

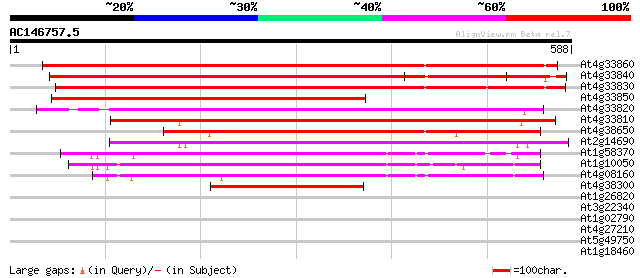
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.5 - phase: 0 /pseudo
(588 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g33860 unknown protein 672 0.0
At4g33840 putative protein 664 0.0
At4g33830 putative protein 644 0.0
At4g33850 putative protein 426 e-119
At4g33820 putative glycosyl hydrolase family 10 protein 395 e-110
At4g33810 beta-xylan endohydrolase -like protein 393 e-109
At4g38650 putative protein 361 e-100
At2g14690 1,4-beta-xylan endohydrolase 360 e-100
At1g58370 unknown protein 238 5e-63
At1g10050 putative xylan endohydrolase 219 3e-57
At4g08160 xylan endohydrolase like proteinchimeric 206 4e-53
At4g38300 unknown protein 174 2e-43
At1g26820 ribonuclease RNS3 32 1.3
At3g22340 hypothetical protein 31 1.7
At1g02790 exopolygalacturonase (At1g02790) 31 1.7
At4g27210 putative protein 31 2.2
At5g49750 putative protein 29 6.4
At1g18460 unknown protein 29 8.3
>At4g33860 unknown protein
Length = 576
Score = 672 bits (1734), Expect = 0.0
Identities = 318/545 (58%), Positives = 409/545 (74%), Gaps = 8/545 (1%)
Query: 35 EAEALSYDYSANVECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKSLGNKFV 94
E++ + YDYSA +ECL P KPQYNGGII +P++ DG GWT FG+A ++ RK + F
Sbjct: 19 ESKVVPYDYSATIECLEIPLKPQYNGGIIVSPDVRDGTLGWTPFGNAKVDFRKIGNHNFF 78
Query: 95 VTHSRNQPHDSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKFGGAIFAEPN 154
V R QP DSVSQK+YL KGL Y+ SAW+QVS+ PV AV K +K G++ AE
Sbjct: 79 VARDRKQPFDSVSQKVYLEKGLLYTFSAWLQVSKGKAPVKAVFKKNGEYKLAGSVVAESK 138
Query: 155 CWSMLKGGLIADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRK 214
CWSMLKGGL D +G AELYFES +T+VEIWVD+VSLQPFT+++W SH E SI+K+RKR
Sbjct: 139 CWSMLKGGLTVDESGPAELYFESEDTTVEIWVDSVSLQPFTQEEWNSHHEQSIQKERKRT 198
Query: 215 VVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEM 274
V +RAVN +G P+P A+IS+ ++ GFPFG + KNIL N AYQ+WF RFTVTTF NEM
Sbjct: 199 VRIRAVNSKGEPIPKATISIEQRKLGFPFGCEVEKNILGNKAYQNWFTQRFTVTTFANEM 258
Query: 275 KWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAV 334
KWY+ E +GK++Y ADAML F ++ G+AVRGHNI W+DP+YQP WV++LS + L +AV
Sbjct: 259 KWYSTEVVRGKEDYSTADAMLRFFKQHGVAVRGHNILWNDPKYQPKWVNALSGNDLYNAV 318
Query: 335 EKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEY 394
++RV S+VSRYKGQL GWDVVNENLHFS+FE K+G S +F D TT+FMNEY
Sbjct: 319 KRRVFSVVSRYKGQLAGWDVVNENLHFSYFEDKMGPKASYNIFKMAQAFDPTTTMFMNEY 378
Query: 395 NTIEDSRDGLSTPPTYIEKIKEIQSVN--SQLPLGIGLESHFPNSPPNLPYMRASLDTLR 452
NT+E+S D S+ Y++K++EI+S+ + LGIGLESHF PN+PYMR++LDTL
Sbjct: 379 NTLEESSDSDSSLARYLQKLREIRSIRVCGNISLGIGLESHF--KTPNIPYMRSALDTLA 436
Query: 453 ATGLPIWITELDVASQPN-QALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNN 511
ATGLPIW+TE+DV + PN QA YFEQVLRE H+HP ++GIV W+ +SP GCYR+CLTD N
Sbjct: 437 ATGLPIWLTEVDVEAPPNVQAKYFEQVLREGHAHPQVKGIVTWSGYSPSGCYRMCLTDGN 496
Query: 512 FKNLPAGDVVDQLINEWG--RAEKSGTTDQNGYFEASLFHGDYEIEINHPIKKKSNFTHH 569
FKN+P GDVVD+L++EWG R + +G TD +GYFEASLFHGDY+++I HP+ S +H
Sbjct: 497 FKNVPTGDVVDKLLHEWGGFRRQTTGVTDADGYFEASLFHGDYDLKIAHPL-TNSKASHS 555
Query: 570 IKVLS 574
K+ S
Sbjct: 556 FKLTS 560
>At4g33840 putative protein
Length = 669
Score = 664 bits (1713), Expect = 0.0
Identities = 318/550 (57%), Positives = 406/550 (73%), Gaps = 15/550 (2%)
Query: 42 DYSANVECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKSLGNKFVVTHSRNQ 101
D++A +CL +P KPQYNGGII NP+L +G QGW+ FG+A ++ R+ GNKFVV RNQ
Sbjct: 119 DFAAKQQCLENPYKPQYNGGIIVNPDLQNGSQGWSQFGNAKVDFREFGGNKFVVATQRNQ 178
Query: 102 PHDSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKFGGAIFAEPNCWSMLKG 161
DS+SQK+YL KG+ Y+ SAW+QVS PV+AV K +K G++ AE CWSMLKG
Sbjct: 179 SSDSISQKVYLEKGILYTFSAWLQVSIGKSPVSAVFKKNGEYKHAGSVVAESKCWSMLKG 238
Query: 162 GLIADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVVVRAVN 221
GL D +G AEL+FES NT VEIWVD+VSLQPFT+++W SH E SI K RK V +R +N
Sbjct: 239 GLTVDESGPAELFFESENTMVEIWVDSVSLQPFTQEEWNSHHEQSIGKVRKGTVRIRVMN 298
Query: 222 EQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEY 281
+G +PNA+IS+ K+ G+PFG A+ NIL N AYQ+WF RFTVTTF NEMKWY+ E
Sbjct: 299 NKGETIPNATISIEQKKLGYPFGCAVENNILGNQAYQNWFTQRFTVTTFGNEMKWYSTER 358
Query: 282 AQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSI 341
+G+++Y ADAML F + GIAVRGHN+ WDDP+YQP WV+SLS + L +AV++RV S+
Sbjct: 359 IRGQEDYSTADAMLSFFKSHGIAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAVKRRVYSV 418
Query: 342 VSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSR 401
VSRYKGQL+GWDVVNENLHFSFFESK G S + H +D +T +FMNEYNT+E +
Sbjct: 419 VSRYKGQLLGWDVVNENLHFSFFESKFGPKASYNTYTMAHAVDPRTPMFMNEYNTLEQPK 478
Query: 402 DGLSTPPTYIEKIKEIQSVN--SQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIW 459
D S+P Y+ K++E+QS+ ++PL IGLESHF S PN+PYMR++LDT ATGLPIW
Sbjct: 479 DLTSSPARYLGKLRELQSIRVAGKIPLAIGLESHF--STPNIPYMRSALDTFGATGLPIW 536
Query: 460 ITELDVASQPN-QALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLPAG 518
+TE+DV + PN +A YFEQVLRE H+HP + G+VMWT +SP GCYR+CLTD NFKNLP G
Sbjct: 537 LTEIDVDAPPNVRANYFEQVLREGHAHPKVNGMVMWTGYSPSGCYRMCLTDGNFKNLPTG 596
Query: 519 DVVDQLINEWG--RAEKSGTTDQNGYFEASLFHGDYEIEINHPI---KKKSNFTHHIKVL 573
DVVD+L+ EWG R++ +G TD NG FEA LFHGDY++ I+HP+ K NFT L
Sbjct: 597 DVVDKLLREWGGLRSQTTGVTDANGLFEAPLFHGDYDLRISHPLTNSKASYNFT-----L 651
Query: 574 SKDEFKKTKQ 583
+ D+ KQ
Sbjct: 652 TSDDDSSQKQ 661
Score = 138 bits (347), Expect = 1e-32
Identities = 65/110 (59%), Positives = 86/110 (78%), Gaps = 5/110 (4%)
Query: 414 IKEIQSV--NSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITELDVASQPN- 470
+KE+QS+ + + L IGLESHF PN+PYMR++LD L ATGL IW+TE+DV + P+
Sbjct: 2 LKELQSIRISGYIRLAIGLESHFKT--PNIPYMRSALDILAATGLLIWLTEIDVEAPPSV 59
Query: 471 QALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLPAGDV 520
QA YFEQVLR+ H+HP ++G+V+W +SP GCYR+CLTD NF+NLP GDV
Sbjct: 60 QAKYFEQVLRDGHAHPQVKGMVVWGGYSPSGCYRMCLTDGNFRNLPTGDV 109
>At4g33830 putative protein
Length = 544
Score = 644 bits (1660), Expect = 0.0
Identities = 315/539 (58%), Positives = 398/539 (73%), Gaps = 9/539 (1%)
Query: 49 CLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKSLGNKFVVTHSRNQPHDSVSQ 108
CL P KPQYNGGII NP++ +G QGW+ F +A + R+ GNKFVV RNQ DSVSQ
Sbjct: 2 CLEIPYKPQYNGGIIVNPDMQNGSQGWSQFENAKVNFREFGGNKFVVATQRNQSSDSVSQ 61
Query: 109 KIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKFGGAIFAEPNCWSMLKGGLIADTT 168
K+YL KG+ Y+ SAW+QVS PV+AV K +K G++ AE CWSMLKGGL D +
Sbjct: 62 KVYLEKGILYTFSAWLQVSTGKAPVSAVFKKNGEYKHAGSVVAESKCWSMLKGGLTVDES 121
Query: 169 GVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVVVRAVNEQGHPLP 228
G AEL+ ES +T+VEIWVD+VSLQPFT+ +W +HQE SI+ RK V +R VN +G +P
Sbjct: 122 GPAELFVESEDTTVEIWVDSVSLQPFTQDEWNAHQEQSIDNSRKGPVRIRVVNNKGEKIP 181
Query: 229 NASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNY 288
NASI++ KR GFPFGSA+ +NIL N AYQ+WF RFTVTTFENEMKWY+ E +G +NY
Sbjct: 182 NASITIEQKRLGFPFGSAVAQNILGNQAYQNWFTQRFTVTTFENEMKWYSTESVRGIENY 241
Query: 289 FDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVSRYKGQ 348
ADAML F + GIAVRGHN+ WD P+YQ WV+SLS + L +AV++RV S+VSRYKGQ
Sbjct: 242 TVADAMLRFFNQHGIAVRGHNVVWDHPKYQSKWVTSLSRNDLYNAVKRRVFSVVSRYKGQ 301
Query: 349 LIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPP 408
L GWDVVNENLH SFFESK G N S +F H ID TT+FMNE+ T+ED D ++P
Sbjct: 302 LAGWDVVNENLHHSFFESKFGPNASNNIFAMAHAIDPSTTMFMNEFYTLEDPTDLKASPA 361
Query: 409 TYIEKIKEIQS--VNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITELDV- 465
Y+EK++E+QS V +PLGIGLESHF S PN+PYMR++LDTL ATGLPIW+TE+DV
Sbjct: 362 KYLEKLRELQSIRVRGNIPLGIGLESHF--STPNIPYMRSALDTLGATGLPIWLTEIDVK 419
Query: 466 ASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLPAGDVVDQLI 525
A +QA YFEQVLRE H+HP ++G+V WTA++P CY +CLTD NFKNLP GDVVD+LI
Sbjct: 420 APSSDQAKYFEQVLREGHAHPHVKGMVTWTAYAP-NCYHMCLTDGNFKNLPTGDVVDKLI 478
Query: 526 NEWG--RAEKSGTTDQNGYFEASLFHGDYEIEINHPIKKKSNFTHHIKVLSKDEFKKTK 582
EWG R++ + TD +G+FEASLFHGDY++ I+HP+ S+ +H+ + S D T+
Sbjct: 479 REWGGLRSQTTEVTDADGFFEASLFHGDYDLNISHPL-TNSSVSHNFTLTSDDSSLHTQ 536
>At4g33850 putative protein
Length = 371
Score = 426 bits (1095), Expect = e-119
Identities = 201/330 (60%), Positives = 249/330 (74%)
Query: 44 SANVECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKSLGNKFVVTHSRNQPH 103
S ++ECL +P KPQYNGGII NPEL +G QGW+ FG+A +E R+ N +VV RNQ
Sbjct: 39 STSLECLENPYKPQYNGGIIVNPELQNGSQGWSKFGNAKVEFREFGDNHYVVARQRNQSF 98
Query: 104 DSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKFGGAIFAEPNCWSMLKGGL 163
DSVSQ +YL K L Y+ SAW+QVSE PV A+ K +K G++ AE CWSMLKGGL
Sbjct: 99 DSVSQTVYLEKELLYTFSAWLQVSEGKAPVRAIFKKNGEYKNAGSVVAESKCWSMLKGGL 158
Query: 164 IADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVVVRAVNEQ 223
D +G AELYFES++T VEIWVD+VSLQPFT+K+W HQE SI K RK V +RAV+ +
Sbjct: 159 TVDESGPAELYFESDDTMVEIWVDSVSLQPFTQKEWNFHQEQSIYKARKGAVRIRAVDSE 218
Query: 224 GHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQ 283
G P+PNA+IS+ KR GFPFG + KNIL N AY++WF RFTVTTF NEMKWY+ E +
Sbjct: 219 GQPIPNATISIQQKRLGFPFGCEVEKNILGNQAYENWFTQRFTVTTFANEMKWYSTEVVR 278
Query: 284 GKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVS 343
GK++Y ADAML F ++ G+AVRGHN+ WDDP+YQP WV+SLS + L +AV++RV S+VS
Sbjct: 279 GKEDYSTADAMLRFFKQHGVAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAVKRRVFSVVS 338
Query: 344 RYKGQLIGWDVVNENLHFSFFESKLGQNFS 373
RYKGQL GWDVVNENLHFSFFE+K+G S
Sbjct: 339 RYKGQLAGWDVVNENLHFSFFENKMGPKAS 368
>At4g33820 putative glycosyl hydrolase family 10 protein
Length = 570
Score = 395 bits (1014), Expect = e-110
Identities = 207/541 (38%), Positives = 309/541 (56%), Gaps = 28/541 (5%)
Query: 29 YITTGFEAEALSYDYSANVECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKS 88
++ +G + S+ +S N EC+ P + G+ LQ + D E K
Sbjct: 19 HVDSGVSIDPFSHSHSLNTECVMKPPRSSETKGL---------LQFSRSLEDDSDEEWKI 69
Query: 89 LGNKFVVTHSRNQPHDSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKF--G 146
GN F+ ++Q+I L +G YS SAW+++ E VV T+ + G
Sbjct: 70 DGNGFI---------REMAQRIQLHQGNIYSFSAWVKLREGNDKKVGVVFRTENGRLVHG 120
Query: 147 GAIFAEPNCWSMLKGGLIADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELS 206
G + A CW++LKGG++ D +G +++FES N +I NV L+ F++++W+ Q+
Sbjct: 121 GEVRANQECWTLLKGGIVPDFSGPVDIFFESENRGAKISAHNVLLKQFSKEEWKLKQDQL 180
Query: 207 IEKDRKRKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFT 266
IEK RK KV E + ISL + F G +N IL + Y+ WFASRF
Sbjct: 181 IEKIRKSKVRFEVTYENKTAVKGVVISLKQTKSSFLLGCGMNFRILQSQGYRKWFASRFK 240
Query: 267 VTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSL- 325
+T+F NEMKWY E A+G++NY AD+ML FAE GI VRGH + WD+P+ QP+WV ++
Sbjct: 241 ITSFTNEMKWYATEKARGQENYTVADSMLKFAEDNGILVRGHTVLWDNPKMQPSWVKNIK 300
Query: 326 SPDQLNDAVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDG 385
P+ + + R+NS++ RYKG+L GWDVVNENLH+ +FE LG N S +N ID
Sbjct: 301 DPNDVMNVTLNRINSVMKRYKGKLTGWDVVNENLHWDYFEKMLGANASTSFYNLAFKIDP 360
Query: 386 QTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSV--NSQLPLGIGLESHFPNSPPNLPY 443
LF+NEYNTIE++++ +TP + ++EI + N + IG + HF + PNL Y
Sbjct: 361 DVRLFVNEYNTIENTKEFTATPIKVKKMMEEILAYPGNKNMKGAIGAQGHFGPTQPNLAY 420
Query: 444 MRASLDTLRATGLPIWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCY 503
+R++LDTL + GLPIW+TE+D+ PNQA Y E +LREA+SHP ++GI+++ G
Sbjct: 421 IRSALDTLGSLGLPIWLTEVDMPKCPNQAQYVEDILREAYSHPAVKGIIIFGGPEVSGFD 480
Query: 504 RICLTDNNFKNLPAGDVVDQLINEWGRAEKSGTTD-----QNGYFEASLFHGDYEIEINH 558
++ L D +F N GDV+D+L+ EW + T+ N E SL HG Y + ++H
Sbjct: 481 KLTLADKDFNNTQTGDVIDKLLKEWQQKSSEIQTNFTADSDNEEEEVSLLHGHYNVNVSH 540
Query: 559 P 559
P
Sbjct: 541 P 541
>At4g33810 beta-xylan endohydrolase -like protein
Length = 536
Score = 393 bits (1009), Expect = e-109
Identities = 196/483 (40%), Positives = 297/483 (60%), Gaps = 16/483 (3%)
Query: 106 VSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKF--GGAIFAEPNCWSMLKGGL 163
++Q+I L +G YS SAW+++ E VV T+ +F GG + A+ CW++LKGG+
Sbjct: 38 MTQRIQLHEGNIYSFSAWVKLREGNNKKVGVVFRTENGRFVHGGEVRAKKRCWTLLKGGI 97
Query: 164 IADTTGVAELYFE-------SNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVV 216
+ D +G +++FE S++ +I +VSL+ F++++W+ Q+ IEK RK KV
Sbjct: 98 VPDVSGSVDIFFEVQQLAIYSDDKEAKISASDVSLKQFSKQEWKLKQDQLIEKIRKSKVR 157
Query: 217 VRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKW 276
+ + A IS+ +P F G A+N IL + Y++WFASRF +T+F NEMKW
Sbjct: 158 FEVTYQNKTAVKGAVISIEQTKPSFLLGCAMNFRILQSEGYRNWFASRFKITSFTNEMKW 217
Query: 277 YTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSL-SPDQLNDAVE 335
YT E +G +NY AD+ML FAE+ GI VRGH + WDDP QP WV + P+ L +
Sbjct: 218 YTTEKERGHENYTAADSMLKFAEENGILVRGHTVLWDDPLMQPTWVPKIEDPNDLMNVTL 277
Query: 336 KRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYN 395
R+NS+++RYKG+L GWDVVNEN+H+ +FE LG N S+ +N +D T+F+NEYN
Sbjct: 278 NRINSVMTRYKGKLTGWDVVNENVHWDYFEKMLGANASSSFYNLAFKLDPDVTMFVNEYN 337
Query: 396 TIEDSRDGLSTPPTYIEKIKEIQSV--NSQLPLGIGLESHFPNSPPNLPYMRASLDTLRA 453
TIE+ + +TP EK++EI + N + IG + HF + PNL YMR++LDTL +
Sbjct: 338 TIENRVEVTATPVKVKEKMEEILAYPGNMNIKGAIGAQGHFRPTQPNLAYMRSALDTLGS 397
Query: 454 TGLPIWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFK 513
GLPIW+TE+D+ PNQ +Y E++LREA+SHP ++GI+++ G ++ L D F
Sbjct: 398 LGLPIWLTEVDMPKCPNQEVYIEEILREAYSHPAVKGIIIFAGPEVSGFDKLTLADKYFN 457
Query: 514 NLPAGDVVDQLINEWGRAEKSG----TTDQNGYFEASLFHGDYEIEINHPIKKKSNFTHH 569
N GDV+D+L+ EW ++ + T +N E SL HG Y + ++HP K + +
Sbjct: 458 NTATGDVIDKLLKEWQQSSEIPKIFMTDSENDEEEVSLLHGHYNVNVSHPWMKNMSTSFS 517
Query: 570 IKV 572
++V
Sbjct: 518 LEV 520
>At4g38650 putative protein
Length = 433
Score = 361 bits (926), Expect = e-100
Identities = 179/406 (44%), Positives = 245/406 (60%), Gaps = 13/406 (3%)
Query: 162 GLIADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIE--------KDRKR 213
GL + L ++ +++ V + SLQPFT++QWR++Q+ I RKR
Sbjct: 4 GLSSKEDSFLILLTSEDDGKIQLQVTSASLQPFTQEQWRNNQDYFINTVIHTLISNARKR 63
Query: 214 KVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENE 273
V + E G + A +++ F GSAI+K IL N YQ+WF RF T FENE
Sbjct: 64 AVTIHVSKENGESVEGAEVTVEQISKDFSIGSAISKTILGNIPYQEWFVKRFDATVFENE 123
Query: 274 MKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDA 333
+KWY E QGK NY AD M+ F I RGHNIFW+DP+Y P+WV +L+ + L A
Sbjct: 124 LKWYATEPDQGKLNYTLADKMMNFVRANRIIARGHNIFWEDPKYNPDWVRNLTGEDLRSA 183
Query: 334 VEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNE 393
V +R+ S+++RY+G+ + WDV NE LHF F+E++LG+N S F ID TLF N+
Sbjct: 184 VNRRIKSLMTRYRGEFVHWDVSNEMLHFDFYETRLGKNASYGFFAAAREIDSLATLFFND 243
Query: 394 YNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRA 453
+N +E D ST YI +++E+Q + GIGLE HF + PN+ MRA LD L
Sbjct: 244 FNVVETCSDEKSTVDEYIARVRELQRYDGVRMDGIGLEGHF--TTPNVALMRAILDKLAT 301
Query: 454 TGLPIWITELDVAS---QPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDN 510
LPIW+TE+D++S +QA+Y EQVLRE SHP + GI++WTA P GCY++CLTD+
Sbjct: 302 LQLPIWLTEIDISSSLDHRSQAIYLEQVLREGFSHPSVNGIMLWTALHPNGCYQMCLTDD 361
Query: 511 NFKNLPAGDVVDQLINEWGRAEKSGTTDQNGYFEASLFHGDYEIEI 556
F+NLPAGDVVDQ + EW E TTD +G F F G+Y + I
Sbjct: 362 KFRNLPAGDVVDQKLLEWKTGEVKATTDDHGSFSFFGFLGEYRVGI 407
>At2g14690 1,4-beta-xylan endohydrolase
Length = 552
Score = 360 bits (925), Expect = e-100
Identities = 190/522 (36%), Positives = 299/522 (56%), Gaps = 41/522 (7%)
Query: 105 SVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFK--FGGAIFAEPNCWSMLKGG 162
+V + + LR+G Y SAW+++ E+ + + K + FGG + A+ CWS+LKGG
Sbjct: 28 NVVEGVDLREGNIYITSAWVKLRNESQRKVGMTFSEKNGRNVFGGEVMAKRGCWSLLKGG 87
Query: 163 LIADTTGVAELYFE-------------SNNTSV-----------EIWVDNVSLQPFTEKQ 198
+ AD +G +++FE S N + EI V NV +Q F + Q
Sbjct: 88 ITADFSGPIDIFFEVKELIVCSFVEISSTNVGIYTKQSDGLAGLEISVQNVRMQRFHKTQ 147
Query: 199 WRSHQELSIEKDRKRKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQ 258
WR Q+ IEK RK KV + + L + IS+ +P F G A+N IL +++Y+
Sbjct: 148 WRLQQDQVIEKIRKNKVRFQMSFKNKSALEGSVISIEQIKPSFLLGCAMNYRILESDSYR 207
Query: 259 DWFASRFTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQ 318
+WF SRF +T+F NEMKWY E +G++NY AD+M+ AE+ I V+GH + WDD +Q
Sbjct: 208 EWFVSRFRLTSFTNEMKWYATEAVRGQENYKIADSMMQLAEENAILVKGHTVLWDDKYWQ 267
Query: 319 PNWVSSLS-PDQLNDAVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMF 377
PNWV +++ P+ L + R+NS++ RYKG+LIGWDV+NEN+HF++FE+ LG N SA ++
Sbjct: 268 PNWVKTITDPEDLKNVTLNRMNSVMKRYKGRLIGWDVMNENVHFNYFENMLGGNASAIVY 327
Query: 378 NEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSV--NSQLPLGIGLESHFP 435
+ +D LF+NE+NT+E +D + +P ++K++EI S N+ + GIG + HF
Sbjct: 328 SLASKLDPDIPLFLNEFNTVEYDKDRVVSPVNVVKKMQEIVSFPGNNNIKGGIGAQGHFA 387
Query: 436 NSPPNLPYMRASLDTLRATGLPIWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWT 495
PNL YMR +LDTL + P+W+TE+D+ P+Q Y E +LREA+SHP ++ I+++
Sbjct: 388 PVQPNLAYMRYALDTLGSLSFPVWLTEVDMFKCPDQVKYMEDILREAYSHPAVKAIILYG 447
Query: 496 AWSPQGCYRICLTDNNFKNLPAGDVVDQLINEWGR------AEKSGTTDQNG------YF 543
G ++ L D +FKN AGD++D+L+ EW + + D+ G
Sbjct: 448 GPEVSGFDKLTLADKDFKNTQAGDLIDKLLQEWKQEPVEIPIQHHEHNDEEGGRIIGFSP 507
Query: 544 EASLFHGDYEIEINHPIKKKSNFTHHIKVLSKDEFKKTKQFI 585
E SL HG Y + + +P K + ++V + + Q +
Sbjct: 508 EISLLHGHYRVTVTNPSMKNLSTRFSVEVTKESGHLQEVQLV 549
>At1g58370 unknown protein
Length = 917
Score = 238 bits (608), Expect = 5e-63
Identities = 160/540 (29%), Positives = 256/540 (46%), Gaps = 51/540 (9%)
Query: 54 QKPQYNGGIIKNPELNDGL-QGWTTFGDAIIE------------HRKSLG------NKFV 94
+ P + I+ N L+D GW + G+ + R SLG +++
Sbjct: 362 ENPAFGVNILTNSHLSDDTTNGWFSLGNCTLSVAEGSPRILPPMARDSLGAHERLSGRYI 421
Query: 95 VTHSRNQPHDSVSQKIY--LRKGLHYSLSAWIQVSE---ETVPVTAVVKTTKGFKFGGAI 149
+ +R Q +Q I L+ L Y +S W++V V + + GG +
Sbjct: 422 LVTNRTQTWMGPAQMITDKLKLFLTYQISVWVKVGSGINSPQNVNVALGIDSQWVNGGQV 481
Query: 150 FAEPNCWSMLKGGL-IADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIE 208
+ W + G I A +Y + ++ +++ V + + P H + +
Sbjct: 482 EINDDRWHEIGGSFRIEKNPSKALVYVQGPSSGIDLMVAGLQIFPVDRLARIKHLKRQCD 541
Query: 209 KDRKRKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVT 268
K RKR V+++ AS+ + R FP G+ I+++ ++N + D+F F
Sbjct: 542 KIRKRDVILKFAGVDSSKFSGASVRVRQIRNSFPVGTCISRSNIDNEDFVDFFLKNFNWA 601
Query: 269 TFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPD 328
F NE+KWY E QGK NY DAD ML I RGH IFW+ W+ +++
Sbjct: 602 VFANELKWYWTEPEQGKLNYQDADDMLNLCSSNNIETRGHCIFWEVQATVQQWIQNMNQT 661
Query: 329 QLNDAVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTT 388
LN+AV+ R+ +++RYKG+ +DV NE LH SF++ KLG++ MF H +D T
Sbjct: 662 DLNNAVQNRLTDLLNRYKGKFKHYDVNNEMLHGSFYQDKLGKDIRVNMFKTAHQLDPSAT 721
Query: 389 LFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASL 448
LF+N+Y+ IED D S P Y E+I ++Q + + GIG++ H + P P + ++L
Sbjct: 722 LFVNDYH-IEDGCDPKSCPEKYTEQILDLQEKGAPVG-GIGIQGHIDS--PVGPIVCSAL 777
Query: 449 DTLRATGLPIWITELDVAS--QPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRIC 506
D L GLPIW TELDV+S + +A E ++ EA HP ++GI++W W
Sbjct: 778 DKLGILGLPIWFTELDVSSVNEHIRADDLEVMMWEAFGHPAVEGIMLWGFWE-----LFM 832
Query: 507 LTDNNFKNLPAGDVVDQLINEWGR----------AEKSGTTDQNGYFEASLFHGDYEIEI 556
DN+ GDV NE G+ + +G DQNG F + G+Y +E+
Sbjct: 833 SRDNSHLVNAEGDV-----NEAGKRFLAVKKDWLSHANGHIDQNGAFPFRGYSGNYAVEV 887
>At1g10050 putative xylan endohydrolase
Length = 1063
Score = 219 bits (558), Expect = 3e-57
Identities = 154/528 (29%), Positives = 255/528 (48%), Gaps = 41/528 (7%)
Query: 62 IIKNPELNDG-LQGWTTFGDAIIEH----------------RKSLG---NKFVVTHSRNQ 101
I+ N L+DG ++GW GD ++ RK+ G ++V+ +R+
Sbjct: 518 IVSNSHLSDGTIEGWFPLGDCHLKVGDGSPRILPPLARDSLRKTQGYLSGRYVLATNRSG 577
Query: 102 ----PHDSVSQKIYLRKGLHYSLSAWIQVSE--ETVP--VTAVVKTTKGFKFGGAIFAEP 153
P +++ K+ L + Y +SAW+++ T P V + + GG + +
Sbjct: 578 TWMGPAQTITDKVKLF--VTYQVSAWVKIGSGGRTSPQDVNIALSVDGNWVNGGKVEVDD 635
Query: 154 NCWSMLKGGL-IADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRK 212
W + G I L+ + + V++ V + + K S+ + RK
Sbjct: 636 GDWHEVVGSFRIEKEAKEVMLHVQGPSPGVDLMVAGLQIFAVDRKARLSYLRGQADVVRK 695
Query: 213 RKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFEN 272
R V ++ L A++ + R FP GS I+++ ++N + D+F + F F
Sbjct: 696 RNVCLKFSGLDPSELSGATVKIRQTRNSFPLGSCISRSNIDNEDFVDFFLNNFDWAVFGY 755
Query: 273 EMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLND 332
E+KWY E QG NY DA+ M+ F E+ I RGH IFW+ WV L+ +L
Sbjct: 756 ELKWYWTEPEQGNFNYRDANEMIEFCERYNIKTRGHCIFWEVESAIQPWVQQLTGSKLEA 815
Query: 333 AVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMN 392
AVE RV +++RY G+ +DV NE LH SF+ +L + A MF H +D TLF+N
Sbjct: 816 AVENRVTDLLTRYNGKFRHYDVNNEMLHGSFYRDRLDSDARANMFKTAHELDPLATLFLN 875
Query: 393 EYNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLR 452
EY+ IED D S+P YI+ + ++Q + + GIG++ H + ++ +R++LD L
Sbjct: 876 EYH-IEDGFDSRSSPEKYIKLVHKLQKKGAPVG-GIGIQGHITSPVGHI--VRSALDKLS 931
Query: 453 ATGLPIWITELDVASQPNQALY---FEQVLREAHSHPGIQGIVMWTAWSP-QGCYRICLT 508
GLPIW TELDV+S N+ + E +L EA +HP ++G+++W W L
Sbjct: 932 TLGLPIWFTELDVSS-TNEHIRGDDLEVMLWEAFAHPAVEGVMLWGFWELFMSREHSHLV 990
Query: 509 DNNFKNLPAGDVVDQLINEWGRAEKSGTTDQNGYFEASLFHGDYEIEI 556
+ + + AG ++ EW + G + G E +HG Y +E+
Sbjct: 991 NADGEVNEAGKRFLEIKREW-LSFVDGEIEDGGGLEFRGYHGSYTVEV 1037
Score = 39.7 bits (91), Expect = 0.005
Identities = 34/156 (21%), Positives = 65/156 (40%), Gaps = 13/156 (8%)
Query: 62 IIKNPELNDGLQGWTTFG--DAIIEHRKSLGNKFVVTHSRNQPHDSVSQKI--YLRKGLH 117
IIKN + +DGL W T G ++ V ++R++ + Q I + G
Sbjct: 184 IIKNHDFSDGLYSWNTNGCDSFVVSSNDCNLESNAVVNNRSETWQGLEQDITDNVSPGFS 243
Query: 118 YSLSAWIQVSEETVPVTAVVKTTK--------GFKFGGAIFAEPNCWSMLKGGL-IADTT 168
Y +SA + VS + V+ T K F+ G +A + W L+G ++
Sbjct: 244 YKVSASVSVSGPVLGSAQVLATLKLEHKSSATEFQLIGKTYASKDIWKTLEGTFEVSGRP 303
Query: 169 GVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQE 204
+ E +++ V +V++ ++ Q+ +E
Sbjct: 304 DRVVFFLEGPPPGIDLLVKSVTIHCESDNQFERSRE 339
>At4g08160 xylan endohydrolase like proteinchimeric
Length = 520
Score = 206 bits (523), Expect = 4e-53
Identities = 141/492 (28%), Positives = 236/492 (47%), Gaps = 26/492 (5%)
Query: 87 KSLGNKFVVTHSRNQ----PHDSVSQKIYLRKGLHYSLSAWIQV-------SEETVPVTA 135
K LG ++V +R Q P ++ KI L L Y +SAW+++ S V
Sbjct: 10 KPLGGNYIVVTNRTQTWMGPAQMITDKIKLF--LTYQISAWVKLGVGVSGSSMSPQNVNI 67
Query: 136 VVKTTKGFKFGGAI-FAEPNCWSMLKGGL-IADTTGVAELYFESNNTSVEIWVDNVSLQP 193
+ + GG + + W + G + +Y + +++ + + + P
Sbjct: 68 ALSVDNQWVNGGQVEVTVGDTWHEIAGSFRLEKQPQNVMVYVQGPGAGIDLMIAALQIFP 127
Query: 194 FTEKQWRSHQELSIEKDRKRKVVVRAV---NEQGHPLPNASISLTMKRPGFPFGSAINKN 250
++ + +++ RKR +V++ +++ L + + FP G+ IN+
Sbjct: 128 VDRRERVRCLKRQVDEVRKRDIVLKFSGLNDDESFDLFPYIVKVKQTYNSFPVGTCINRT 187
Query: 251 ILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNI 310
++N + D+F F F NE+KWY E +GK NY DAD ML I VRGH I
Sbjct: 188 DIDNEDFVDFFTKNFNWAVFGNELKWYATEAERGKVNYQDADDMLDLCIGNNINVRGHCI 247
Query: 311 FWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQ 370
FW+ WV L+ L +AV+KR+ +++RYKG+ +DV NE LH SF++ +LG+
Sbjct: 248 FWEVESTVQPWVRQLNKTDLMNAVQKRLTDLLTRYKGKFKHYDVNNEMLHGSFYQDRLGK 307
Query: 371 NFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGL 430
A MFN H +D LF+N+Y+ +ED D S+P YI+ + ++++ + + GIG+
Sbjct: 308 GVRALMFNIAHKLDPSPLLFVNDYH-VEDGDDPRSSPEKYIKLVLDLEAQGATVG-GIGI 365
Query: 431 ESHFPNSPPNLPYMRASLDTLRATGLPIWITELDVASQPN--QALYFEQVLREAHSHPGI 488
+ H + P + ++LD L G PIW TELDV+S + E +L EA +HP +
Sbjct: 366 QGHIDS--PVGAIVCSALDMLSVLGRPIWFTELDVSSSNEYVRGEDLEVMLWEAFAHPSV 423
Query: 489 QGIVMWTAWS-PQGCYRICLTDNNFKNLPAGDVVDQLINEWGRAEKSGTTDQNGYFEASL 547
+GI++W W L + + AG ++ EW + G + F
Sbjct: 424 EGIMLWGFWELSMSRENANLVEGEGEVNEAGKRFLEVKQEW-LSHAYGIINDESEFTFRG 482
Query: 548 FHGDYEIEINHP 559
+HG Y +EI P
Sbjct: 483 YHGTYAVEICTP 494
>At4g38300 unknown protein
Length = 181
Score = 174 bits (440), Expect = 2e-43
Identities = 77/161 (47%), Positives = 103/161 (63%)
Query: 211 RKRKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTF 270
RKR V + E G + A +++ FP GSAI+K IL N YQ+WF RF T F
Sbjct: 12 RKRAVTIHVSKENGESVEGAEVTVEQISKDFPIGSAISKTILGNIPYQEWFVKRFDATVF 71
Query: 271 ENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQL 330
ENE+KWY E QGK NY AD M+ I RGHNIFW+DP+Y P+WV +L+ + L
Sbjct: 72 ENELKWYATESDQGKLNYTLADKMMNLVRANRIIARGHNIFWEDPKYNPDWVRNLTGEDL 131
Query: 331 NDAVEKRVNSIVSRYKGQLIGWDVVNENLHFSFFESKLGQN 371
AV +R+ S+++RY+G+ + WDV NE LHF F+ES+LG+N
Sbjct: 132 RSAVNRRIKSLMTRYRGEFVHWDVSNEMLHFDFYESRLGKN 172
>At1g26820 ribonuclease RNS3
Length = 222
Score = 31.6 bits (70), Expect = 1.3
Identities = 28/102 (27%), Positives = 39/102 (37%), Gaps = 33/102 (32%)
Query: 487 GIQGIVMWTAWSPQGCYRICLTDNNFKNLPAGDVVDQLINEWGRA--------------- 531
GI G+ W + G + C D+ F +L D++ L EW
Sbjct: 55 GIHGL--WPNYKTGGWPQNCNPDSRFDDLRVSDLMSDLQREWPTLSCPSNDGMKFWTHEW 112
Query: 532 EKSGT-----TDQNGYFEASLFHGDYEIEINHPIKKKSNFTH 568
EK GT DQ+ YFEA L +K+K+N H
Sbjct: 113 EKHGTCAESELDQHDYFEAGL-----------KLKQKANLLH 143
>At3g22340 hypothetical protein
Length = 220
Score = 31.2 bits (69), Expect = 1.7
Identities = 23/93 (24%), Positives = 38/93 (40%), Gaps = 8/93 (8%)
Query: 94 VVTHSRNQPHDSVSQKIYLRKGLHYSLSAWIQVSEETVPVTAVVKTTKGFKFGGAIFAEP 153
VV + + P Q I LR +L+AW T P+T T + GG+ P
Sbjct: 15 VVRYLKGNP----GQGILLRADAELTLTAWCDADHATCPLTRRSLTAWFIQLGGS----P 66
Query: 154 NCWSMLKGGLIADTTGVAELYFESNNTSVEIWV 186
W K ++ ++ AE ++ S +W+
Sbjct: 67 ISWKTRKQDTVSRSSAEAEYRSMADTVSEILWL 99
>At1g02790 exopolygalacturonase (At1g02790)
Length = 422
Score = 31.2 bits (69), Expect = 1.7
Identities = 25/57 (43%), Positives = 31/57 (53%), Gaps = 8/57 (14%)
Query: 120 LSAWIQVSEETVPVTAVVKTTKG-FKFGGAIFAEPNCWSMLK----GGLIADTTGVA 171
L+ WIQV + VP T +V KG F G IFA P C S + G +IA T+G A
Sbjct: 72 LNTWIQVCDSPVPATLLV--PKGTFLAGPVIFAGP-CKSKVTVNVIGTIIATTSGYA 125
>At4g27210 putative protein
Length = 1318
Score = 30.8 bits (68), Expect = 2.2
Identities = 48/205 (23%), Positives = 77/205 (37%), Gaps = 44/205 (21%)
Query: 359 LHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQ 418
LH FE + R V D ++N+ I D + +P T EK+K
Sbjct: 43 LHMRLFELQ-------RRLQNVSKRDKTMDAYLNDLKNICDQLASVGSPVT--EKMKIFA 93
Query: 419 SVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITELDVASQPNQALYFEQV 478
++N G+G E P + S+DT QP +L E V
Sbjct: 94 ALN-----GLGREYE-----PIKTTIENSMDT-----------------QPGPSL--ENV 124
Query: 479 LREAHSHPG-IQGIVMWTAWSPQGCYRICLTDNNFKNL--PAGDVVDQLINEW---GRAE 532
+ + + +QG + T SP + I +D++++N DV DQ NEW A
Sbjct: 125 IPKLTGYDDRLQGYLEETTISPYVAFNITTSDDSYRNSYNRGKDVTDQHGNEWLPDSAAT 184
Query: 533 KSGTTDQNGYFEASLFHGDYEIEIN 557
T ++ +HG I ++
Sbjct: 185 AHVTNSPRSLQQSQPYHGTDAIMVD 209
>At5g49750 putative protein
Length = 349
Score = 29.3 bits (64), Expect = 6.4
Identities = 27/113 (23%), Positives = 49/113 (42%), Gaps = 10/113 (8%)
Query: 351 GWDVVNENLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTY 410
G D++ + HF F ++KL + ++FN N+ LF N T + LS T
Sbjct: 148 GLDMLTQTQHFHFGKNKLSGHIPEKLFNS--NMSLIHVLFNNNQFT-GKIPESLSLVTTL 204
Query: 411 IEKIKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITEL 463
+ + ++ +P + N+ +L + S +TL + +P WI L
Sbjct: 205 LVLRLDTNRLSGDIPPSL-------NNLTSLNQLDVSNNTLEFSLVPSWIVSL 250
>At1g18460 unknown protein
Length = 701
Score = 28.9 bits (63), Expect = 8.3
Identities = 15/55 (27%), Positives = 28/55 (50%), Gaps = 4/55 (7%)
Query: 380 VHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHF 434
+H + + +Y++ +D DG+STPP+ + + S+ S L ESH+
Sbjct: 81 IHELSVDSDTSSLDYSSGDDDSDGMSTPPSPLSQ----SSLRSWASLPANYESHW 131
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,974,576
Number of Sequences: 26719
Number of extensions: 627710
Number of successful extensions: 1368
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1313
Number of HSP's gapped (non-prelim): 21
length of query: 588
length of database: 11,318,596
effective HSP length: 105
effective length of query: 483
effective length of database: 8,513,101
effective search space: 4111827783
effective search space used: 4111827783
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146757.5


