

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.11 - phase: 0 /pseudo
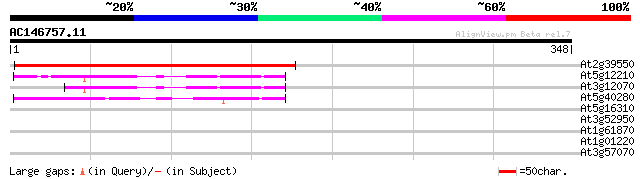
(348 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g39550 putative geranylgeranyl transferase type I beta subunit 206 1e-53
At5g12210 Rab geranylgeranyltransferase, beta subunit 52 4e-07
At3g12070 unknown protein 49 4e-06
At5g40280 farnesyltransferase beta subunit (ERA1) 43 3e-04
At5g16310 ubiquitin C-terminal hydrolase-like protein 28 5.7
At3g52950 unknown protein 28 5.7
At1g61870 unknown protein 28 5.7
At1g01220 unknown protein 28 7.4
At3g57070 unknown protein 28 9.7
>At2g39550 putative geranylgeranyl transferase type I beta subunit
Length = 375
Score = 206 bits (525), Expect = 1e-53
Identities = 103/174 (59%), Positives = 125/174 (71%)
Query: 4 SEEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVE 63
S S + +KD H+ + +MMY LLP Y+SQEIN LTLA+F+IS L L + V+
Sbjct: 24 SPPVQSSPSANFEKDRHLMYLEMMYELLPYHYQSQEINRLTLAHFIISGLHFLGARDRVD 83
Query: 64 KEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALA 123
K+ VA WVLSFQ +G+FYGF GS++SQFP DENG HN SHLASTYCALA
Sbjct: 84 KDVVAKWVLSFQAFPTNRVSLKDGEFYGFFGSRSSQFPIDENGDLKHNGSHLASTYCALA 143
Query: 124 ILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
ILK++G+DLS++DS+S+ SM NLQQ DGSFMPIHIGGETDLRFVYCA I M
Sbjct: 144 ILKVIGHDLSTIDSKSLLISMINLQQDDGSFMPIHIGGETDLRFVYCAAAICYM 197
Score = 34.3 bits (77), Expect = 0.10
Identities = 29/100 (29%), Positives = 45/100 (45%), Gaps = 27/100 (27%)
Query: 43 LTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPP 102
L Y + +L+S ++KE+ N++L+ Q G GF
Sbjct: 185 LRFVYCAAAICYMLDSWSGMDKESAKNYILNCQSYDG-----------GF---------- 223
Query: 103 DENGVFHHNNSHLASTYCALAILKIVGY---DLSSLDSES 139
G+ + SH +TYCA+A L+++GY DL S DS S
Sbjct: 224 ---GLIPGSESHGGATYCAIASLRLMGYIGVDLLSNDSSS 260
>At5g12210 Rab geranylgeranyltransferase, beta subunit
Length = 321
Score = 52.4 bits (124), Expect = 4e-07
Identities = 50/171 (29%), Positives = 80/171 (46%), Gaps = 31/171 (18%)
Query: 3 SSEEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTL--AYFVISSLDILNSLH 60
SS +S M DK HV + +M +ES ++HL + AY+ +++LD+L+ L
Sbjct: 2 SSTSSSQMVQLVADK--HVRYI-LMAEKKKESFESVVMDHLRMNGAYWGLTTLDLLDKLG 58
Query: 61 LVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYC 120
V +E V +W+++ Q + G GF G+ ++ H+ T
Sbjct: 59 CVSEEEVISWLMTCQHESG-----------GFAGNT-------------GHDPHILYTLS 94
Query: 121 ALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
A+ IL + ++ LD +SS + LQ DGSF + GE D RF Y A
Sbjct: 95 AVQILALFD-KINILDIGKVSSYVAKLQNEDGSFSG-DMWGEIDTRFSYIA 143
>At3g12070 unknown protein
Length = 317
Score = 48.9 bits (115), Expect = 4e-06
Identities = 42/139 (30%), Positives = 68/139 (48%), Gaps = 28/139 (20%)
Query: 35 YESQEINHLTL--AYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGF 92
+ES ++HL + AY+ +++L +L+ L V ++ V +WV++ Q + G GF
Sbjct: 26 FESVVMDHLRMNGAYWGLTTLALLDKLGSVSEDEVVSWVMTCQHESG-----------GF 74
Query: 93 HGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDG 152
G+ ++ H+ T A+ IL + L+ LD E +S+ + LQ DG
Sbjct: 75 AGNT-------------GHDPHVLYTLSAVQILALFD-KLNILDVEKVSNYIAGLQNEDG 120
Query: 153 SFMPIHIGGETDLRFVYCA 171
SF I GE D RF Y A
Sbjct: 121 SFSG-DIWGEVDTRFSYIA 138
>At5g40280 farnesyltransferase beta subunit (ERA1)
Length = 482
Score = 42.7 bits (99), Expect = 3e-04
Identities = 42/171 (24%), Positives = 73/171 (42%), Gaps = 26/171 (15%)
Query: 3 SSEEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLV 62
+S+ ++ I +D + + L + S + N L Y+++ S+ +L V
Sbjct: 69 ASDVSTQKYMMEIQRDKQLDYLMKGLRQLGPQFSSLDANRPWLCYWILHSIALLGET--V 126
Query: 63 EKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCAL 122
+ E +N + +G+ G +G Q P HLA+TY A+
Sbjct: 127 DDELESNAIDFLGRCQGSE---------GGYGGGPGQLP------------HLATTYAAV 165
Query: 123 AILKIVGYD--LSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
L +G D LSS++ E MS ++ ++ G F +H GE D+R Y A
Sbjct: 166 NALVTLGGDKALSSINREKMSCFLRRMKDTSGGFR-MHDMGEMDVRACYTA 215
>At5g16310 ubiquitin C-terminal hydrolase-like protein
Length = 334
Score = 28.5 bits (62), Expect = 5.7
Identities = 24/95 (25%), Positives = 42/95 (43%), Gaps = 17/95 (17%)
Query: 99 QFPPDENGVFHHNNSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDG-----S 153
+FPP+ G+ +NN + + + A D SS+ + ++ KNL + D S
Sbjct: 112 EFPPELKGLAINNNEAIRAAHNTFA-----RPDPSSIMEDEELAAAKNLDEDDDVYHYIS 166
Query: 154 FMPI-----HIGG--ETDLRFVYCAGWITGMEWTR 181
++P+ + G E + C G G+EW R
Sbjct: 167 YLPVDGILYELDGLKEGPISLGQCLGEPEGIEWLR 201
>At3g52950 unknown protein
Length = 556
Score = 28.5 bits (62), Expect = 5.7
Identities = 35/164 (21%), Positives = 62/164 (37%), Gaps = 28/164 (17%)
Query: 1 MSSSEEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLH 60
M+ + E +S+ETT +D LH HL P ++ + + +D+L H
Sbjct: 302 MTPNPECASLETTILDA-LHTMHDGKFLHL---PIIDKDGS-------AAACVDVLQITH 350
Query: 61 LVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDE-------NGVFHHNNS 113
+ + G ND N F S + PPD+ + + HH++
Sbjct: 351 AA--------ISMVENSSGAVNDMANTMMQKFWDSALALEPPDDSDTQSEMSAMMHHSDI 402
Query: 114 HLASTYCALAILKIVGYDLSSLDS--ESMSSSMKNLQQPDGSFM 155
S+Y +L + + L +S +NL++ G M
Sbjct: 403 GKLSSYPSLGLGNSFSFKFEDLKGRVHRFTSGAENLEELMGIVM 446
>At1g61870 unknown protein
Length = 408
Score = 28.5 bits (62), Expect = 5.7
Identities = 12/41 (29%), Positives = 20/41 (48%)
Query: 42 HLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTN 82
+ TL Y++ D +L L ++ NWV SF + + N
Sbjct: 330 YFTLIYYLCKGGDFETALSLCKESMEKNWVPSFSIMKSLVN 370
>At1g01220 unknown protein
Length = 1053
Score = 28.1 bits (61), Expect = 7.4
Identities = 14/56 (25%), Positives = 24/56 (42%)
Query: 129 GYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGMEWTRKKS 184
G+ L+S S S ++ PD F P E +R + GW W+ +++
Sbjct: 683 GFREHLLESSGKSHSENHISHPDRVFQPRRTKVELPVRVDFVGGWSDTPPWSLERA 738
>At3g57070 unknown protein
Length = 417
Score = 27.7 bits (60), Expect = 9.7
Identities = 18/53 (33%), Positives = 27/53 (49%), Gaps = 3/53 (5%)
Query: 101 PPDENGVFHHNNSHLASTYCALAILKIVGYDLSSLDSESM---SSSMKNLQQP 150
PP G HH S +++Y +L +L + G SS D +++ S S KN P
Sbjct: 51 PPARKGDSHHLVSLTSTSYGSLLLLDLDGSKNSSSDQQTLPRISISGKNTPDP 103
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.335 0.144 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,099,249
Number of Sequences: 26719
Number of extensions: 275798
Number of successful extensions: 981
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 970
Number of HSP's gapped (non-prelim): 15
length of query: 348
length of database: 11,318,596
effective HSP length: 100
effective length of query: 248
effective length of database: 8,646,696
effective search space: 2144380608
effective search space used: 2144380608
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146757.11


