

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.4 - phase: 0 /pseudo
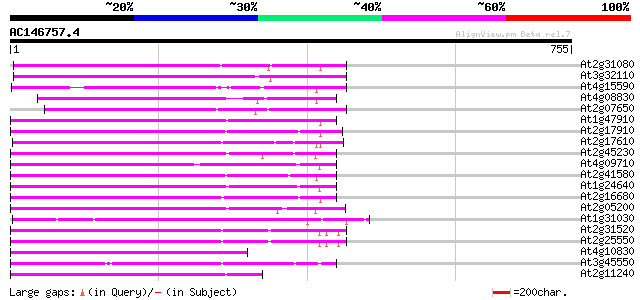
(755 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g31080 putative non-LTR retroelement reverse transcriptase 327 2e-89
At3g32110 non-LTR reverse transcriptase, putative 325 8e-89
At4g15590 reverse transcriptase like protein 273 2e-73
At4g08830 putative protein 253 4e-67
At2g07650 putative non-LTR retrolelement reverse transcriptase 248 7e-66
At1g47910 reverse transcriptase, putative 244 1e-64
At2g17910 putative non-LTR retroelement reverse transcriptase 234 1e-61
At2g17610 putative non-LTR retroelement reverse transcriptase 232 5e-61
At2g45230 putative non-LTR retroelement reverse transcriptase 227 2e-59
At4g09710 RNA-directed DNA polymerase -like protein 218 1e-56
At2g41580 putative non-LTR retroelement reverse transcriptase 217 2e-56
At1g24640 hypothetical protein 213 2e-55
At2g16680 putative non-LTR retroelement reverse transcriptase 209 6e-54
At2g05200 putative non-LTR retroelement reverse transcriptase 206 3e-53
At1g31030 F17F8.5 197 2e-50
At2g31520 putative non-LTR retroelement reverse transcriptase 196 4e-50
At2g25550 putative non-LTR retroelement reverse transcriptase 196 5e-50
At4g10830 putative protein 192 8e-49
At3g45550 putative protein 190 2e-48
At2g11240 pseudogene 187 2e-47
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 327 bits (837), Expect = 2e-89
Identities = 174/457 (38%), Positives = 262/457 (57%), Gaps = 12/457 (2%)
Query: 6 IMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQI 65
++WNGE+TD+F RG+RQGDPLSPYLFV+C++RL H+I V + W P+ G ++
Sbjct: 480 VLWNGERTDSFVPARGLRQGDPLSPYLFVLCLERLCHLIEASVGKREWKPIAVSCGGSKL 539
Query: 66 SHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLREDIL 125
SH+ FADDL+LFAEAS+ Q + L+ FC+ASGQK++ K+ ++FS NV ++ + I
Sbjct: 540 SHVCFADDLILFAEASVAQIRIIRRVLERFCEASGQKVSLEKSKIFFSHNVSREMEQLIS 599
Query: 126 HHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRITLAKSV 185
+G LG YLG + R + F ++ ++ ++L GWK + L+LAGRITL K+V
Sbjct: 600 EESGIGCTKELGKYLGMPILQKRMNKETFGEVLERVSARLAGWKGRSLSLAGRITLTKAV 659
Query: 186 ISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADGGLGFR 245
+S+IP + M +P + D +D+ R F+WG + + +K HL+SW C PKA+GG+G R
Sbjct: 660 LSSIPVHVMSAILLPVSTLDTLDRYSRTFLWGSTMEKKKQHLLSWRKICKPKAEGGIGLR 719
Query: 246 QTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKY--GRNNDLIASINAHPYDSPLWKAL-V 302
MN+A + K+ W L+++ E LW RV+R KY G D + + P S W+++ V
Sbjct: 720 SARDMNKALVAKVGWRLLQDKESLWARVVRKKYKVGGVQD-TSWLKPQPRWSSTWRSVAV 778
Query: 303 NIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMD---ITLSVRDVLSPSG 359
+ V W GDG FWLD+W+ E L+ +G + I ++ D P
Sbjct: 779 GLREVVVKGVGWVPGDGCTIRFWLDRWL-LQEPLVELGTDMIPEGERIKVAA-DYWLPGS 836
Query: 360 DWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSLQHQDC--- 416
WN++ L LP ++L++ + D I W GT FTV+SAYSL D
Sbjct: 837 GWNLEILGLYLPETVKRRLLSVVVQVFLGNGDEISWKGTQDGAFTVRSAYSLLQGDVGDR 896
Query: 417 PILEGDWKSIWKWHGPHRIQTFIWLDTHERILTNFQR 453
P + + IWK P R++ FIWL + I+TN +R
Sbjct: 897 PNMGSFFNRIWKLITPERVRVFIWLVSQNVIMTNVER 933
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 325 bits (832), Expect = 8e-89
Identities = 167/458 (36%), Positives = 265/458 (57%), Gaps = 14/458 (3%)
Query: 6 IMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQI 65
++WNGEKTD F RG+RQGDPLSPYLFV+C++RL H+I + AK W P++ + GP++
Sbjct: 1156 LLWNGEKTDAFKPLRGLRQGDPLSPYLFVLCIERLCHLIESSIAAKKWKPIKISQSGPRL 1215
Query: 66 SHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLREDIL 125
SH+ FADDL+LFAEASI+Q + L+ FC ASGQK++ K+ +YFSKNV L + I
Sbjct: 1216 SHICFADDLILFAEASIDQIRVLRGVLEKFCGASGQKVSLEKSKIYFSKNVLRDLGKRIS 1275
Query: 126 HHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRITLAKSV 185
+G LG YLG + R + F ++ + S+L GWK + L+ AGR+TL K+V
Sbjct: 1276 EESGMKATKDLGKYLGVPILQKRINKETFGEVIKRFSSRLAGWKGRMLSFAGRLTLTKAV 1335
Query: 186 ISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADGGLGFR 245
+++I + M K+P++ D +DK+ R F+WG + + +K HL++W CLP+ +GGLG R
Sbjct: 1336 LTSILVHTMSTIKLPQSTLDGLDKVSRAFLWGSTLEKKKQHLVAWTRVCLPRREGGLGIR 1395
Query: 246 QTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKY--GRNNDLIASINAHPYDSPLWKALVN 303
MN+A + K+ W ++ + LW +V+RSKY G +D ++ + S V
Sbjct: 1396 SATAMNKALIAKVGWRVLNDGSSLWAQVVRSKYKVGDVHDRNWTVAKSNWSSTWRSVGVG 1455
Query: 304 IWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITL-----SVRDVLSPS 358
+ + W IGDGR FW D+W+ S I + + + ++L + RD+
Sbjct: 1456 LRDVIWREQHWVIGDGRQIRFWTDRWL----SETPIADDSIVPLSLAQMLCTARDLWRDG 1511
Query: 359 GDWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYS-LQHQDCP 417
W++ + + N +LA+ + + D + WG T+ FTV+SA++ L + D P
Sbjct: 1512 TGWDMSQIAPFVTDNKRLDLLAVIVDSVTGAHDRLAWGMTSDGRFTVKSAFAMLTNDDSP 1571
Query: 418 --ILEGDWKSIWKWHGPHRIQTFIWLDTHERILTNFQR 453
+ + +WK P R++ F+WL ++ I+TN +R
Sbjct: 1572 RQDMSSLYGRVWKVQAPERVRVFLWLVVNQAIMTNSER 1609
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 273 bits (699), Expect = 2e-73
Identities = 157/454 (34%), Positives = 241/454 (52%), Gaps = 34/454 (7%)
Query: 3 EYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYG 62
E ++WNGEKTD+F RG+RQGDP+SPYLFV+C++RL H I V W + + G
Sbjct: 429 EMSLLWNGEKTDSFTPERGLRQGDPISPYLFVLCIERLCHQIETAVGRGDWKSISISQGG 488
Query: 63 PQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLRE 122
P++SH+ FADDL+LFAEAS+ QK++ K+ ++FS NV L
Sbjct: 489 PKVSHVCFADDLILFAEASV-----------------AQKVSLEKSKIFFSNNVSRDLEG 531
Query: 123 DILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRITLA 182
I TG LG YLG + R + F ++ ++ S+L+GWK + L+LAGRITL
Sbjct: 532 LITAETGIGSTRELGKYLGMPVLQKRINKDTFGEVLERVSSRLSGWKSRSLSLAGRITLT 591
Query: 183 KSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADGGL 242
K+V+ +IP + M +P ++ +++DK+ R F+WG + + RK HL+SW C PKA GGL
Sbjct: 592 KAVLMSIPIHTMSSILLPASLLEQLDKVSRNFLWGSTVEKRKQHLLSWKKVCRPKAAGGL 651
Query: 243 GFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSPLWKALV 302
G R + MN A L K+ W L+ + LW RVLR KY + D+ +DS W
Sbjct: 652 GLRASKDMNRALLAKVGWRLLNDKVSLWARVLRRKY-KVTDV--------HDSS-WLVPK 701
Query: 303 NIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVLSPSGDWN 362
W+ + + +G + L + + + + + ++ V + + W+
Sbjct: 702 ATWSSTWRSIGVGLREGVAKGWILHEPLCTRATCL----LSPEELNARVEEFWTEGVGWD 757
Query: 363 IDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSL---QHQDCPIL 419
+ L LP + +++ A+ I D I W GT+ FTV SAY L + + P +
Sbjct: 758 MVKLGQCLPRSVTDRLHAVVIKGVLGLRDRISWQGTSDGDFTVGSAYVLLTQEEESKPCM 817
Query: 420 EGDWKSIWKWHGPHRIQTFIWLDTHERILTNFQR 453
E +K IW P R++ F+WL + I+TN +R
Sbjct: 818 ESFFKRIWGVIAPERVRVFLWLVGQQVIMTNVER 851
>At4g08830 putative protein
Length = 947
Score = 253 bits (645), Expect = 4e-67
Identities = 139/410 (33%), Positives = 220/410 (52%), Gaps = 32/410 (7%)
Query: 38 DRLSHMIADQVEAKYWIPMRAGRYGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQ 97
+RL HMI V K W + + GP+ISH+ FADDL+LFAEAS+ Q + L+ FC
Sbjct: 326 ERLCHMIDRAVAVKEWKSIGLSQGGPKISHICFADDLILFAEASVSQIRVIRRILETFCI 385
Query: 98 ASGQKINRNKTCVYFSKNVDTQLREDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHI 157
ASGQK++ +K+ ++FSKNV L + I +G LG YLG + R + F +
Sbjct: 386 ASGQKVSLDKSKIFFSKNVSRDLEKLISKESGIKSTRELGKYLGMPILQRRINKDTFGEV 445
Query: 158 VNKIQSKLNGWKQQCLTLAGRITLAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWG 217
+ ++ S+L GWK + L+ AGR+TL KSV+S IP + M +P++ + +DK+ R F+ G
Sbjct: 446 LERVSSRLAGWKGRSLSFAGRLTLTKSVLSLIPIHTMSTISLPQSTLEGLDKLARVFLLG 505
Query: 218 DSDQGRKAHLISWDVCCLPKADGGLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSK 277
S + +K HL++WD CLPK++GGLG R + MN+A + K+ W LI + LW R+LRSK
Sbjct: 506 SSAEKKKLHLVAWDRVCLPKSEGGLGIRTSKCMNKALVSKVGWRLINDRYSLWARILRSK 565
Query: 278 YGRNNDLIASINAHPYDSPLWKALVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPN----N 333
Y + S + W +G+GR+ FW D W+ + N
Sbjct: 566 YRVGLREVVSRGSR----------------------WVVGNGRDILFWSDNWLSHEALIN 603
Query: 334 ESLMSIGNQTYMDITLSVRDVLSPSGDWNIDFLINNLPTNTVNQILALSIPNDDDGPDTI 393
+++ I N + L V+D+ + W +D + + +T ++ A+ + + D +
Sbjct: 604 RAVIEIPNS---EKELRVKDLWANGLGWKLDKIEPYISYHTRLELAAVVVDSVTGARDRL 660
Query: 394 GWGGTNTHHFTVQSAYSL---QHQDCPILEGDWKSIWKWHGPHRIQTFIW 440
WG + FTV+SAY L H P + + +W+ R++TF+W
Sbjct: 661 SWGYSADGVFTVKSAYRLLTEDHDPRPNMAAFFDRLWRVVALERVKTFLW 710
>At2g07650 putative non-LTR retrolelement reverse transcriptase
Length = 732
Score = 248 bits (634), Expect = 7e-66
Identities = 141/415 (33%), Positives = 223/415 (52%), Gaps = 12/415 (2%)
Query: 48 VEAKYWIPMRAGRYGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNK 107
+ K W P+ + GP++SH+ FADDL+LFAEAS+ Q + L+ FC ASGQK++ K
Sbjct: 184 IARKDWKPISLSQGGPKLSHICFADDLILFAEASVAQIRVIRRVLERFCVASGQKVSLEK 243
Query: 108 TCVYFSKNVDTQLREDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNG 167
+ ++FS+NV L + I +G + LG YLG + R + F I+ K+ ++L G
Sbjct: 244 SKIFFSENVSRDLGKLISDESGISSTRELGKYLGMPVLQRRINKDTFGDILEKLTTRLAG 303
Query: 168 WKQQCLTLAGRITLAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHL 227
WK + L+LAGR+TL K+V+S+IP + M +PK+ D +DK+ R F+WG S RK HL
Sbjct: 304 WKGRFLSLAGRVTLTKAVLSSIPVHTMSTIALPKSTLDGLDKVSRSFLWGSSVTQRKQHL 363
Query: 228 ISWDVCCLPKADGGLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLI-- 285
ISW C P+++GGLG R+ MN+A L K+ W LI++ LW R++R Y R D+
Sbjct: 364 ISWKRVCKPRSEGGLGIRKAQDMNKALLSKVGWRLIQDYHSLWARIMRCNY-RVQDVRDG 422
Query: 286 ASINAHPYDSPLWKAL-VNIWNDFKGHVVWNIGDGRNTNFWLDKW---VPNNESLMSIGN 341
A S W+++ + + + W IGDGR FW+DKW +P E L+
Sbjct: 423 AWTKVRSVCSSTWRSVALGMREVVIPGLSWVIGDGREILFWMDKWLTNIPLAELLVQEPP 482
Query: 342 QTYMDITLSVRDVLSPSGDWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTH 401
+ + RD+ W++ + + + Q+ + + + D I WG +
Sbjct: 483 AGWKG--MRARDLRRNGIGWDMATIAPYISSYNRLQLQSFVLDDITGARDRISWGESQDG 540
Query: 402 HFTVQSAYS-LQHQDCP--ILEGDWKSIWKWHGPHRIQTFIWLDTHERILTNFQR 453
F V + YS L + P ++ + +W P R++ F+WL + I+TN +R
Sbjct: 541 RFKVSTTYSFLTRDEAPRQDMKRFFDRVWSVTAPERVRLFLWLVAQQAIMTNQER 595
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 244 bits (624), Expect = 1e-64
Identities = 142/448 (31%), Positives = 228/448 (50%), Gaps = 11/448 (2%)
Query: 2 IEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRY 61
++Y ++ NG+ RG+RQGDPLSPYLF++C + L I ++
Sbjct: 397 VQYKVLINGQPKGLIIPERGLRQGDPLSPYLFILCTEVLIANIRKAERQNLITGIKVATP 456
Query: 62 GPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLR 121
P +SHLLFADD L F +A+ EQ +L L + SGQ+IN +K+ + F V+ ++
Sbjct: 457 SPAVSHLLFADDSLFFCKANKEQCGIILEILKQYESVSGQQINFSKSSIQFGHKVEDSIK 516
Query: 122 EDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRITL 181
DI G + + +G YLG + G S F+ + +++QS++NGW + L+ G+ +
Sbjct: 517 ADIKLILGIHNLGGMGSYLGLPESLGGSKTKVFSFVRDRLQSRINGWSAKFLSKGGKEVM 576
Query: 182 AKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADGG 241
KSV +T+P Y M ++PK I ++ F W + R H ++WD C K+DGG
Sbjct: 577 IKSVAATLPRYVMSCFRLPKAITSKLTSAVAKFWWSSNGDSRGMHWMAWDKLCSSKSDGG 636
Query: 242 LGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSPLWKAL 301
LGFR N A L K LW LI P+ L+ +V + +Y R ++ + SI ++ S W+++
Sbjct: 637 LGFRNVDDFNSALLAKQLWRLITAPDSLFAKVFKGRYFRKSNPLDSIKSYS-PSYGWRSM 695
Query: 302 VNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVL-SPSGD 360
++ + ++ +G G + + W D W+P + + +D +L V+ ++ S S
Sbjct: 696 ISARSLVYKGLIKRVGSGASISVWNDPWIPAQFPRPAKYGGSIVDPSLKVKSLIDSRSNF 755
Query: 361 WNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSLQHQDC---- 416
WNID L V I AL I N + DT+GW T ++TV+S Y D
Sbjct: 756 WNIDLLKELFDPEDVPLISALPIGN-PNMEDTLGWHFTKAGNYTVKSGYHTARLDLNEGT 814
Query: 417 ----PILEGDWKSIWKWHGPHRIQTFIW 440
P L IWK P +++ F+W
Sbjct: 815 TLIGPDLTTLKAYIWKVQCPPKLRHFLW 842
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 234 bits (597), Expect = 1e-61
Identities = 137/459 (29%), Positives = 226/459 (48%), Gaps = 15/459 (3%)
Query: 1 NIEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGR 60
++ Y ++ NG+ TRGIRQGDPLSP LFV+C + L H++ +A ++
Sbjct: 590 SVSYSVLINGQPYGHIIPTRGIRQGDPLSPALFVLCTEALIHILNKAEQAGKITGIQFQD 649
Query: 61 YGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQL 120
++HLLFADD LL +A+ ++ ++ CL + Q SGQ IN NK+ + F KNVD Q+
Sbjct: 650 KKVSVNHLLFADDTLLMCKATKQECEELMQCLSQYGQLSGQMINLNKSAITFGKNVDIQI 709
Query: 121 REDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRIT 180
++ I +G + G YLG S R F I K+QS+L GW + L+ G+
Sbjct: 710 KDWIKSRSGISLEGGTGKYLGLPECLSGSKRDLFGFIKEKLQSRLTGWYAKTLSQGGKEV 769
Query: 181 LAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADG 240
L KS+ +P Y M K+PK +C ++ + F W Q RK H +SW LPK G
Sbjct: 770 LLKSIALALPVYVMSCFKLPKNLCQKLTTVMMDFWWNSMQQKRKIHWLSWQRLTLPKDQG 829
Query: 241 GLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSPLWKA 300
G GF+ N+A L K W +++ L+ RV +S+Y N+D +++ S W++
Sbjct: 830 GFGFKDLQCFNQALLAKQAWRVLQEKGSLFSRVFQSRYFSNSDFLSATRG-SRPSYAWRS 888
Query: 301 LVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVLSP-SG 359
++ + IG+G+ T W DKW+ + + + + ++++ L V ++ P S
Sbjct: 889 ILFGRELLMQGLRTVIGNGQKTFVWTDKWLHDGSNRRPLNRRRFINVDLKVSQLIDPTSR 948
Query: 360 DWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSLQHQDC--- 416
+WN++ L + P V IL + D+ W ++ ++V++ Y +
Sbjct: 949 NWNLNMLRDLFPWKDVEIILKQRPLFFKE--DSFCWLHSHNGLYSVKTGYEFLSKQVHHR 1006
Query: 417 --------PILEGDWKSIWKWHGPHRIQTFIWLDTHERI 447
P + + IW H +I+ F+W H I
Sbjct: 1007 LYQEAKVKPSVNSLFDKIWNLHTAPKIRIFLWKALHGAI 1045
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 232 bits (592), Expect = 5e-61
Identities = 141/458 (30%), Positives = 221/458 (47%), Gaps = 15/458 (3%)
Query: 4 YIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGP 63
Y I+ NG+ RGIRQGDP+SPYL+++C + LS +I ++AK +A R GP
Sbjct: 70 YSILINGQPVRRIIPKRGIRQGDPISPYLYLLCTEGLSALIQASIKAKQLHGFKASRNGP 129
Query: 64 QISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLRED 123
ISHLLFA D L+F +A++E+ +++ L ++ +ASGQ +N K+ + F K +D + E
Sbjct: 130 AISHLLFAHDSLVFCKATLEECMTLVNVLKLYEKASGQAVNFQKSAILFGKGLDFRTSEQ 189
Query: 124 ILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRITLAK 183
+ G + G YLG GR+ F+ I + K++ W + L+ AG+ L K
Sbjct: 190 LSQLLGIYKTEGFGRYLGLPEFVGRNKTNAFSFIAQTMDQKMDNWYNKLLSPAGKEVLIK 249
Query: 184 SVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADGGLG 243
S+++ IP Y M +P + +I R F W ++ K ++W PK GGL
Sbjct: 250 SIVTAIPTYSMSCFLLPMRLIHQITSAMRWFWWSNTKVKHKIPWVAWSKLNDPKKMGGLA 309
Query: 244 FRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSPLWKALVN 303
R N A L K W +++ P L RV ++KY L+ A S WK++++
Sbjct: 310 IRDLKDFNIALLAKQSWRILQQPFSLMARVFKAKYFPKERLL-DAKATSQSSYAWKSILH 368
Query: 304 IWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVLSPSGDWNI 363
+ + G+G N W D W+P N +G + L V D+L G WN
Sbjct: 369 GTKLISRGLKYIAGNGNNIQLWKDNWLPLNPPRPPVGTCDSIYSQLKVSDLLI-EGRWNE 427
Query: 364 DFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSL-------QHQDC 416
D L + N + I A+ P+ D I W T+ +++V+S Y L QH
Sbjct: 428 DLLCKLIHQNDIPHIRAIR-PSITGANDAITWIYTHDGNYSVKSGYHLLRKLSQQQHASL 486
Query: 417 P-----ILEGDWKSIWKWHGPHRIQTFIWLDTHERILT 449
P + + +IWK + P +I+ F W H + T
Sbjct: 487 PSPNEVSAQTVFTNIWKQNAPPKIKHFWWRSAHNALPT 524
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 227 bits (579), Expect = 2e-59
Identities = 140/460 (30%), Positives = 227/460 (48%), Gaps = 23/460 (5%)
Query: 1 NIEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGR 60
++ Y ++ NG +RG+RQGDPLSPYLFVIC + L M+ + ++ R
Sbjct: 612 SVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLFVICTEMLVKMLQSAEQKNQITGLKVAR 671
Query: 61 YGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQL 120
P ISHLLFADD + + + + E ++ ++ + ASGQ++N K+ +YF K++ +
Sbjct: 672 GAPPISHLLFADDSMFYCKVNDEALGQIIRIIEEYSLASGQRVNYLKSSIYFGKHISEER 731
Query: 121 REDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRIT 180
R + G + G+YLG + S +++ +++ K+ GW+ L+ G+
Sbjct: 732 RCLVKRKLGIEREGGEGVYLGLPESFQGSKVATLSYLKDRLGKKVLGWQSNFLSPGGKEI 791
Query: 181 LAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADG 240
L K+V +P Y M KIPKTIC +I+ + F W + +GR H +W PKA G
Sbjct: 792 LLKAVAMALPTYTMSCFKIPKTICQQIESVMAEFWWKNKKEGRGLHWKAWCHLSRPKAVG 851
Query: 241 GLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKY-GRNNDLIASINAHPYDSPLWK 299
GLGF++ N A L K LW +I + L +V +S+Y +++ L A + + P S WK
Sbjct: 852 GLGFKEIEAFNIALLGKQLWRMITEKDSLMAKVFKSRYFSKSDPLNAPLGSRP--SFAWK 909
Query: 300 ALVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMS-------IGNQTYMDITLSVR 352
++ K + IG+G N W D W+ + + + +Q + V+
Sbjct: 910 SIYEAQVLIKQGIRAVIGNGETINVWTDPWIGAKPAKAAQAVKRSHLVSQYAANSIHVVK 969
Query: 353 DVLSPSG-DWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAY-- 409
D+L P G DWN + + P NT ILAL P + D W + + H++V+S Y
Sbjct: 970 DLLLPDGRDWNWNLVSLLFPDNTQENILALR-PGGKETRDRFTWEYSRSGHYSVKSGYWV 1028
Query: 410 ---------SLQHQDCPILEGDWKSIWKWHGPHRIQTFIW 440
+ Q P L+ ++ IWK P +I F+W
Sbjct: 1029 MTEIINQRNNPQEVLQPSLDPIFQQIWKLDVPPKIHHFLW 1068
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 218 bits (554), Expect = 1e-56
Identities = 129/446 (28%), Positives = 221/446 (48%), Gaps = 18/446 (4%)
Query: 2 IEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRY 61
+ Y + NG + +RG+RQGDPLSPYLF++C + LS + E + +R R
Sbjct: 554 VSYSFLINGSPQGSVVPSRGLRQGDPLSPYLFILCTEVLSGLCRKAQEKGVMVGIRVARG 613
Query: 62 GPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLR 121
PQ++HLLFADD + F + + + + L + ASGQ IN K+ + FS ++
Sbjct: 614 SPQVNHLLFADDTMFFCKTNPTCCGALSNILKKYELASGQSINLAKSAITFSSKTPQDIK 673
Query: 122 EDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRITL 181
+ + +G YLG GR R F+ IV++I+ + + W + L+ AG+ L
Sbjct: 674 RRVKLSLRIDNEGGIGKYLGLPEHFGRRKRDIFSSIVDRIRQRSHSWSIRFLSSAGKQIL 733
Query: 182 AKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADGG 241
K+V+S++P Y M K+P ++C +I + F W RK +SWD LP +GG
Sbjct: 734 LKAVLSSMPSYAMMCFKLPASLCKQIQSVLTRFWWDSKPDKRKMAWVSWDKLTLPINEGG 793
Query: 242 LGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSPLWKAL 301
LGFR+ K+ W ++K P L RVL KY + + + + S W+ +
Sbjct: 794 LGFREIE-------AKLSWRILKEPHSLLSRVLLGKYCNTSSFMDCSASPSFASHGWRGI 846
Query: 302 VNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVLSPS-GD 360
+ + + + W+IG G + N W + W+ + IG T + LSV D++
Sbjct: 847 LAGRDLLRKGLGWSIGQGDSINVWTEAWLSPSSPQTPIGPPTETNKDLSVHDLICHDVKS 906
Query: 361 WNIDFLINNLP--TNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSLQHQD--- 415
WN++ + +LP + + +I ++P D++ W + +T ++ Y+L +
Sbjct: 907 WNVEAIRKHLPQYEDQIRKITINALPLQ----DSLVWLPVKSGEYTTKTGYALAKLNSFP 962
Query: 416 CPILEGDW-KSIWKWHGPHRIQTFIW 440
L+ +W K+IWK H +++ F+W
Sbjct: 963 ASQLDFNWQKNIWKIHTSPKVKHFLW 988
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 217 bits (552), Expect = 2e-56
Identities = 133/452 (29%), Positives = 221/452 (48%), Gaps = 15/452 (3%)
Query: 1 NIEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGR 60
++ Y ++ NG+ RG+RQGDPLSP+LFV+C + L H++ + ++
Sbjct: 341 SVSYSVLINGQPFGHITPHRGLRQGDPLSPFLFVLCTEALIHILNQAEKIGKISGIQFNG 400
Query: 61 YGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQL 120
GP ++HLLFADD LL +AS + ++HCL + SGQ IN K+ + F V+ +
Sbjct: 401 TGPSVNHLLFADDTLLICKASQLECAEIMHCLSQYGHISGQMINSEKSAITFGAKVNEET 460
Query: 121 REDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRIT 180
++ I++ +G G YLG S + F I K+QS+L+GW + L+ G+
Sbjct: 461 KQWIMNRSGIQTEGGTGKYLGLPECFQGSKQVLFGFIKEKLQSRLSGWYAKTLSQGGKDI 520
Query: 181 LAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADG 240
L KS+ P Y M ++ KT+C ++ + F W +K H I LPK G
Sbjct: 521 LLKSIAMAFPVYAMTCFRLSKTLCTKLTSVMMDFWWNSVQDKKKIHWIGAQKLMLPKFLG 580
Query: 241 GLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSPLWKA 300
G GF+ N+A L K L + + L ++L+S+Y N+D +++ S W++
Sbjct: 581 GFGFKDLQCFNQALLAKQASRLHTDSDSLLSQILKSRYYMNSDFLSATKG-TRPSYAWQS 639
Query: 301 LVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVLSP-SG 359
++ + IG+G NT W+D W+ +++ Q +DI L V ++ P S
Sbjct: 640 ILYGRELLVSGLKKIIGNGENTYVWMDNWIFDDKPRRPESLQIMVDIQLKVSQLIDPFSR 699
Query: 360 DWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSL-------- 411
+WN++ L + P + QI+ P D+ W GTN +TV+S Y L
Sbjct: 700 NWNLNMLRDLFPWKEI-QIICQQRPM-ASRQDSFCWFGTNHGLYTVKSEYDLCSRQVHKQ 757
Query: 412 ---QHQDCPILEGDWKSIWKWHGPHRIQTFIW 440
+ ++ P L + IW + +I+ F+W
Sbjct: 758 MFKEAEEQPSLNPLFGKIWNLNSAPKIKVFLW 789
>At1g24640 hypothetical protein
Length = 1270
Score = 213 bits (543), Expect = 2e-55
Identities = 136/453 (30%), Positives = 217/453 (47%), Gaps = 17/453 (3%)
Query: 1 NIEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGR 60
++ Y ++ N RG+RQGDPLSP+LFV+C + L+H++ ++
Sbjct: 575 SVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGLTHLLNKAQWEGALEGIQFSE 634
Query: 61 YGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQL 120
GP + HLLFADD L +AS EQ+ + L ++ A+GQ IN NK+ + F + VD QL
Sbjct: 635 NGPMVHHLLFADDSLFLCKASREQSLVLQKILKVYGNATGQTINLNKSSITFGEKVDEQL 694
Query: 121 REDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRIT 180
+ I G G YLG S +++ ++++ KL+ W +CL+ G+
Sbjct: 695 KGTIRTCLGIFTEGGAGTYLGLPECFSGSKVDMLHYLKDRLKEKLDVWFTRCLSQGGKEV 754
Query: 181 LAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADG 240
L KSV +P + M K+P T C+ ++ F W D RK H SW+ CLPK G
Sbjct: 755 LLKSVALAMPVFAMSCFKLPITTCENLESAMASFWWDSCDHSRKIHWQSWERLCLPKDSG 814
Query: 241 GLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLI-ASINAHPYDSPLWK 299
GLGFR N+A L K W L+ P+ L R+L+S+Y D + A+++ P S W+
Sbjct: 815 GLGFRDIQSFNQALLAKQAWRLLHFPDCLLSRLLKSRYFDATDFLDAALSQRP--SFGWR 872
Query: 300 ALVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVLSP-S 358
+++ + +GDG + W+D W+ +N D+TL V+ +L+P +
Sbjct: 873 SILFGRELLSKGLQKRVGDGASLFVWIDPWIDDNGFRAPWRKNLIYDVTLKVKALLNPRT 932
Query: 359 GDWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSLQHQD--- 415
G W+ + L + + +I A+ D W + F+V+SAY L +Q
Sbjct: 933 GFWDEEVLHDLFLPEDILRIKAIKPVISQ--ADFFVWKLNKSGDFSVKSAYWLAYQTKSQ 990
Query: 416 --------CPILEGDWKSIWKWHGPHRIQTFIW 440
P G +W +I+ F+W
Sbjct: 991 NLRSEVSMQPSTLGLKTQVWNLQTDPKIKIFLW 1023
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 209 bits (531), Expect = 6e-54
Identities = 127/454 (27%), Positives = 211/454 (45%), Gaps = 19/454 (4%)
Query: 1 NIEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGR 60
++ Y ++ NG+ RGIRQGDP+SP+LFV+C + L H++ +K ++
Sbjct: 564 SVTYSVLINGQHFGHITPERGIRQGDPISPFLFVLCTEALIHILQQAENSKKVSGIQFNG 623
Query: 61 YGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQL 120
GP ++HLLF DD L A+ ++ CL + SGQ IN K+ + F VD
Sbjct: 624 SGPSVNHLLFVDDTQLVCRATKSDCEQMMLCLSQYGHISGQLINVEKSSITFGVKVDEDT 683
Query: 121 REDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRIT 180
+ I + +G + G YLG S + F +I K+QS L+GW + L+ G+
Sbjct: 684 KRWIKNRSGIHLEGGTGKYLGLPENLSGSKQDLFGYIKEKLQSHLSGWYDKTLSQGGKEI 743
Query: 181 LAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADG 240
L KS+ +P Y M ++PK +C ++ + F W + K H I LPK+ G
Sbjct: 744 LLKSIALALPVYIMTCFRLPKGLCTKLTSVMMDFWWNSMEFSNKIHWIGGKKLTLPKSLG 803
Query: 241 GLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSP--LW 298
G GF+ N+A L K W L + + + ++ +S+Y N D +NA P W
Sbjct: 804 GFGFKDLQCFNQALLAKQAWRLFSDSKSIVSQIFKSRYFMNTDF---LNARQGTRPSYTW 860
Query: 299 KALVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVLSP- 357
++++ G + IG+G TN W+DKW+ + S + + M+I + V ++ P
Sbjct: 861 RSILYGRELLNGGLKRLIGNGEQTNVWIDKWLFDGHSRRPMNLHSLMNIHMKVSHLIDPL 920
Query: 358 SGDWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSLQHQDC- 416
+ +WN+ L V I+ + D+ W GTN +TV+S Y ++
Sbjct: 921 TRNWNLKKLTELFHEKDVQLIMHQRPLISSE--DSYCWAGTNNGLYTVKSGYERSSRETF 978
Query: 417 ----------PILEGDWKSIWKWHGPHRIQTFIW 440
P + + +W +I+ F+W
Sbjct: 979 KNLFKEADVYPSVNPLFDKVWSLETVPKIKVFMW 1012
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 206 bits (525), Expect = 3e-53
Identities = 134/460 (29%), Positives = 222/460 (48%), Gaps = 14/460 (3%)
Query: 1 NIEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGR 60
++ Y + NG TRG+RQGDPLSP LF++C + LS + + +R
Sbjct: 507 SVSYSFLINGTPQGKVVPTRGLRQGDPLSPCLFILCTEVLSGLCTRAQRLRQLPGVRVSI 566
Query: 61 YGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQL 120
GP+++HLLFADD + F+++ E + + L + +ASGQ IN +K+ V FS +
Sbjct: 567 NGPRVNHLLFADDTMFFSKSDPESCNKLSEILSRYGKASGQSINFHKSSVTFSSKTPRSV 626
Query: 121 REDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRIT 180
+ + + G YLG GR R F I++KI+ K + W + L+ AG+
Sbjct: 627 KGQVKRILKIRKEGGTGKYLGLPEHFGRRKRDIFGAIIDKIRQKSHSWASRFLSQAGKQV 686
Query: 181 LAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADG 240
+ K+V++++P Y M K+P +C +I + F W RK ++W PK G
Sbjct: 687 MLKAVLASMPLYSMSCFKLPSALCRKIQSLLTRFWWDTKPDVRKTSWVAWSKLTNPKNAG 746
Query: 241 GLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLI-ASINAHPYDSPLWK 299
GLGFR + N++ L K+ W L+ +PE L R+L KY ++ + + + P S W+
Sbjct: 747 GLGFRDIERCNDSLLAKLGWRLLNSPESLLSRILLGKYCHSSSFMECKLPSQP--SHGWR 804
Query: 300 ALVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVLSPS- 358
+++ K + W I +G + W D W+ ++ L+ IG L V +++ +
Sbjct: 805 SIIAGREILKEGLGWLITNGEKVSIWNDPWLSISKPLVPIGPALREHQDLRVSALINQNT 864
Query: 359 --GDWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAY---SLQH 413
DWN +I LP N N I L P+ G D + W + +T +S Y S+
Sbjct: 865 LQWDWNKIAVI--LP-NYENLIKQLPAPS-SRGVDKLAWLPVKSGQYTSRSGYGIASVAS 920
Query: 414 QDCPILEGDWKS-IWKWHGPHRIQTFIWLDTHERILTNFQ 452
P + +W+S +WK +I+ +W E + Q
Sbjct: 921 IPIPQTQFNWQSNLWKLQTLPKIKHLMWKAAMEALPVGIQ 960
>At1g31030 F17F8.5
Length = 872
Score = 197 bits (500), Expect = 2e-50
Identities = 132/492 (26%), Positives = 230/492 (45%), Gaps = 17/492 (3%)
Query: 4 YIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEA-KYWIPMRAGRYG 62
+ + NG+ F S RG+RQG LSPYLFVICMD LS M+ K+ + R G
Sbjct: 285 FSVQVNGDLVGYFQSKRGLRQGCSLSPYLFVICMDVLSKMLDKAAGVRKFGFHPKCQRLG 344
Query: 63 PQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLRE 122
++HL FADDL++ ++ +L D FC+ SG +I+ K+ +Y + V +++
Sbjct: 345 --LTHLSFADDLMVLSDGKTRSIEGILEVFDEFCKRSGLRISLEKSTLYMA-GVSPIIKQ 401
Query: 123 DILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRITLA 182
+I F+ YLG L R ++ ++ +I+ ++ W + + AGR L
Sbjct: 402 EIAAKFLFDVGQLPVRYLGLPLVTKRLTSADYSPLLEQIKKRIATWTFRFFSFAGRFNLI 461
Query: 183 KSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADGGL 242
KSV+ +I + + ++P+ EIDK+ F+W S+ ISWD+ C PKA+GGL
Sbjct: 462 KSVLWSICNFWLAAFRLPRQCIREIDKLCSSFLWSGSEMSSHKAKISWDIVCKPKAEGGL 521
Query: 243 GFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSPLWKALV 302
G R + N+ +K++W +I N LW + + R + + + S +W+ ++
Sbjct: 522 GLRNLKEANDVSCLKLVWRIISNSNSLWTKWVAEYLIRKKSIWSLKQSTSMGSWIWRKIL 581
Query: 303 NIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVLSPSGDWN 362
I + K +G+G + +FW D W + + ++G++ +D+ + ++ +
Sbjct: 582 KIRDVAKSFSRVEVGNGESASFWYDHWSAHGRLIDTVGDKGTIDLGIPREASVADAWTRR 641
Query: 363 IDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTN---THHFTVQSAYSLQHQDCPIL 419
N + +++A + D DT+ W G N HF+ + + L +
Sbjct: 642 SRRRHRTSLLNEIEEMMAYQRIHHSDAEDTVLWRGKNDVFKPHFSTRDTWHLIKATSSTV 701
Query: 420 EGDW-KSIWKWHGPHRIQTFIWLDTHERILTNFQ----RSNGVV--EYLLIARIVEMKMK 472
W K +W H + WL H R+ T + S+G V +L +
Sbjct: 702 --SWHKGVWFRHATPKYALCTWLAIHNRLPTGDRMLKWNSSGSVSGNCVLCTNNSKTLEH 759
Query: 473 LFFMCFAIASTL 484
LFF C + AST+
Sbjct: 760 LFFSC-SYASTV 770
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 196 bits (498), Expect = 4e-50
Identities = 136/471 (28%), Positives = 214/471 (44%), Gaps = 21/471 (4%)
Query: 1 NIEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGR 60
++ Y ++ NG TRGIRQGDPLSPYLF++C D LSH+I + + +R G
Sbjct: 768 SVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGN 827
Query: 61 YGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQL 120
P I+HL FADD L F +A++ + D++ SGQKIN K+ + F V
Sbjct: 828 GAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGST 887
Query: 121 REDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRIT 180
+ + G YLG GR + F +I+++++ + + W + L+ AG+
Sbjct: 888 QSKLKQILEIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEI 947
Query: 181 LAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADG 240
+ KSV +P Y M K+PK I EI+ + F W + R ++W K +G
Sbjct: 948 MLKSVALAMPVYAMSCFKLPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEG 1007
Query: 241 GLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSPLWKA 300
GLGFR K N+A L K W LI+ P L+ RV++++Y ++ ++ S W +
Sbjct: 1008 GLGFRDLAKFNDALLAKQAWRLIQYPNSLFARVMKARYFKDVSIL-DAKVRKQQSYGWAS 1066
Query: 301 LVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVLSPSGD 360
L++ K IGDG+N LD V ++ +TY ++T + ++ G
Sbjct: 1067 LLDGIALLKKGTRHLIGDGQNIRIGLDNIVDSHPPRPLNTEETYKEMT--INNLFERKGS 1124
Query: 361 WNI--DFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSLQHQD--- 415
+ D I+ + + + PD I W T +TV+S Y L D
Sbjct: 1125 YYFWDDSKISQFVDQSDHGFIHRIYLAKSKKPDKIIWNYNTTGEYTVRSGYWLLTHDPST 1184
Query: 416 -CPILEGDWKS------IWKWHGPHRIQTFIW------LDTHERILTNFQR 453
P + S IW +++ F+W L T ER+ T R
Sbjct: 1185 NIPAINPPHGSIDLKTRIWNLPIMPKLKHFLWRALSQALATTERLTTRGMR 1235
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 196 bits (497), Expect = 5e-50
Identities = 136/471 (28%), Positives = 214/471 (44%), Gaps = 21/471 (4%)
Query: 1 NIEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGR 60
++ Y ++ NG TRGIRQGDPLSPYLF++C D LSH+I + + +R G
Sbjct: 994 SVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGN 1053
Query: 61 YGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQL 120
P I+HL FADD L F +A++ + D++ SGQKIN K+ + F V
Sbjct: 1054 GAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGST 1113
Query: 121 REDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRIT 180
+ + G YLG GR + F +I+++++ + + W + L+ AG+
Sbjct: 1114 QSRLKQILEIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEI 1173
Query: 181 LAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADG 240
+ KSV +P Y M K+PK I EI+ + F W + R ++W K +G
Sbjct: 1174 MLKSVALAMPVYAMSCFKLPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEG 1233
Query: 241 GLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSPLWKA 300
GLGFR K N+A L K W LI+ P L+ RV++++Y ++ ++ S W +
Sbjct: 1234 GLGFRDLAKFNDALLAKQAWRLIQYPNSLFARVMKARYFKDVSIL-DAKVRKQQSYGWAS 1292
Query: 301 LVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITLSVRDVLSPSGD 360
L++ K IGDG+N LD V ++ +TY ++T + ++ G
Sbjct: 1293 LLDGIALLKKGTRHLIGDGQNIRIGLDNIVDSHPPRPLNTEETYKEMT--INNLFERKGS 1350
Query: 361 WNI--DFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSLQHQD--- 415
+ D I+ + + + PD I W T +TV+S Y L D
Sbjct: 1351 YYFWDDSKISQFVDQSDHGFIHRIYLAKSKKPDKIIWNYNTTGEYTVRSGYWLLTHDPST 1410
Query: 416 -CPILEGDWKS------IWKWHGPHRIQTFIW------LDTHERILTNFQR 453
P + S IW +++ F+W L T ER+ T R
Sbjct: 1411 NIPAINPPHGSIDLKTRIWNLPIMPKLKHFLWRALSQALATTERLTTRGMR 1461
>At4g10830 putative protein
Length = 1294
Score = 192 bits (487), Expect = 8e-49
Identities = 105/320 (32%), Positives = 168/320 (51%), Gaps = 1/320 (0%)
Query: 1 NIEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGR 60
++ Y ++ NG T RGIRQGDPLSPYLF++C D L+H+I ++V +R G
Sbjct: 974 SVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHLIKNRVAEGDIRGIRIGN 1033
Query: 61 YGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQL 120
P ++HL FADD L F ++++ + D++ SGQKIN +K+ + F V
Sbjct: 1034 GVPGVTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSGQKINMSKSMITFGSRVHGTT 1093
Query: 121 REDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRIT 180
+ + + G G YLG GR R FN+I+ +++ + + W + L+ AG+
Sbjct: 1094 QNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYIIERVKKRTSSWSAKYLSPAGKEI 1153
Query: 181 LAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADG 240
+ KSV ++P Y M K+P I EI+ + F W + + R+ I+W K +G
Sbjct: 1154 MLKSVAMSMPVYAMSCFKLPLNIVSEIEALLMNFWWEKNAKKREIPWIAWKRLQYSKKEG 1213
Query: 241 GLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSPLWKA 300
GLGFR K N+A L K +W +I NP L+ R+++++Y R D I Y S W +
Sbjct: 1214 GLGFRDLAKFNDALLAKQVWRMINNPNSLFARIMKARYFR-EDSILDAKRQRYQSYGWTS 1272
Query: 301 LVNIWNDFKGHVVWNIGDGR 320
++ + K + +GDG+
Sbjct: 1273 MLAGLDVIKKGSRFIVGDGK 1292
>At3g45550 putative protein
Length = 851
Score = 190 bits (483), Expect = 2e-48
Identities = 132/443 (29%), Positives = 215/443 (47%), Gaps = 10/443 (2%)
Query: 1 NIEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGR 60
++ Y ++ NG TRGIRQGDPLSPYLF++C D LSH+I + + +R G
Sbjct: 233 SVHYSVLINGSPHGYISPTRGIRQGDPLSPYLFILCGDILSHLIKVKASSGDIRGVRIGN 292
Query: 61 YGPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNV--DT 118
P I+HL FADD L F +A++ + D++ SGQKIN K+ + F V T
Sbjct: 293 GAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSLITFGSRVYGST 352
Query: 119 QLREDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGR 178
Q R L + NQ G YLG GR + FN+I+++++ + W + L+ AG+
Sbjct: 353 QTRLKTLLNIP-NQGGG-GKYLGLPEQFGRKKKEMFNYIIDRVKERTASWSAKFLSPAGK 410
Query: 179 ITLAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKA 238
L KSV +P Y M K+P+ I EI+ + F W + R ++W K
Sbjct: 411 EILLKSVALAMPVYAMSCFKLPQGIVSEIESLLMNFWWEKASNKRGIPWVAWKRLQYSKK 470
Query: 239 DGGLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLIASINAHPYDSPLW 298
+GGLGFR K N+A L K W +I+ P L+ RV++++Y ++N +I S W
Sbjct: 471 EGGLGFRDLAKFNDALLAKQAWRIIQYPNSLFARVMKARYFKDNSII-DAKTRSQQSYGW 529
Query: 299 KALVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYMDITL-SVRDVLSP 357
+L++ + + IGDG+ +D V ++ + ++ + ++L ++
Sbjct: 530 SSLLSGIALLRKGTRYVIGDGKTIRLGIDNVVDSHPPRPLLTDEQHNGLSLDNLFQHRGH 589
Query: 358 SGDWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSLQHQDCP 417
S W+ L + + + I + + + D + W +T +TV+S Y L D
Sbjct: 590 SRCWDNAKLQTFVDQSDHDYIKRIYL-STRSKTDRLIWSYNSTGDYTVRSGYWLSTHD-- 646
Query: 418 ILEGDWKSIWKWHGPHRIQTFIW 440
++ K HG ++T IW
Sbjct: 647 -PSNTIPTMAKPHGSVDLKTKIW 668
>At2g11240 pseudogene
Length = 1044
Score = 187 bits (475), Expect = 2e-47
Identities = 105/340 (30%), Positives = 171/340 (49%), Gaps = 3/340 (0%)
Query: 2 IEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRY 61
+ Y + NG RG+RQGDPLSP+LF+IC + LS + + +R +
Sbjct: 439 VSYSFLLNGSAQGAVTPERGLRQGDPLSPFLFIICSEVLSGLCRKAQLDGSLLGLRVSKG 498
Query: 62 GPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLR 121
P+++HLLFADD + F + ++ L L + +ASGQ IN++K+ + FS+ ++
Sbjct: 499 NPRVNHLLFADDTIFFCRSDLKSCKTFLCILKKYEEASGQMINKSKSAITFSRKTPDHIK 558
Query: 122 EDILHHTGFNQVNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRITL 181
+ G V LG YLG GR R FN IV++I+ + W + L+ AG+ T+
Sbjct: 559 TEAQQILGIQLVGGLGKYLGLPKMFGRKKRDLFNQIVDRIRQRSLSWSSRFLSTAGKTTM 618
Query: 182 AKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADGG 241
KSV++++P Y M K+ ++C I F W S +K I+W K +GG
Sbjct: 619 LKSVLASMPTYTMSCFKLLVSLCKRIQSALTHFWWDSSADKKKMCWIAWSKMAKNKKEGG 678
Query: 242 LGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLI-ASINAHPYDSPLWKA 300
LGF+ N+A L K+ W ++++P + R+L KY R + + S+ A S W+
Sbjct: 679 LGFKDITNFNDALLAKLSWRIVQSPSCVLVRILLGKYCRTSSFLDCSVTA--ASSHGWRG 736
Query: 301 LVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIG 340
+ + K + IG G +T W + W+ + S +G
Sbjct: 737 ICTGKDLIKSQLGKVIGSGLDTLVWNEPWLSLSTSSTPMG 776
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.338 0.149 0.503
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,375,897
Number of Sequences: 26719
Number of extensions: 692123
Number of successful extensions: 2367
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 2116
Number of HSP's gapped (non-prelim): 115
length of query: 755
length of database: 11,318,596
effective HSP length: 107
effective length of query: 648
effective length of database: 8,459,663
effective search space: 5481861624
effective search space used: 5481861624
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146757.4


