (52), Expect = 9.2
Identities = 14/57 (24%), Positives = 27/57 (46%), Gaps = 2/57 (3%)
Query: 2 VLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHW 58
++ ++ ++ IK+++DKFF +F+ V L +G EW S+ W
Sbjct: 1 MVEAEVEVDVEIKSTADKFF-MFSRRSQHASKATRYVQGCDLLEG-EWGEVGSILLW 55
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.129 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,554,686
Number of Sequences: 26719
Number of extensions: 48871
Number of successful extensions: 171
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 152
Number of HSP's gapped (non-prelim): 22
length of query: 74
length of database: 11,318,596
effective HSP length: 50
effective length of query: 24
effective length of database: 9,982,646
effective search space: 239583504
effective search space used: 239583504
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146758.2


