

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146758.11 + phase: 0 /pseudo
(1309 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
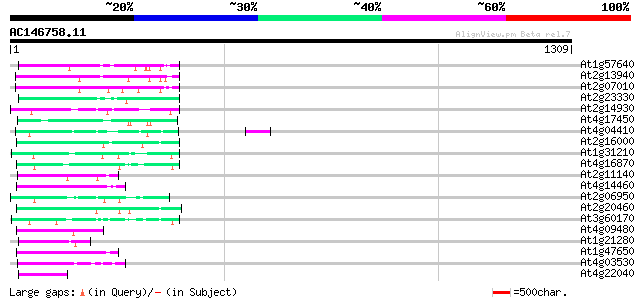
Score E
Sequences producing significant alignments: (bits) Value
At1g57640 106 8e-23
At2g13940 putative retroelement pol polyprotein 97 6e-20
At2g07010 putative retroelement pol polyprotein 92 2e-18
At2g23330 putative retroelement pol polyprotein 91 3e-18
At2g14930 pseudogene 91 3e-18
At4g17450 retrotransposon like protein 89 2e-17
At4g04410 putative polyprotein 87 8e-17
At2g16000 putative retroelement pol polyprotein 86 1e-16
At1g31210 putative reverse transcriptase 82 2e-15
At4g16870 retrotransposon like protein 82 3e-15
At2g11140 putative retroelement pol polyprotein 81 5e-15
At4g14460 retrovirus-related like polyprotein 79 2e-14
At2g06950 putative retroelement pol polyprotein 79 2e-14
At2g20460 putative retroelement pol polyprotein 78 3e-14
At3g60170 putative protein 74 4e-13
At4g09480 unknown protein 72 2e-12
At1g21280 unknown protein (At1g21280) 72 2e-12
At1g47650 hypothetical protein 69 1e-11
At4g03530 putative reverse transcriptase 65 2e-10
At4g22040 LTR retrotransposon like protein 64 4e-10
>At1g57640
Length = 1444
Score = 106 bits (265), Expect = 8e-23
Identities = 99/446 (22%), Positives = 192/446 (42%), Gaps = 86/446 (19%)
Query: 20 NYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAA-LEEWETKDAQIITWILSTINPQMIN 78
NY W F+ ++ + + L+G P + + LE+W T +A +++W+ TI+ +++
Sbjct: 42 NYEEWACGFKTALRSRKKFGFLDGTIPQPLDGSPDLEDWLTINALLVSWMKMTIDSELLT 101
Query: 79 NLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEHSA 138
N+ A+++W +++ ++ N K +++ ++A KQ +V+ YY +W ++
Sbjct: 102 NISHRDVARDLWEQIRKRFSVSNGPKNQKMKADLATCKQEGMTVEGYYGKLNKIWDNINS 161
Query: 139 -----IIHADVPNIYLAAVQEVYNTSKQ-DQFLMIL-RPEFEVVRGSLLNRNVVPSLDTC 191
I L QE Y Q+L L +F +R SL +R +P L+
Sbjct: 162 YRPLRICKCGRCICNLGTDQEKYREDDMVHQYLYGLNETKFHTIRSSLTSRVPLPGLEE- 220
Query: 192 VSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSC 251
V ++R+E+ ++ + + + + A+A Q R + + + +F + +
Sbjct: 221 VYNIVRQEEDMVNNRSSNEE----RTDVTAFAVQMRPRSEVISE-------KFANSEKLQ 269
Query: 252 SKKFCNYCKQRGHIITECYV---RPPPSTQSPM*ALHAYSTTN--------ATNGGVSQS 300
+KK C +C + GH C+V P P ++ +T+ NGG +
Sbjct: 270 NKKLCTHCNRGGHSPENCFVLIGYPEWWGDRPRGKSNSNGSTSRGRGRFGPGFNGGQPRP 329
Query: 301 EMIQQMVISALPSI------------------------GI-----QGKSSNASH------ 325
+ ++ PS G+ G+S N S+
Sbjct: 330 TYVNVVMTGPFPSSEHVNRVITDSDRDAVSGLTDEQWRGVVKLLNAGRSDNKSNAHETQS 389
Query: 326 -------PWFLDSGASYHMTGSLEYLHNLHSY--------DGNKKIQIADGNTLSITDVG 370
W LD+GAS+HMTG+LE L ++ S DGNK++ +++G T+ +
Sbjct: 390 GTCSLFTSWILDTGASHHMTGNLELLSDMRSMSPVLIILADGNKRVAVSEG-TVRLGS-- 446
Query: 371 DINSDFRDVLVSPGLASNLLSVGQLV 396
+ + V L S+L+SVGQ++
Sbjct: 447 --HLILKSVFYVKELESDLISVGQMM 470
Score = 30.8 bits (68), Expect = 5.3
Identities = 17/52 (32%), Positives = 30/52 (57%), Gaps = 3/52 (5%)
Query: 557 YLRNF*HMLKLNFKQVLKFF-TDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
+L++F +++ F +K +D+G E++ +EY HKGI + SC TP
Sbjct: 594 HLKDFIALVERQFDTEIKIVRSDNGTEFLC--MREYFLHKGIAHETSCVGTP 643
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 97.1 bits (240), Expect = 6e-20
Identities = 106/442 (23%), Positives = 182/442 (40%), Gaps = 74/442 (16%)
Query: 14 VRFSGKNYSAWEFQFRMYVKGERLWSHLNG-VSKAPTEKAALEEWETKDAQIITWILSTI 72
V +G NY+ W + ++ +R +NG + + P E W ++ I+ WI ++I
Sbjct: 47 VELNGDNYNQWATEMLNALQAKRKTGFINGTIPRPPPNDPNYENWTAVNSMIVGWIRTSI 106
Query: 73 NPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNL 132
P++ + S A +W LK+ ++ N + Q+ ++++ +Q +V EYY NL
Sbjct: 107 EPKVKATVTFISDAHLLWKDLKQRFSVGNKVRIHQIRAQLSSCRQDGQAVIEYYGRLSNL 166
Query: 133 WTEHSAIIHADVPNIYLAAVQEVYNTSKQ------DQFLMIL-RPEFEVVRGSLLNRNVV 185
W E++ V L +K+ QF++ L F + +L+N + +
Sbjct: 167 WEEYNIYKPVTVCTCGLCRCGATSEPTKEREEEKIHQFVLGLDESRFGGLCATLINMDPL 226
Query: 186 PSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFG 245
PSL S ++REEQRL + + + V + A+R H + S +
Sbjct: 227 PSLGEIYSRVIREEQRLASVHVREQ-----KEEAVGFLARREQLDHHSRVDASSSRSEHT 281
Query: 246 HIARSCS----KKFCNYCKQRGHIITECY--VRPPP------------------------ 275
+RS S + C+ C + GH EC+ V P
Sbjct: 282 GGSRSNSIIKGRVTCSNCGRTGHEKKECWQIVGFPDWWSERNGGRGSNGRGRGGRGSNGG 341
Query: 276 STQSPM*ALHAYSTTNATNGGVSQSEM--IQQMVISALPSIGIQGKSSNASH------PW 327
Q + A HA S+ ++ ++ M + Q+V S +S+
Sbjct: 342 RGQGQVMAAHATSSNSSVFPEFTEEHMRVLSQLVKEKSNSGSTSNNNSDRLSGKTKLGDI 401
Query: 328 FLDSGASYHMTGSLEYLHNLHSY--------DGNKKIQIADG-----NTLSITDVGDINS 374
LDSGAS+HMTG+L L N+ DG+K ++ G NT+S+T
Sbjct: 402 ILDSGASHHMTGTLSSLTNVVPVPPCPVGFADGSKAFALSVGVLTLSNTVSLT------- 454
Query: 375 DFRDVLVSPGLASNLLSVGQLV 396
+VL P L L+SV +L+
Sbjct: 455 ---NVLFVPSLNCTLISVSKLL 473
>At2g07010 putative retroelement pol polyprotein
Length = 1413
Score = 92.0 bits (227), Expect = 2e-18
Identities = 103/422 (24%), Positives = 175/422 (41%), Gaps = 46/422 (10%)
Query: 14 VRFSGKNYSAWEFQFRMYVKGERLWSHLNG-VSKAPTEKAALEEWETKDAQIITWILSTI 72
V +G NY+ W + ++ +R +NG +SK P + E W+ ++ I+ WI ++I
Sbjct: 42 VMLTGDNYNEWSTKMLNALQAKRKTGFINGSISKPPLDNPDYENWQAVNSMIVGWIRASI 101
Query: 73 NPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNL 132
P++ + + A ++W+ LK+ ++ N Q++ ++A +Q V +YY L
Sbjct: 102 EPKVKSTVTFICDAHQLWSELKQRFSVGNKVHVHQIKTQLAACRQDGQPVIDYYGRLCKL 161
Query: 133 WTEHSAIIHADVPNIYLAAVQEVYNTSKQ------DQFLMIL-RPEFEVVRGSLLNRNVV 185
W E V L SK+ QF++ L F + +L+ +
Sbjct: 162 WEEFQIYKPITVCKCGLCTCGATLEPSKEREEEKIHQFVLGLDDSRFGGLSATLIAMDPF 221
Query: 186 PSLDTCVSELLREEQRLLTQ--GAMSRDALIFESTTVAYAAQRRG-----KGHDMQQVQC 238
PSL S ++REEQRL + + A+ F + A R K D + V C
Sbjct: 222 PSLGEIYSRVVREEQRLASVQIREQQQSAIGFLTRQSEVTADGRTDSSIIKSRD-RSVLC 280
Query: 239 FSCKQFGHIARSCSK--KFCNYCKQ------RGHIITECYVRPPPSTQSPM*ALHAYSTT 290
C + GH + C + F ++ + RG R S S +
Sbjct: 281 SHCGRSGHEKKDCWQIVGFPDWWTERTNGGGRGSSSRGRGGRSSGSNNSGR-GRGQVTAA 339
Query: 291 NATNGGVS--------QSEMIQQMVISALPSIGIQGKSSNASHPWFLDSGASYHMTGSLE 342
+AT +S Q +I QM+ + + LD+GAS+HMTG L
Sbjct: 340 HATTSNLSPFPEFTPDQLRVITQMIQNKNNGTSDKLSGKMKLGDVILDTGASHHMTGQLS 399
Query: 343 YLHNLHSY--------DGNKKIQIADGNTLSITDVGDINSDFRDVLVSPGLASNLLSVGQ 394
L N+ + DG K I+ G T +++ ++ +VL P L +L+SV +
Sbjct: 400 LLTNIVTIPSCSVGFADGRKTFAISMG-TFKLSETVSLS----NVLYVPALNCSLISVSK 454
Query: 395 LV 396
LV
Sbjct: 455 LV 456
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 91.3 bits (225), Expect = 3e-18
Identities = 90/422 (21%), Positives = 168/422 (39%), Gaps = 66/422 (15%)
Query: 20 NYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALEEWETKDAQIITWILSTINPQMINN 79
NY+ W + + +++ ++ ++G P L W ++ I+ WI ++I+P + +
Sbjct: 43 NYAEWSEELQNFLRAKQKLGFIDGSIPKPAADPELSLWIAINSMIVGWIRTSIDPTIRST 102
Query: 80 LRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEHSAI 139
+ S A ++W L+R ++ N ++ L+ EIA Q V YY + LW E
Sbjct: 103 VGFVSEASQLWENLRRRFSVGNGVRKTLLKDEIAACTQDGQPVLAYYGRLIKLWEELQNY 162
Query: 140 IHADVPNIYLAA-VQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTCVSELLRE 198
A+ +++ + +FL+ L F +R S+ + +P L S ++RE
Sbjct: 163 KSGRECKCEAASDIEKEREDDRVHKFLLGLDSRFSSIRSSITDIEPLPDLYQVYSRVVRE 222
Query: 199 EQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSCSKKFCNY 258
EQ L SR + ++ + ++ VQ + +F R S FC +
Sbjct: 223 EQNLNA----SRTKDVVKTEAIGFS------------VQSSTTPRF----RDKSTLFCTH 262
Query: 259 CKQRGHIITECYV--------------------------------------RPPPSTQSP 280
C ++GH +T+C++ P+T+
Sbjct: 263 CNRKGHEVTQCFLVHGYPDWWLEQNPQENQPSTRGRGSNGRGSSSGRGGNRSSAPTTRGR 322
Query: 281 M*ALHAYSTTNATNG-GVSQSEMIQQMVISALPSIGIQGKSSNAS-HPWFLDSGASYHMT 338
A +A + +G G Q + ++ + PS + S N +D+GAS+HMT
Sbjct: 323 GRANNAQAAAPTVSGDGNDQIAQLISLLQAQRPSSSSERLSGNTCLTDGVIDTGASHHMT 382
Query: 339 GSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDI----NSDFRDVLVSPGLASNLLSVGQ 394
G L ++ + + DG T G + + DVL P L+SV +
Sbjct: 383 GDCSILVDVFDITPS-PVTKPDGKASQATKCGTLLLHDSYKLHDVLFVPDFDCTLISVSK 441
Query: 395 LV 396
L+
Sbjct: 442 LL 443
Score = 35.4 bits (80), Expect = 0.22
Identities = 23/51 (45%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Query: 558 LRNF*HMLKLNF-KQVLKFFTDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
++NF M + F KQV F TD+G E+M Y Q GIL Q SC +TP
Sbjct: 605 IKNFCAMSERQFGKQVKAFRTDNGTEFMC--LTPYFQTHGILHQTSCVDTP 653
>At2g14930 pseudogene
Length = 1149
Score = 91.3 bits (225), Expect = 3e-18
Identities = 105/427 (24%), Positives = 174/427 (40%), Gaps = 77/427 (18%)
Query: 1 YTMDSQKQIDNMCVRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPT-------EKAA 53
YT+ S + + V+ + +NY W+ QF ++ G+ L +NG APT +
Sbjct: 8 YTLPSLNISNCVTVKLTDRNYILWKSQFESFLSGQGLLGFVNGAYAAPTGTVSGPQDAGV 67
Query: 54 LEEWETKDAQIITWILS---TINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLEL 110
E D Q W S ++ +++ + + E+W L + +N+ + ++ F+L+
Sbjct: 68 TEAIPNPDYQ--AWFRSDQVVMSEDILSVVVGSKTSHEVWMNLAKHFNRISSSRIFELQR 125
Query: 111 EIANYKQGNSSVQEYYSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQDQFLMI--L 168
+ + + +++EY +L + A + + V K F M+ L
Sbjct: 126 RLHSLSKEGKTMEEYLR-YLKTICDQLASVGSPV-------------AEKMKIFAMVHGL 171
Query: 169 RPEFEVVRGSL---LNRNVVPSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQ 225
E+E + SL L+ PS + V L + RL QG D + ++
Sbjct: 172 TREYEPLITSLEGTLDAFPGPSYEDVVYRLKNFDDRL--QGYTVTDVSPHLAFNTFRSSN 229
Query: 226 R-------RGKGHDMQQVQCFSCKQFGHIARSCS---KKFCNYCKQRGHIITECYVRPPP 275
R RGKG+ + + F +QF + S S K C C +RGH +C+ R
Sbjct: 230 RGRGGRNNRGKGNFSTRGRGFQ-QQFSSSSSSVSASEKPMCQICGKRGHYALQCWHRFDD 288
Query: 276 STQSPM*ALHAYSTTNATNGGVSQSEMIQQMVISALPSIGIQGKSSNASHPWFLDSGASY 335
S Q A A+S + T+ VS W DS A+
Sbjct: 289 SYQHSEAAAAAFSALHITD--VSDDS------------------------GWVPDSAATA 322
Query: 336 HMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDI-------NSDFRDVLVSPGLASN 388
H+T + L + Y GN + +DGN L IT +G N +DVLV P +A +
Sbjct: 323 HITNNSSRLQQMQPYLGNDTVMASDGNFLPITHIGSANLPSTSGNLPLKDVLVCPNIAKS 382
Query: 389 LLSVGQL 395
LLSV +L
Sbjct: 383 LLSVSKL 389
Score = 38.1 bits (87), Expect = 0.033
Identities = 19/54 (35%), Positives = 31/54 (57%)
Query: 554 CFLYLRNF*HMLKLNFKQVLKFFTDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
C L+++ + L ++ F +D GGE+ S+ F ++LQ GI SCP+TP
Sbjct: 549 CSLFMKFQSFVENLLQTKIGTFQSDGGGEFTSNRFLQHLQESGIQHYISCPHTP 602
>At4g17450 retrotransposon like protein
Length = 1433
Score = 88.6 bits (218), Expect = 2e-17
Identities = 103/445 (23%), Positives = 165/445 (36%), Gaps = 103/445 (23%)
Query: 18 GKNYSAWEFQFRMYVKGERLWSHLNGVSKAP-TEKAALEEWETKDAQIITWILSTINPQM 76
G NY+ W RM + + S ++G P + W ++ + TW+L+ +
Sbjct: 87 GTNYNNWSIAMRMSLDAKNKLSFVDGSLPRPDVSDRMFKIWSRCNSMVKTWLLNVVT--- 143
Query: 77 INNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEH 136
EMWN L + N +++QLE I KQGN + YY+ LW +
Sbjct: 144 -----------EMWNDLFSRFRVSNLPRKYQLEQSIHTLKQGNLDLSTYYTKKKTLWEQL 192
Query: 137 SAIIHADVPNIYLAAVQEVYN---TSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTCVS 193
+ V V+E+ TS+ QFLM L F +RG +LN P L +
Sbjct: 193 ANTRVLTVRKCNCEHVKELLEEAETSRIIQFLMGLNDNFAHIRGQILNMKPRPGLTEIYN 252
Query: 194 ELLREE-QRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSCS 252
L ++E QRL+ +S F+ Q Q S
Sbjct: 253 MLDQDESQRLVGNPTLSNPTAAFQV-----------------QASPIIDSQVNMAQGSYK 295
Query: 253 KKFCNYCKQRGHIITECYVR---PP---------------------PSTQSP-------- 280
K C+YC + GH++ +CY + PP P ++P
Sbjct: 296 KPKCSYCNKLGHLVDKCYKKHGYPPGSKWTKGQTIGSTNLASTQLQPVNETPNEKTDSYE 355
Query: 281 ------M*ALHAYSTTNATNGGVSQSEMIQQMVISALPSIGIQGK--------------- 319
+ + +Y +T S ISA PS+ + +
Sbjct: 356 EFSTDQIQTMISYLSTKLHIASASPMPTTSSASISASPSVPMISQISGTFLSLFSNAYYD 415
Query: 320 ---SSNASHP------WFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVG 370
SS + P W +DSGA++H+T + + N S + N +++ + T+ I +G
Sbjct: 416 MLISSVSQEPAVSPRGWVIDSGATHHVTHNRDLYLNFRSLE-NTFVRLPNDCTVKIAGIG 474
Query: 371 DIN-SD---FRDVLVSPGLASNLLS 391
I SD +VL P NL+S
Sbjct: 475 FIQLSDAISLHNVLYIPEFKFNLIS 499
>At4g04410 putative polyprotein
Length = 1017
Score = 86.7 bits (213), Expect = 8e-17
Identities = 90/418 (21%), Positives = 166/418 (39%), Gaps = 73/418 (17%)
Query: 14 VRFSGKNYSAWEFQFRMYVKGERLWSHLNGVS--------------KAPTEKAALEE--- 56
V G+NY W + + G LWSH+ K E+A +E+
Sbjct: 11 VVLKGENYLLWMRTTKTVLCGRGLWSHVETKHVQFKATKEDGDEGFKREEEEAEIEKEAK 70
Query: 57 WETKDAQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIY-NQDNPAKRFQLELEIANY 115
W +D ++ + ++ ++ A+E+W L+ +Y N N + F+++ I N
Sbjct: 71 WFQEDQNVLAILQHSLESSILEAYSYCETARELWETLENVYGNVSNLTRVFEVKKAINNL 130
Query: 116 KQGNSSVQEYYSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQDQFLMILRPEFEVV 175
Q + +++ F +LW E + + V A + E K L+ L P F +
Sbjct: 131 SQVDLEFTKHFGKFRSLWAELEMLRPSTVDP---AILNERKEQEKVFGLLLTLNPAFNDL 187
Query: 176 RGSLLNRNVVPSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQ 235
LL + +PSL+ S++ +E+ L G S + + Y + R
Sbjct: 188 IKHLLRADKLPSLENVCSQVQKEQGSL---GLFSGKGELITAHKGIYKGEER-------- 236
Query: 236 VQCFSCKQFGHIARSCSKKFCNYCKQRGHIITECYVRPPPSTQSPM*ALHAYSTTNATNG 295
C++CK++GH+ +C++ P + A + +
Sbjct: 237 ----------------KVWVCDHCKKKGHMKDKCWILHPHLKPAKFKANISQEVASDQGE 280
Query: 296 GVSQSEMIQQMVISALPSIGIQGKS-------------------SNASHPWFLDSGASYH 336
V +S++ + +I ++ S+ G S S S +DSGAS+H
Sbjct: 281 VVRKSDL--ESLIRSIASLKESGTSFLTYEPNKMLKESGTSFFTSEPSKTLVIDSGASHH 338
Query: 337 MTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDIN--SDFRDVLVSPGLASNLLSV 392
M + + N+ GN + IA+G+ + + ++G++N L P SNLLSV
Sbjct: 339 MINNPSLIDNIKPALGN--VVIANGDKVPVKEIGELNLFDKKSKALYMPSFTSNLLSV 394
Score = 45.8 bits (107), Expect = 2e-04
Identities = 26/60 (43%), Positives = 35/60 (58%), Gaps = 2/60 (3%)
Query: 550 PSLKCFL-YLRNF*HMLKLNFKQVLKFF-TDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
PS C + NF + + F +K +D+GGEY SH+F+EYL GI+ Q SCP TP
Sbjct: 468 PSKDCVMDAFINFQNYVTNYFNAKIKVLRSDNGGEYTSHKFKEYLAKHGIIHQTSCPYTP 527
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 85.9 bits (211), Expect = 1e-16
Identities = 93/432 (21%), Positives = 170/432 (38%), Gaps = 58/432 (13%)
Query: 15 RFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAA-LEEWETKDAQIITWILSTIN 73
R NY W + + + ++G P E W ++ + +W+L++++
Sbjct: 78 RLDETNYGDWSVAMLISLDAKNKTGFIDGTLSRPLESDLNFRLWSRCNSMVKSWLLNSVS 137
Query: 74 PQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLW 133
PQ+ ++ + A ++W L +N N + + L EI +++QG S+ EYY+ LW
Sbjct: 138 PQIYRSILRMNDASDIWRDLNSRFNVTNLPRTYNLTQEIQDFRQGTLSLSEYYTRLKTLW 197
Query: 134 TEHSAIIHADVPNIYLAA--VQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTC 191
+ + D P A +Q+ +K +FL L + +VR ++ + +PSL
Sbjct: 198 DQLDSTEALDEPCTCGKAMRLQQKAEQAKIVKFLAGLNESYAIVRRQIIAKKALPSLGEV 257
Query: 192 VSELLREEQRLLTQGAMSRDALIFES---------TTVAYAAQRRGKGHDMQQVQCFSCK 242
L ++ + ++ A S TV Y KG + C
Sbjct: 258 YHILDQDNSQQSFSNVVAPPAAFQVSEITQSPSMDPTVCYVQNGPNKGRPI----CSFYN 313
Query: 243 QFGHIARSCSKK--FCNYCKQRGHIITECYVRPPPSTQSPM*ALHAYSTTNATNGGVSQS 300
+ GHIA C KK F +G E +P P + + ++ + G +S+
Sbjct: 314 RVGHIAERCYKKHGFPPGFTPKGK-AGEKLQKPKPLAANVAESSEVNTSLESMVGNLSK- 371
Query: 301 EMIQQMVI-------------------------------SALPSIGI--QGKSSNASHPW 327
E +QQ + S IGI + + +S W
Sbjct: 372 EQLQQFIAMFSSQLQNTPPSTYATASTSQSDNLGICFSPSTYSFIGILTVARHTLSSATW 431
Query: 328 FLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVG--DINSD--FRDVLVSP 383
+DSGA++H++ +L + + + G T+ I+ VG +N D ++VL P
Sbjct: 432 VIDSGATHHVSHDRSLFSSLDT-SVLSAVNLPTGPTVKISGVGTLKLNDDILLKNVLFIP 490
Query: 384 GLASNLLSVGQL 395
NL+S+ L
Sbjct: 491 EFRLNLISISSL 502
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 82.4 bits (202), Expect = 2e-15
Identities = 91/431 (21%), Positives = 167/431 (38%), Gaps = 81/431 (18%)
Query: 4 DSQKQIDNMCVRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAAL--------- 54
D+ ++ ++ + NY W+ QF + ++L +NG AP++ +
Sbjct: 9 DNVHVTSSVTLKLTDSNYLLWKTQFESLLSSQKLIGFVNGAVNAPSQSRLVVNGEVTSEE 68
Query: 55 -----EEWETKDAQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLE 109
E W D + +W+ T++ +++ ++ + S ++++W L +N+ + A+ F L
Sbjct: 69 PNPLYESWFCTDQLVRSWLFGTLSEEVLGHVHNLSTSRQIWVSLAENFNKSSVAREFSLR 128
Query: 110 LEIANYKQGNSSVQEYYSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQDQFLMILR 169
+ + Y F + S+I + V + K FL L
Sbjct: 129 QNLQLLSKKEKPFSVYCREFKTICDALSSI------------GKPVDESMKIFGFLNGLG 176
Query: 170 PEFE---VVRGSLLNRNVVPSLDTCVSELLREEQRLLT--QGAMSRDALIF-----ESTT 219
+++ V S L++ P+ + VSE+ + +L + + A L F ES +
Sbjct: 177 RDYDPITTVIQSSLSKLPTPTFNDVVSEVQGFDSKLQSYEEAASVTPHLAFNIERSESGS 236
Query: 220 VAYAAQRRGKGHDMQQV--QCFSCKQFGHIARSCS------KKFCNYCKQRGHIITECYV 271
Y ++G+G Q +S + G S + C C + GH +CY
Sbjct: 237 PQYNPNQKGRGRSGQNKGRGGYSTRGRGFSQHQSSPQVSGPRPVCQICGRTGHTALKCYN 296
Query: 272 RPPPSTQSPM*ALHAYSTTNATNGGVSQSEMIQQMVISALPSIGIQGKSSNASHPWFLDS 331
R + Q+ + A+ST S + W DS
Sbjct: 297 RFDNNYQA---EIQAFSTLRV---------------------------SDDTGKEWHPDS 326
Query: 332 GASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGD--INSD-----FRDVLVSPG 384
A+ H+T S L + Y+G+ + + DG L IT G I S +VLV P
Sbjct: 327 AATAHVTSSTNGLQSATEYEGDDAVLVGDGTYLPITHTGSTTIKSSNGKIPLNEVLVVPN 386
Query: 385 LASNLLSVGQL 395
+ +LLSV +L
Sbjct: 387 IQKSLLSVSKL 397
Score = 33.9 bits (76), Expect = 0.63
Identities = 17/42 (40%), Positives = 27/42 (63%), Gaps = 1/42 (2%)
Query: 566 KLNFKQVLKFFTDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
+LN K + F +D GGE++S++ + +L GI + SCP TP
Sbjct: 570 QLNTK-IKVFQSDGGGEFVSNKLKTHLSEHGIHHRISCPYTP 610
>At4g16870 retrotransposon like protein
Length = 1474
Score = 81.6 bits (200), Expect = 3e-15
Identities = 84/409 (20%), Positives = 157/409 (37%), Gaps = 62/409 (15%)
Query: 15 RFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALEE----------WETKDAQI 64
+ + NY W Q + G L HL+G + P W+ +D I
Sbjct: 33 KLTSNNYLMWSLQIHALLDGYELAGHLDGSIETPAPTLTTNNVVSANPQYTLWKRQDRLI 92
Query: 65 ITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQE 124
+ ++ I+P + + + A ++W L Y + + QL +I K+G ++ E
Sbjct: 93 FSALIGAISPPVQPLVSRATKASQIWKTLTNTYAKSSYDHIKQLRTQIKQLKKGTKTIDE 152
Query: 125 YYSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNV 184
Y + H + + + + + + ++ L L +++ V + ++
Sbjct: 153 Y------------VLSHTTLLDQLAILGKPMEHEEQVERILEGLPEDYKTVVDQIEGKDN 200
Query: 185 VPSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQF 244
PS+ L+ E +LL+ A+S +L + + ++ + +
Sbjct: 201 TPSITEIHERLINHEAKLLSTAALSSSSLPMSANVAQQRHHNNNRNNNQNKNRTQGNTYT 260
Query: 245 GHIARSCSKKF-----------CNYCKQRGHIITECYVRPPPSTQSPM*ALHAYSTTNAT 293
+ S + K C C +GH C P Q A+ S+++A+
Sbjct: 261 NNWQPSANNKSGQRPFKPYLGKCQICNVQGHSARRC-----PQLQ----AMQPSSSSSAS 311
Query: 294 NGGVSQSEMIQQMVISALPSIGIQGKSSNASHPWFLDSGASYHMTGSLEYLHNLHSYDGN 353
Q P + + ++ W LDSGA++H+T L L Y+G+
Sbjct: 312 TFTPWQ------------PRANLAMGAPYTANNWLLDSGATHHITSDLNALALHQPYNGD 359
Query: 354 KKIQIADGNTLSITDVGD--INSDFRD-----VLVSPGLASNLLSVGQL 395
+ IADG +L IT G + S+ RD VL P + NL+SV +L
Sbjct: 360 -DVMIADGTSLKITKTGSTFLPSNARDLTLNKVLYVPDIQKNLVSVYRL 407
>At2g11140 putative retroelement pol polyprotein
Length = 411
Score = 80.9 bits (198), Expect = 5e-15
Identities = 57/251 (22%), Positives = 112/251 (43%), Gaps = 23/251 (9%)
Query: 18 GKNYSAWEFQFRMYVKGERLWSHLNGVSKAPT-EKAALEEWETKDAQIITWILSTINPQM 76
G NY++W R+ + + ++G P+ + + W ++ + +WIL+ +N ++
Sbjct: 86 GSNYNSWSIAMRISLDAKNKLGFVDGSLLRPSVDDSTFRIWSRCNSMVKSWILNVVNKEI 145
Query: 77 INNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLW--- 133
+++ + A EMW L + +N +++QLE + KQG+ ++ Y++ LW
Sbjct: 146 YDSILYYEDAVEMWTDLFTRFRVNNLPRKYQLEQAVMTLKQGSLNLSTYFTKKKTLWEQL 205
Query: 134 --TEHSAIIHADVPNIYLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTC 191
T+ ++ D + + E TS+ QFLM L +F + +LN P L+
Sbjct: 206 LNTKTRSVKKCDCDQV--KELLEDAETSRVIQFLMGLNDDFNTIMSQILNMKPRPGLNEI 263
Query: 192 VSELLREE-QRLL--------TQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCK 242
+ L ++E QRL+ + A L+ E + A + ++ +C C
Sbjct: 264 YNMLDQDESQRLVGHASKPTPSPAAFQTQGLLTEQNPILMAQ------GNFKKPKCTHCN 317
Query: 243 QFGHIARSCSK 253
+ GH C K
Sbjct: 318 RIGHTVDKCYK 328
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 79.0 bits (193), Expect = 2e-14
Identities = 53/261 (20%), Positives = 115/261 (43%), Gaps = 20/261 (7%)
Query: 17 SGKNYSAWEFQFRMYVKGERLWSHLNG-VSKAPTEKAALEEWETKDAQIITWILSTINPQ 75
+ ++ +W M + +NG ++K P + W + + TW++++++ +
Sbjct: 53 TASDFHSWRRSILMALNVRNKLGFINGTITKPPEDHRDFGAWSRCNDIVSTWLMNSVDKK 112
Query: 76 MINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTE 135
+ +L + Q +WN L + QD+ + F +E +++ +QG+ + YY+ L LW E
Sbjct: 113 IGQSLLYIATVQGIWNNLLSRFKQDDAPRIFDIEQKLSKIEQGSMDISTYYTALLTLWEE 172
Query: 136 HSAIIHADVPNIYL----AAV--QEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLD 189
H + V AAV + + S+ +FL L F+ R +L +P++
Sbjct: 173 HRNYVELPVCTCGRCECDAAVKWEHLQQRSRVTKFLKELNEGFDQTRRHILMLKPIPTIK 232
Query: 190 TCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIAR 249
+ + ++E++ + D++ F++T++ + V ++ R
Sbjct: 233 EAFNMVTQDERQRNVKPLTRVDSVAFQNTSMINEDENA-------YVAAYNT------VR 279
Query: 250 SCSKKFCNYCKQRGHIITECY 270
K C +C + GH I +CY
Sbjct: 280 PNQKPICTHCGKVGHTIQKCY 300
Score = 34.3 bits (77), Expect = 0.48
Identities = 30/109 (27%), Positives = 43/109 (38%), Gaps = 16/109 (14%)
Query: 324 SHPWFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDINSDFRDVLVSP 383
S W +DSGAS H+ L L S G T+ IT +VL P
Sbjct: 479 SDAWIIDSGASSHVCSDLAMFRELKSVSG----------TVHITQ----KLILHNVLHVP 524
Query: 384 GLASNLLSVGQLVVILMLIFHVLVVLC--KSRCRGKLSRRGLKWEDCFL 430
NL+SV LV + H V C + +G + RG + + ++
Sbjct: 525 DFKFNLMSVSSLVKTISCSAHFYVDCCLIQELSQGLMIGRGRLYHNLYI 573
>At2g06950 putative retroelement pol polyprotein
Length = 405
Score = 78.6 bits (192), Expect = 2e-14
Identities = 88/416 (21%), Positives = 160/416 (38%), Gaps = 84/416 (20%)
Query: 1 YTMDSQKQIDNMCVRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALEE---- 56
Y++ S + + V + KNY W+ QF ++ G+ L + G AP++ + + +
Sbjct: 14 YSVPSLNISNCVTVTLTAKNYILWKSQFESFLDGQGLLGFVTGSIPAPSQTSVVSDIDGS 73
Query: 57 -----------WETKDAQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKR 105
W D + +W+L + +++ + + + + E+W + +N+ + ++
Sbjct: 74 TSASPNPEYYTWFKTDRVVKSWLLGSFLEDILSVVVNCNTSHEVWISVANHFNRVSSSRL 133
Query: 106 FQLELEIANYKQGNSSVQEYYSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQDQFL 165
F+L+ + N + + S+ EY + + LA+V T K F
Sbjct: 134 FELQRRLQNVSKRDKSMDEYLKDLKTICDQ-------------LASVGSPV-TEKMKIFA 179
Query: 166 MI--LRPEFEVVRGSLLNRNVV---PSLDTCVSELLREEQRLLTQGAMSRDA----LIFE 216
+ L E+E ++ ++ N PSL+ + +L + RL QG + A + F
Sbjct: 180 ALNGLGREYEPIKTTIENSMDALPGPSLEDVIPKLTGYDDRL--QGYLEETAVSPHVAFN 237
Query: 217 STTV-------AYAAQRRGKGHDMQQVQCFSCKQFGHIARSCSKK------------FCN 257
TT + A RGKG + FS + G + S C
Sbjct: 238 ITTSDDSNASGYFNAYNRGKGKSNRGRNSFSTRGRGFHQQISSTNSSSGSQSGGTSVVCQ 297
Query: 258 YCKQRGHIITECYVRPPPSTQSPM*ALHAYSTTNATNGGVSQSEMIQQMVISALPSIGIQ 317
C + GH +C+ R N E+ + AL ++ I
Sbjct: 298 ICGKMGHPALKCWHR--------------------FNNSYQYEELPR-----ALAAMRIT 332
Query: 318 GKSSNASHPWFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDIN 373
+ + W DS A+ H+T S L Y G+ + +ADGN L IT G N
Sbjct: 333 DITDQHGNEWLPDSAATAHVTNSPRSLQQSQPYHGSDAVMVADGNFLPITHTGSTN 388
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 78.2 bits (191), Expect = 3e-14
Identities = 94/437 (21%), Positives = 166/437 (37%), Gaps = 53/437 (12%)
Query: 15 RFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAA-LEEWETKDAQIITWILSTIN 73
R Y W R+ + + ++G P E W ++ + +W+L++++
Sbjct: 82 RLDETTYGDWSVAMRISLDAKNKLGFVDGSLPRPLESDPNFRLWSRCNSMVKSWLLNSVS 141
Query: 74 PQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLW 133
PQ+ ++ + A ++W L +N N + + L EI + +QG S+ EYY+ LW
Sbjct: 142 PQIYRSILRLNDATDIWRDLFDRFNLTNLPRTYNLTQEIQDLRQGTMSLSEYYTLLKTLW 201
Query: 134 TEHSAIIHADVPNIYLAAVQEVYNTSKQD--QFLMILRPEFEVVRGSLLNRNVVPSLDTC 191
+ + D P AV+ K +FL L + +VR ++ + +PSL
Sbjct: 202 DQLDSTEALDDPCTCGKAVRLYQKAEKAKIMKFLAGLNESYAIVRRQIIAKKALPSLAEV 261
Query: 192 VSELLREEQR-----LLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGH 246
L ++ + ++ A + + + S + G + + C C + GH
Sbjct: 262 YHILDQDNSQKGFFNVVAPPAAFQVSEVSHSPITSPEIMYVQSGPNKGRPTCSFCNRVGH 321
Query: 247 IARSCSKKF---------------------------CNYCKQRGHIITECYVRPPPSTQ- 278
IA C KK + K G + T P Q
Sbjct: 322 IAERCYKKHGFPPGFTPKGKSSDKPPKPQAVAAQVTLSPDKMTGQLETLAGNFSPDQIQN 381
Query: 279 ------SPM*ALHAYSTTNATNGGVSQSEMIQQMVISALPS----IGIQGKSSN--ASHP 326
S + T ++ S S+ + I PS IGI S N +S
Sbjct: 382 LIALFSSQLQPQIVSPQTASSQHEASSSQSVAPSGILFSPSTYCFIGILAVSHNSLSSDT 441
Query: 327 WFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGD--INSD--FRDVLVS 382
W +DSGA++H++ + L + + + G + I+ VG IN D ++VL
Sbjct: 442 WVIDSGATHHVSHDRKLFQTLDT-SIVSFVNLPTGPNVRISGVGTVLINKDIILQNVLFI 500
Query: 383 PGLASNLLSVGQLVVIL 399
P NL+S+ L L
Sbjct: 501 PEFRLNLISISSLTTDL 517
>At3g60170 putative protein
Length = 1339
Score = 74.3 bits (181), Expect = 4e-13
Identities = 93/414 (22%), Positives = 163/414 (38%), Gaps = 66/414 (15%)
Query: 5 SQKQIDNMCVRFSGKNYSAWEFQFRMYVKGERLWSHLNG------VSKAPTEKA---ALE 55
S+K + RF G Y W +++ LW + V P +A A+E
Sbjct: 4 SEKFVQPAIPRFDGY-YDFWSMTMENFLRSRELWRLVEEGIPAIVVGTTPVSEAQRSAVE 62
Query: 56 EWETKDAQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQL-----EL 110
E + KD ++ ++ I+ +++ + S ++ +W +K+ Y KR QL E
Sbjct: 63 EAKLKDLKVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEF 122
Query: 111 EIANYKQGNSSVQEYYSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQDQFLMILRP 170
E+ K+G + + L V N + + ++ + L L P
Sbjct: 123 ELLAMKEGEK-IDTFLGRTLT------------VVNKMKTNGEVMEQSTIVSKILRSLTP 169
Query: 171 EFEVVRGSLLNRNVVP--SLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRG 228
+F V S+ N + S+D LL EQRL G + + + + T +Q RG
Sbjct: 170 KFNYVVCSIEESNDLSTLSIDELHGSLLVHEQRL--NGHVQEEQAL-KVTHEERPSQGRG 226
Query: 229 KGHDMQQVQCFSCKQFGHIARSCSKKFCNYCKQRGHIITECYVRPPPSTQSPM*ALHAYS 288
+G + + + G + + C C GH EC P +
Sbjct: 227 RG--VFRGSRGRGRGRGRSGTNRAIVECYKCHNLGHFQYEC-----PEWEK--------- 270
Query: 289 TTNATNGGVSQSEMIQQMVISALPSIGIQGKSSNASHPWFLDSGASYHMTGSLEYLHNLH 348
N ++ E +++++ A ++ +N WFLDSG S HMTGS E+ L
Sbjct: 271 -----NANYAELEEEEELLLMAY----VEQNQANRDEVWFLDSGCSNHMTGSKEWFSELE 321
Query: 349 SYDGNKKIQIADGNTLSITDVGDINSDFR-------DVLVSPGLASNLLSVGQL 395
N+ +++ + +S+ G + +V P L +NLLS+GQL
Sbjct: 322 E-GFNRTVKLGNDTRMSVVGKGSVKVKVNGVTQVIPEVYYVPELRNNLLSLGQL 374
Score = 30.0 bits (66), Expect = 9.1
Identities = 14/31 (45%), Positives = 19/31 (61%)
Query: 577 TDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
TD GGE+ S+EF E+ + GI Q + TP
Sbjct: 563 TDRGGEFTSNEFGEFCRSHGISRQLTAAFTP 593
>At4g09480 unknown protein
Length = 301
Score = 72.4 bits (176), Expect = 2e-12
Identities = 41/210 (19%), Positives = 98/210 (46%), Gaps = 7/210 (3%)
Query: 17 SGKNYSAWEFQFRMYVKGERLWSHLNG-VSKAPTEKAALEEWETKDAQIITWILSTINPQ 75
SG + +W RM + ++G + + P+ W + + TW++++++ +
Sbjct: 45 SGSEFHSWRRSVRMALNVRNKLGFIDGTIPQPPSTHRDAGSWSRCNDMVATWLMNSVSKK 104
Query: 76 MINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTE 135
+ +L S A+ +W L + QD+ + F++E + + +QG+ V YY+ + LW E
Sbjct: 105 IGQSLLFMSTAESIWKNLLSRFKQDDAPRVFEIEQRLGSLQQGSMDVSTYYTELVTLWEE 164
Query: 136 HSAIIHADVPNI------YLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLD 189
+ I + A +++ S+ +FLM L +E + +L +P+++
Sbjct: 165 YKNYIELPLCTCGRCECNASALWEKMQQRSRVTKFLMGLNEAYEATQRHILMLKPIPTIE 224
Query: 190 TCVSELLREEQRLLTQGAMSRDALIFESTT 219
+ + ++E++ + D + F+S++
Sbjct: 225 DVFNMVAQDERQKSIKPVAKMDNVAFQSSS 254
>At1g21280 unknown protein (At1g21280)
Length = 237
Score = 72.4 bits (176), Expect = 2e-12
Identities = 44/180 (24%), Positives = 88/180 (48%), Gaps = 13/180 (7%)
Query: 20 NYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAAL-EEWETKDAQIITWILSTINPQMIN 78
NY AW+ +FR +++ + + ++G P + L + WE +A ++ W+++++ +++
Sbjct: 41 NYVAWKIRFRSFLRVTKKFGFIDGTLPKPDPFSPLYQPWEQCNAMVMYWLMNSMTDKLLE 100
Query: 79 NLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEHSA 138
++ A +MW L+R++ K +QL +A +QG SV+EY+ +W E S
Sbjct: 101 SVMYAETAHKMWEDLRRVFVPCVDLKIYQLRRRLATLRQGGDSVEEYFGKLSKVWMELSE 160
Query: 139 IIHADVPNIYLAA--------VQEVYNTSKQDQFLMILR--PEFEVVRGSLLNRNVVPSL 188
+A +P +E ++ +FLM L+ FE V ++ + PSL
Sbjct: 161 --YAPIPECKCGGCNCECTKRAEEAREKEQRYEFLMGLKLNQGFEAVTTKIMFQKPPPSL 218
>At1g47650 hypothetical protein
Length = 1409
Score = 69.3 bits (168), Expect = 1e-11
Identities = 52/243 (21%), Positives = 99/243 (40%), Gaps = 12/243 (4%)
Query: 15 RFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAA-LEEWETKDAQIITWILSTIN 73
R NY W + + + ++G P E W + + +W+L++++
Sbjct: 81 RLDETNYGDWNVAMLISLDAKNKSGFIDGTLPRPLESDKNFRLWSRCKSMVKSWLLNSVS 140
Query: 74 PQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLW 133
PQ+ ++ + A ++W L +N N + + L EI +++QG S+ EYY+ LW
Sbjct: 141 PQIYRSILRMNDASDIWRDLHSRFNVTNLPRTYNLTQEIQDFRQGTLSLSEYYTRLKTLW 200
Query: 134 TEHSAIIHADVPNIYLAA--VQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTC 191
+ + D P I A +Q+ +K + L L + ++R ++ + +PSL
Sbjct: 201 DQLESTEELDDPCICGKAMRLQQKAEQAKIVKLLAGLNDSYAIIRRQIIAKKALPSLAE- 259
Query: 192 VSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSC 251
V +L ++ + F+ + A V + GHIA C
Sbjct: 260 VYHILDQDNSQQGFSNVVAPPAAFQVSEAMMATN--------NDVNMLCSEWVGHIAERC 311
Query: 252 SKK 254
KK
Sbjct: 312 YKK 314
>At4g03530 putative reverse transcriptase
Length = 374
Score = 65.5 bits (158), Expect = 2e-10
Identities = 51/256 (19%), Positives = 109/256 (41%), Gaps = 23/256 (8%)
Query: 18 GKNYSAWEFQFRMYVKGERLWSHLNG-VSKAPTEKAALEEWETKDAQIITWILSTINPQM 76
G +Y W ++ + ++G + K + + W ++ + +W+L++++ ++
Sbjct: 89 GNSYGTWIIAMTTSIEAKNKLGFVDGSIPKPDDDDPYCKIWRRCNSMVKSWLLNSVSKEI 148
Query: 77 INNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEH 136
++ F A +W L +++ + + ++L +I + +QGN + Y++ LW E
Sbjct: 149 YTSILYFPTAAAIWKDLYTRFHKSSLPRLYKLRQQIHSLRQGNLDLSSYHTRKQTLWEEL 208
Query: 137 SAI--IHADVPNIYLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTCVSE 194
+++ I V ++ + T++ FLM L ++ VR +L + +PSL
Sbjct: 209 TSLQAIPRTVEDLLIER-----ETNRVIDFLMGLNDCYDAVRSQILMKKTLPSLS----- 258
Query: 195 LLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSCSKK 254
E ++ Q + R A I STT + + Q G + +
Sbjct: 259 ---EVFNMIDQDEIQRSARI--STTPGMTSSVFAVSNQSSQSVLN-----GDTYQKKERP 308
Query: 255 FCNYCKQRGHIITECY 270
C YC + GH+ CY
Sbjct: 309 VCTYCSRPGHVEDTCY 324
>At4g22040 LTR retrotransposon like protein
Length = 1109
Score = 64.3 bits (155), Expect = 4e-10
Identities = 27/115 (23%), Positives = 62/115 (53%), Gaps = 1/115 (0%)
Query: 20 NYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAA-LEEWETKDAQIITWILSTINPQMIN 78
NY W F+ ++ + + L+G P + + LE+W T +A +++W+ TI+ +++
Sbjct: 42 NYEEWACGFKTALRSRKKFGFLDGTIPQPLDGSPDLEDWLTINALLVSWMKMTIDSELLT 101
Query: 79 NLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLW 133
N+ A+++W +++ ++ N K +++ ++A KQ +V+ YY +W
Sbjct: 102 NISHRDVARDLWEQIRKRFSVSNGPKNQKMKADLATCKQEGMTVEGYYGKLNKIW 156
Score = 38.1 bits (87), Expect = 0.033
Identities = 28/88 (31%), Positives = 47/88 (52%), Gaps = 17/88 (19%)
Query: 333 ASYHMTGSLEYLHNLHSY--------DGNKKIQIADGNTLSITDVGDINSDFRDVLVSPG 384
AS+HMTG+LE L + S DGNK++ +++G T+ + + + V
Sbjct: 169 ASHHMTGNLELLSGMRSMSPVLIILADGNKRVAVSEG-TVRLGS----HLILKSVFYVKE 223
Query: 385 LASNLLSVGQLV----VILMLIFHVLVV 408
L S+L+SVGQ++ ++ L H LV+
Sbjct: 224 LESDLISVGQMMDENHCVVQLADHFLVI 251
Score = 30.0 bits (66), Expect = 9.1
Identities = 17/52 (32%), Positives = 30/52 (57%), Gaps = 3/52 (5%)
Query: 557 YLRNF*HMLKLNFKQVLKFF-TDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
+L++F +++ F +K +D+G E++ +EY HKGI + SC TP
Sbjct: 394 HLKDFMALVERQFDTEIKTVRSDNGTEFLC--MREYFLHKGIAHETSCVGTP 443
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.347 0.154 0.508
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,921,440
Number of Sequences: 26719
Number of extensions: 1026966
Number of successful extensions: 4932
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 40
Number of HSP's that attempted gapping in prelim test: 4521
Number of HSP's gapped (non-prelim): 299
length of query: 1309
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1198
effective length of database: 8,352,787
effective search space: 10006638826
effective search space used: 10006638826
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146758.11


