

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146756.6 - phase: 0
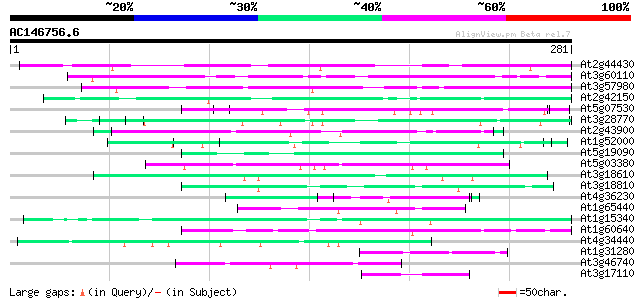
(281 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g44430 unknown protein 83 2e-16
At3g60110 unknown protein 77 8e-15
At3g57980 unknown protein 64 1e-10
At2g42150 unknown protein 57 1e-08
At5g07530 glycine-rich protein atGRP-7 53 2e-07
At3g28770 hypothetical protein 53 2e-07
At2g43900 putative inositol polyphosphate 5'-phosphatase 50 1e-06
At1g52000 50 1e-06
At5g19090 unknown protein 50 1e-06
At5g03380 farnesylated protein - like 50 1e-06
At3g18610 unknown protein 50 1e-06
At3g18810 protein kinase, putative 49 3e-06
At4g36230 putative glycine-rich cell wall protein 47 1e-05
At1g65440 47 1e-05
At1g15340 unknown protein 47 1e-05
At1g60640 unknown protein 46 2e-05
At4g34440 serine/threonine protein kinase - like 46 3e-05
At1g31280 unknown protein 46 3e-05
At3g46740 chloroplast import-associated channel protein homolog 45 3e-05
At3g17110 hypothetical protein 45 4e-05
>At2g44430 unknown protein
Length = 646
Score = 82.8 bits (203), Expect = 2e-16
Identities = 86/295 (29%), Positives = 120/295 (40%), Gaps = 86/295 (29%)
Query: 6 QAQSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVK--PSTTTFSHKGDHK 63
Q S + K+D+ ++ LSR K P++VC+KR S+ K PS+++FS K D
Sbjct: 419 QEASGMRSGKADAETSDSSLSR----QKSSGPLVVCKKRRSVSAKASPSSSSFSQKDD-- 472
Query: 64 PIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGAR-SRRSNKNLSSNASNKKP 122
T EE + K+ TG R SRR+NK + A+N K
Sbjct: 473 ------------------------TKEETLSEEKDNIATGVRSSRRANKVAAVVANNTK- 507
Query: 123 PSNSTPRTGSSANKPAETPKLKNKAEGVSDKK-------------KNNGAAGFLNRIKKN 169
TG NK +T N + S K+ K A FL R+KKN
Sbjct: 508 -------TGKGRNKQKQTESKTNSSNDNSSKQDTGKTEKKTVSADKKKSVADFLKRLKKN 560
Query: 170 ESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGS 229
+ + + GG N K K K R R S S
Sbjct: 561 SPQKEAKDQNKSGG-----------------------NVKKDSKTKPRELR------SSS 591
Query: 230 GDKRNKNIESNS-KRNVGRPPKKAAETVAST--KRGRESSASGGKDKRPKKRSKK 281
K+ +E+ KR GRP KK AE AS KRGR++ ++G +K+PKKR +K
Sbjct: 592 VGKKKAEVENTPVKRAPGRPQKKTAEATASASGKRGRDTGSTGKDNKQPKKRIRK 646
>At3g60110 unknown protein
Length = 644
Score = 77.4 bits (189), Expect = 8e-15
Identities = 77/255 (30%), Positives = 112/255 (43%), Gaps = 31/255 (12%)
Query: 30 SLSKHKPPILV---CRKRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYS 86
S+S+ K +L C+K+SS K S ++ S + D K +E+ + T S
Sbjct: 418 SVSRQKSSVLSLVPCKKKSSALKKTSPSSSSRQKDEKKSQEVSEEKIVTTTATTSARSSR 477
Query: 87 ETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNK 146
T +E AK+ ++ R+ N+ KK T + ++ E K + K
Sbjct: 478 RTSKEIAVVAKD-----TKTGRAKNNI------KKQTDTKTESSDDDDDEKEENSKTEKK 526
Query: 147 AEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVV 206
V+DKKK+ A FL RIKKN + G S K G QKK
Sbjct: 527 T--VADKKKS--VADFLKRIKKNSPQK------GKETTSKNQKKNDGNVKKENDHQKKSD 576
Query: 207 NNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESS 266
N K E K + G + N N +S+SKR + K+ AE VA+ KRGRES
Sbjct: 577 GNVKKENSKVKPRELRSSTGKKKVEVENNNSKSSSKR---KQTKETAE-VATGKRGRES- 631
Query: 267 ASGGKDKRPKKRSKK 281
G DK+P+KRS++
Sbjct: 632 --GKDDKQPRKRSRR 644
>At3g57980 unknown protein
Length = 650
Score = 63.5 bits (153), Expect = 1e-10
Identities = 66/250 (26%), Positives = 101/250 (40%), Gaps = 29/250 (11%)
Query: 37 PILVCRKRSSIPVKPS---TTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEP 93
PI+ CRKRSS+ V+ T T K P +++K+ + +P+ D++E
Sbjct: 425 PIIACRKRSSLAVRSPASVTETLKKKTRVVPTVDEKQVSEEEEGRPS--------DKDEK 476
Query: 94 PKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGV--S 151
P +K GA + K S N K + G S N + K + +G+ S
Sbjct: 477 PIVSKKMARGAAPSTAKKVGSRNV--KTSLNAGISNRGRSPNGSSVLKKSVQQKKGINTS 534
Query: 152 DKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKG 211
K AA FL R+K S E + + K G EQ+K +N K
Sbjct: 535 GGSKKQSAASFLKRMKGVSSSETVVE----------TVKAESSNGKRGAEQRK--SNSKS 582
Query: 212 EKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGK 271
EK A + G +G + S +K+N G K+ ++ + + S G
Sbjct: 583 EKVD--AVKLPAGQKRLTGKRPTIEKGSPTKKNSGVASKRGTASLMAKRDSETSEKETGS 640
Query: 272 DKRPKKRSKK 281
RPKKRSK+
Sbjct: 641 STRPKKRSKR 650
>At2g42150 unknown protein
Length = 631
Score = 56.6 bits (135), Expect = 1e-08
Identities = 67/269 (24%), Positives = 103/269 (37%), Gaps = 35/269 (13%)
Query: 18 SLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDV 77
++P + P+S +S P I+ CRKRS++ KP P DKK + + V
Sbjct: 393 AIPSSKPVSSKPRMSV--PNIVACRKRSALAAKPLLLL--------PPGPDKKAKKTDHV 442
Query: 78 KPTLKPSYSETDEEEPPKAKE-----KPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGS 132
+ S+ D E K + K +T R+ + K + N N+ S
Sbjct: 443 VDYDEKPVSDKDGEASGKDDDDSLIVKIMTRGRTSSTGKVANRNDKNRD----------S 492
Query: 133 SANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGG 192
S N K+K E K AA FL R+K S + L+ S ++G KGG
Sbjct: 493 SLNVDDSKDKVKKTDEEKKGGSKKKRAASFLRRMKVGSSDDTLKRSSAADSSTTG--KGG 550
Query: 193 GGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKA 252
G EQ+K N+ K + K + S KR+ N NS+R
Sbjct: 551 GA------EQRK-NNSNKADNKKTPIPKIRQTNKKASPVKRSNN-GRNSEREAAPSSSSY 602
Query: 253 AETVASTKRGRESSASGGKDKRPKKRSKK 281
++ E + R KKR+++
Sbjct: 603 PILAKRSRDAGEKEEASSYSPRLKKRARR 631
>At5g07530 glycine-rich protein atGRP-7
Length = 543
Score = 52.8 bits (125), Expect = 2e-07
Identities = 51/198 (25%), Positives = 82/198 (40%), Gaps = 32/198 (16%)
Query: 111 KNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAE-----------------GVSDK 153
+ + + PP+ P +G+ A K K+KA+ G
Sbjct: 141 RRMGVKPKDNPPPAGLPPNSGAGAGGAQSLIK-KSKAKSKGGLKAWCKKMLKSKFGGKKG 199
Query: 154 KKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNT-------KEQKKVV 206
K G + F + K+E E + SG G GS G GG GG + + K +KK
Sbjct: 200 KSGGGKSKFGGKGGKSEGEEGMSSGDEGMSGSEGGMSGGEGGKSKSGKGKLKAKLEKKKG 259
Query: 207 NNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESS 266
+G E E ++GG SG G ++K+ +S K +G+ K + ++ ++ G S
Sbjct: 260 MSGGSE--SEEGMSGSEGGMSGGGGSKSKSKKSKLKAKLGK-KKGMSGGMSGSEEGMSGS 316
Query: 267 ----ASGGKDKRPKKRSK 280
+SGG K K+SK
Sbjct: 317 EGGMSSGGGSKSKSKKSK 334
Score = 52.0 bits (123), Expect = 4e-07
Identities = 52/202 (25%), Positives = 85/202 (41%), Gaps = 28/202 (13%)
Query: 87 ETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNK-KPPSNSTPRTGSSANKPAETPKLKN 145
+++ EE + ++ ++G+ S + S K K + + G S +E
Sbjct: 214 KSEGEEGMSSGDEGMSGSEGGMSGGEGGKSKSGKGKLKAKLEKKKGMSGGSESE------ 267
Query: 146 KAEGVSDKKKN-NGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKG----------GGG 194
EG+S + +G G ++ KK++ L G GG SGS +G GGG
Sbjct: 268 --EGMSGSEGGMSGGGGSKSKSKKSKLKAKLGKKKGMSGGMSGSEEGMSGSEGGMSSGGG 325
Query: 195 GGTNTKEQKKVVNNGK------GEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRP 248
+ +K+ K GK G G E ++GG SG G ++K+ +S K N+G+
Sbjct: 326 SKSKSKKSKLKAKLGKKKSMSGGMSGSEEGMSGSEGGMSGGGGGKSKSRKSKLKANLGK- 384
Query: 249 PKKAAETVASTKRGRESSASGG 270
KK S G S + GG
Sbjct: 385 -KKCMSGGMSGSEGGMSRSEGG 405
Score = 42.0 bits (97), Expect = 4e-04
Identities = 46/180 (25%), Positives = 65/180 (35%), Gaps = 18/180 (10%)
Query: 103 GARSRRSNKNLSSNASNKKPPSN---------STPRTGSSANKPAETPKLKNKAEGVSDK 153
G++S+ L + KK S S G S ++ K+K + K
Sbjct: 325 GSKSKSKKSKLKAKLGKKKSMSGGMSGSEEGMSGSEGGMSGGGGGKSKSRKSKLKANLGK 384
Query: 154 KK--NNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKK--VVNNG 209
KK + G +G + ++E + G GG GS GG GG K KK + +G
Sbjct: 385 KKCMSGGMSGSEGGMSRSEG-GISGGGMSGGSGSKHKIGGGKHGGLGGKFGKKRGMSGSG 443
Query: 210 KGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASG 269
G G E ++G SG G S SK +G KRG S G
Sbjct: 444 GGMSGSEGGVSGSEGSMSGGGMSGG----SGSKHKIGGGKHGGLRGKFGKKRGMSGSEGG 499
Score = 40.0 bits (92), Expect = 0.001
Identities = 39/140 (27%), Positives = 57/140 (39%), Gaps = 17/140 (12%)
Query: 103 GARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGA--A 160
G +S+ L +N KK S + ++ + G K K G
Sbjct: 368 GGKSKSRKSKLKANLGKKKCMSGGMSGSEGGMSRSEGGISGGGMSGGSGSKHKIGGGKHG 427
Query: 161 GFLNRIKKNESVEVLRSGSGGG--------GGSSGSSKGGG-GGGTNTKEQKKVVNNG-- 209
G + K + SGSGGG GS GS GGG GG+ +K + +G
Sbjct: 428 GLGGKFGKKRGM----SGSGGGMSGSEGGVSGSEGSMSGGGMSGGSGSKHKIGGGKHGGL 483
Query: 210 KGEKGKERASRYNDGGGSGS 229
+G+ GK+R ++GG SGS
Sbjct: 484 RGKFGKKRGMSGSEGGMSGS 503
Score = 28.1 bits (61), Expect = 5.5
Identities = 22/72 (30%), Positives = 29/72 (39%), Gaps = 11/72 (15%)
Query: 155 KNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKG 214
K+ G G + + E SGS GG SG S GGG K + GK + G
Sbjct: 479 KHGGLRGKFGKKRGMSGSEGGMSGSEGGMSESGMSGSGGG--------KHKIGGGKHKFG 530
Query: 215 KERASRYNDGGG 226
++ GGG
Sbjct: 531 ---GGKHGGGGG 539
>At3g28770 hypothetical protein
Length = 2081
Score = 52.8 bits (125), Expect = 2e-07
Identities = 51/226 (22%), Positives = 84/226 (36%), Gaps = 22/226 (9%)
Query: 68 DKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNST 127
D K+ S++ + K E ++E+ ++K+ A+ + N + +N NK+ S
Sbjct: 803 DNKKLSSTENRDEAKERSGEDNKEDKEESKDYQSVEAKEKNENGGVDTNVGNKED-SKDL 861
Query: 128 PRTGSSANKPAETPKLKNKAEGV--SDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGS 185
S K + +K K E V +DK F N N ++V + G G
Sbjct: 862 KDDRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDFAN----NMDIDVQK-----GSGE 912
Query: 186 SGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRN----------K 235
S K N +E K +N +KGK++ + + S K K
Sbjct: 913 SVKYKKDEKKEGNKEENKDTINTSSKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELK 972
Query: 236 NIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRSKK 281
E N K K E K +ES S K++ K+ +K
Sbjct: 973 KQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEK 1018
Score = 46.2 bits (108), Expect = 2e-05
Identities = 59/271 (21%), Positives = 97/271 (35%), Gaps = 34/271 (12%)
Query: 29 DSLSKHKPPILVCRKRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSET 88
DS SK+ R++ K S T K + K + K+E S+ + + K E
Sbjct: 1004 DSASKN-------REKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKK----EK 1052
Query: 89 DEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAE 148
+E KAK+K K S N +KK S K + + K E
Sbjct: 1053 EESRDLKAKKK-----EEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEE 1107
Query: 149 GVSDKKK-------------NNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGG 195
S KK+ +N N KK++ V++++ S K
Sbjct: 1108 SKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKE 1167
Query: 196 GTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAET 255
++K QK V+ + + K++ + K KN E K+ KK ET
Sbjct: 1168 IESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKET 1227
Query: 256 VASTKRGRE-----SSASGGKDKRPKKRSKK 281
+ ++ + SGGK + + SK+
Sbjct: 1228 KKEKNKPKDDKKNTTKQSGGKKESMESESKE 1258
Score = 42.7 bits (99), Expect = 2e-04
Identities = 46/229 (20%), Positives = 85/229 (37%), Gaps = 20/229 (8%)
Query: 59 KGDHKPII---NDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSS 115
K H ++ +DKKE+ ++ K K S ++ KEK + + ++ K +
Sbjct: 1140 KSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKE 1199
Query: 116 NASNKKPPSNSTPRTGSSA---NKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESV 172
+ K + + +S K ET K KNK + DKK +G ++ES
Sbjct: 1200 SEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPK--DDKKNTTKQSGGKKESMESESK 1257
Query: 173 EVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDK 232
E + K ++ E K + + + D S +
Sbjct: 1258 E-----------AENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEIL 1306
Query: 233 RNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRSKK 281
+ ++ ++RN +K +VA K+ +E+ K K KK + K
Sbjct: 1307 MQADSQATTQRN-NEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTK 1354
Score = 41.2 bits (95), Expect = 6e-04
Identities = 55/245 (22%), Positives = 88/245 (35%), Gaps = 25/245 (10%)
Query: 46 SIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGAR 105
S+ K +K ++K IN ++ D K K S + +++ KE +
Sbjct: 913 SVKYKKDEKKEGNKEENKDTINTSSKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELK 972
Query: 106 SRRSNKNLSS----------NASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKK 155
+ NK ++ N NK+ + + + K E K K K E +KKK
Sbjct: 973 KQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKK 1032
Query: 156 NNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGK 215
+ KK E + S S K TKE KK N K +K +
Sbjct: 1033 SQD--------KKREEKDSEERKSKKEKEESRDLK-AKKKEEETKE-KKESENHKSKKKE 1082
Query: 216 ERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRP 275
++ ++ DK+ K SK KK E K ++S +DK
Sbjct: 1083 DKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDME-----KLEDQNSNKKKEDKNE 1137
Query: 276 KKRSK 280
KK+S+
Sbjct: 1138 KKKSQ 1142
Score = 38.5 bits (88), Expect = 0.004
Identities = 51/234 (21%), Positives = 83/234 (34%), Gaps = 34/234 (14%)
Query: 54 TTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNL 113
TT S K D KE+ S+ + K + EE+ K KE+ + + K
Sbjct: 981 TTKSENSKLKEENKDNKEKKESEDSAS-KNREKKEYEEKKSKTKEEAKKEKKKSQDKKRE 1039
Query: 114 SSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVE 173
++ +K A K E K K ++E KKK + N+ K E
Sbjct: 1040 EKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEE-- 1097
Query: 174 VLRSGSGGGGGSSGSSKGGGGGGTNTKEQKK--VVNNGKGEKGKERASRYNDGGGSGSGD 231
+ KE+KK + K E+ K+ + D + +
Sbjct: 1098 ------------------------DKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKE 1133
Query: 232 KRNKNIESNS----KRNVGRPPKKAAETVASTKRGRESSASGGK-DKRPKKRSK 280
+N+ +S K+ + KK E + TK S + + DK+ KK SK
Sbjct: 1134 DKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSK 1187
Score = 31.6 bits (70), Expect = 0.50
Identities = 41/204 (20%), Positives = 74/204 (36%), Gaps = 20/204 (9%)
Query: 67 NDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPV---TGARSRRSNKNLSSNASNKKPP 123
NDK +++ + K + ET E K + + G +NL + N++
Sbjct: 402 NDKMVNATTNDEDHKKENKEETHENNGESVKGENLENKAGNEESMKGENLENKVGNEELK 461
Query: 124 SNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNR-IKKNESVEVLRSGSGGG 182
N A+ A+T +K E + +++N ++N+ K E+V + G
Sbjct: 462 GN--------ASVEAKTNNESSKEEKREESQRSNEV--YMNKETTKGENVNIQGESIGD- 510
Query: 183 GGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSK 242
S+K + K N G KER G + DK NI ++ +
Sbjct: 511 -----STKDNSLENKEDVKPKVDANESDGNSTKERHQEAQVNNGVSTEDKNLDNIGADEQ 565
Query: 243 RNVGRPPKKAAETVASTKRGRESS 266
+ + + TK RE +
Sbjct: 566 KKNDKSVEVTTNDGDHTKEKREET 589
Score = 30.8 bits (68), Expect = 0.85
Identities = 33/158 (20%), Positives = 60/158 (37%), Gaps = 21/158 (13%)
Query: 132 SSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSG---- 187
+S + ++ K++ EG + K + G +N I ++ + GG S+G
Sbjct: 1688 NSTKEGSKDDKIEEGMEGKENSTKESSKDGKINEIHGDKEATMEEGSKDGGTNSTGKDSK 1747
Query: 188 ---SSKGGGGGGTNTKEQKK-----VVNNGKGEKGKERA--------SRYNDGGGSGSGD 231
S + G + K+ K +NNGK + K+ S N +GD
Sbjct: 1748 DSKSVEINGVKDDSLKDDSKNGDINEINNGKEDSVKDNVTEIQGNDNSLTNSTSSEPNGD 1807
Query: 232 KRNKNIESNSKRNV-GRPPKKAAETVASTKRGRESSAS 268
K + N +S + + T T+ +ES+ S
Sbjct: 1808 KLDTNKDSMKNNTMEAQGGSNGDSTNGETEETKESNVS 1845
Score = 30.4 bits (67), Expect = 1.1
Identities = 40/204 (19%), Positives = 69/204 (33%), Gaps = 19/204 (9%)
Query: 90 EEEPPKAKEKPVT---GARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNK 146
+E+ K VT G ++ + N N + K E+ K
Sbjct: 563 DEQKKNDKSVEVTTNDGDHTKEKREETQGNNGESVKNENLENKEDKKELKDDESVGAKTN 622
Query: 147 AEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTN-------- 198
E ++K+ G N I + GG S+ + G TN
Sbjct: 623 NETSLEEKREQTQKGHDNSINSK-----IVDNKGGNADSNKEKEVHVGDSTNDNNMESKE 677
Query: 199 -TKEQKKVV-NNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETV 256
TK + +V N+G EKG+E D + + +S ++V ++ A+
Sbjct: 678 DTKSEVEVKKNDGSSEKGEEGKENNKDSMEDKKLENKESQTDSKDDKSVD-DKQEEAQIY 736
Query: 257 ASTKRGRESSASGGKDKRPKKRSK 280
+ +S + GK K K+ K
Sbjct: 737 GGESKDDKSVEAKGKKKESKENKK 760
Score = 30.0 bits (66), Expect = 1.5
Identities = 38/236 (16%), Positives = 83/236 (35%), Gaps = 23/236 (9%)
Query: 54 TTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNL 113
TT G + + ++ KE + + S+ + E + +++ +
Sbjct: 1241 TTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDE 1300
Query: 114 SSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVE 173
S N + S +T + + ++ +T +NK + + ++KN N K++
Sbjct: 1301 SKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTKQS---- 1356
Query: 174 VLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKE-------RASRYNDGGG 226
GG S + N ++ + E E +A ++D
Sbjct: 1357 ---------GGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQA 1407
Query: 227 SGSGDKRNKNIESNSKRNVGR---PPKKAAETVASTKRGRESSASGGKDKRPKKRS 279
K ++++S+ R +K +VA K+ +E+ K K KK +
Sbjct: 1408 DSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNT 1463
Score = 27.3 bits (59), Expect = 9.4
Identities = 43/198 (21%), Positives = 63/198 (31%), Gaps = 15/198 (7%)
Query: 75 SDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSA 134
S ++ L+ E E K +T N S N+K S
Sbjct: 287 SAIEKNLESKEDVKSEVEAAKNDGSSMTENLGEAQGNNGVSTIDNEKEVEGQGESIEDSD 346
Query: 135 NKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGG 194
+ K K+E + K + G L ++N V + S G G
Sbjct: 347 IEKNLESKEDVKSEVEAAKNAGSSMTGKLEEAQRNNGVST--------NETMNSENKGSG 398
Query: 195 GGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNK--NIES----NSKRNVGRP 248
TN K N+ +K + + N+G + NK N ES N + VG
Sbjct: 399 ESTNDKMVNATTNDEDHKKENKEETHENNGESVKGENLENKAGNEESMKGENLENKVGNE 458
Query: 249 PKKAAETVASTKRGRESS 266
K +V K ESS
Sbjct: 459 ELKGNASV-EAKTNNESS 475
>At2g43900 putative inositol polyphosphate 5'-phosphatase
Length = 1305
Score = 50.4 bits (119), Expect = 1e-06
Identities = 49/194 (25%), Positives = 82/194 (42%), Gaps = 21/194 (10%)
Query: 52 STTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNK 111
ST T H+ + + K R S+ + S + + + K+ +K + S+ S K
Sbjct: 1080 STETVCHRVHVRHCFSAKNLRIDSNPSNSKSQSLKKNEGDSNSKSSKKSDGDSNSKSSKK 1139
Query: 112 NLSSNASNKKPPSNSTPRTGSSANKPAE---TPKLKNKAEGVSDKKKNNGAAGFLNRIKK 168
S SN K S + S ++K ++ K K++G S+ K + KK
Sbjct: 1140 --SDGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSS----------KK 1187
Query: 169 NESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSG 228
++ +S G S S+ G TN+K QKK G G+ ++ + NDG S
Sbjct: 1188 SDGDSNSKSSKKSDGDSCSKSQKKSDGDTNSKSQKK----GDGD-SSSKSHKKNDGDSSS 1242
Query: 229 SGDKRNKNIESNSK 242
K+N +S+SK
Sbjct: 1243 KSHKKNDG-DSSSK 1255
Score = 41.2 bits (95), Expect = 6e-04
Identities = 44/208 (21%), Positives = 83/208 (39%), Gaps = 9/208 (4%)
Query: 43 KRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVT 102
K I PS + ++ N K + S + S+ D K +
Sbjct: 1097 KNLRIDSNPSNSKSQSLKKNEGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDS 1156
Query: 103 GARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGF 162
++S + + S++ S+KK +S ++ ++ + + K K++G S K + G
Sbjct: 1157 NSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSNS-KSSKKSDGDSCSKSQKKSDGD 1215
Query: 163 LN--RIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASR 220
N KK + +S G SS S G +++K KK + + K+
Sbjct: 1216 TNSKSQKKGDGDSSSKSHKKNDGDSSSKSHKKNDGDSSSKSHKKSDGDSSSKSHKK---- 1271
Query: 221 YNDGGGSGSGDKRNKNI-ESNSKRNVGR 247
++G S S +R I ++++ R VGR
Sbjct: 1272 -SEGDSSLSLTRRTMEILQAHTSRRVGR 1298
Score = 34.7 bits (78), Expect = 0.059
Identities = 25/124 (20%), Positives = 49/124 (39%), Gaps = 10/124 (8%)
Query: 166 IKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKE-------RA 218
+KKNE +S G S+ S G +N+K KK + + K+ ++
Sbjct: 1113 LKKNEGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKS 1172
Query: 219 SRYNDGGGSGSGDKR---NKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRP 275
S+ +DG + K+ + N +S+ K + K ++ T + G +
Sbjct: 1173 SKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSCSKSQKKSDGDTNSKSQKKGDGDSSSKS 1232
Query: 276 KKRS 279
K++
Sbjct: 1233 HKKN 1236
Score = 32.7 bits (73), Expect = 0.22
Identities = 31/131 (23%), Positives = 51/131 (38%), Gaps = 4/131 (3%)
Query: 154 KKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEK 213
KKN G + + KK++ +S G S+ S G +N+K KK + +
Sbjct: 1114 KKNEGDSNSKSS-KKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKS 1172
Query: 214 GKERASRYNDGGGSGS-GDKRNKNIESNSKRNVGRPPKKA--AETVASTKRGRESSASGG 270
K+ N S GD +K+ + + + + KK+ S K+G S+S
Sbjct: 1173 SKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSCSKSQKKSDGDTNSKSQKKGDGDSSSKS 1232
Query: 271 KDKRPKKRSKK 281
K S K
Sbjct: 1233 HKKNDGDSSSK 1243
>At1g52000
Length = 730
Score = 50.4 bits (119), Expect = 1e-06
Identities = 56/252 (22%), Positives = 84/252 (33%), Gaps = 27/252 (10%)
Query: 50 KPSTTTFSHKGDHKPIINDKKERPSSDVKPT-----LKPSYSETDEEEPPK--AKEKPVT 102
KPS+ + + N + E+ + KP+ P S E K KP +
Sbjct: 172 KPSSGSAGTNPGASAVGNGETEKNAGGSKPSSGSAGTNPGASAGGNGETEKNVGGSKPSS 231
Query: 103 GARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETP--------KLKNKAEGVSDKK 154
G N N +K S +GS+ P + + + K
Sbjct: 232 GKAGTNPGANAGGNGGTEKNAGGSKSSSGSARTNPGASAGGNGETVSNIGDTESNAGGSK 291
Query: 155 KNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKG 214
N+GA + I+ N G+GG GG + GG TN+ N+G G
Sbjct: 292 SNDGANNGASGIESNAGSTGTNFGAGGTGGIGDTESDAGGSKTNSGNGG--TNDGASGIG 349
Query: 215 KERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAE-------TVASTKRGRESSA 267
S G G+G + NIE G+ A + ++ G ES+A
Sbjct: 350 SNDGS---TGTNPGAGGGTDSNIEGTENNVGGKETNPGASGIGNSDGSTGTSPEGTESNA 406
Query: 268 SGGKDKRPKKRS 279
G K K S
Sbjct: 407 DGTKTNTGGKES 418
Score = 43.9 bits (102), Expect = 1e-04
Identities = 43/169 (25%), Positives = 65/169 (38%), Gaps = 31/169 (18%)
Query: 106 SRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNR 165
S +N +K S P +GS+ P + A G + +KN G +
Sbjct: 151 SAGTNPGADGTRETEKNAGGSKPSSGSAGTNPGAS------AVGNGETEKNAGGS----- 199
Query: 166 IKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGG 225
+ SG G + G+S GG G E +K V K GK + + G
Sbjct: 200 ----------KPSSGSAGTNPGASAGGNG------ETEKNVGGSKPSSGKAGTNPGANAG 243
Query: 226 GSGSGDKR---NKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGK 271
G+G +K +K+ +++ N G ETV S ES+A G K
Sbjct: 244 GNGGTEKNAGGSKSSSGSARTNPGASAGGNGETV-SNIGDTESNAGGSK 291
Score = 40.8 bits (94), Expect = 8e-04
Identities = 42/185 (22%), Positives = 56/185 (29%), Gaps = 33/185 (17%)
Query: 83 PSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPK 142
P T E E KP +G+ + N +K S P +GS+ P +
Sbjct: 156 PGADGTRETEKNAGGSKPSSGSAGTNPGASAVGNGETEKNAGGSKPSSGSAGTNPGAS-- 213
Query: 143 LKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQ 202
A G + +KN G G S K G G N
Sbjct: 214 ----AGGNGETEKNVG------------------------GSKPSSGKAGTNPGANAGGN 245
Query: 203 KKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRG 262
N G K ++R N G +G + NI G A AS G
Sbjct: 246 GGTEKNAGGSKSSSGSARTNPGASAGGNGETVSNIGDTESNAGGSKSNDGANNGAS---G 302
Query: 263 RESSA 267
ES+A
Sbjct: 303 IESNA 307
Score = 40.0 bits (92), Expect = 0.001
Identities = 36/146 (24%), Positives = 54/146 (36%), Gaps = 23/146 (15%)
Query: 128 PRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSG 187
P GS+ KP A G+ + G+AG E GS GS+G
Sbjct: 128 PPPGSTGAKPG--------ASGIGSDSGSIGSAGTNPGADGTRETEKNAGGSKPSSGSAG 179
Query: 188 SSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGR 247
++ G G E+ N G K ++ N G +G + KN+ SK + G+
Sbjct: 180 TNPGASAVGNGETEK-----NAGGSKPSSGSAGTNPGASAGGNGETEKNV-GGSKPSSGK 233
Query: 248 PPKKAAETVASTKRGRESSASGGKDK 273
A T G + +GG +K
Sbjct: 234 ---------AGTNPGANAGGNGGTEK 250
Score = 29.3 bits (64), Expect = 2.5
Identities = 29/123 (23%), Positives = 37/123 (29%), Gaps = 18/123 (14%)
Query: 110 NKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEG----VSDKKKNNGAAGFLNR 165
+K S N S GS+ P + EG V K+ N GA+G N
Sbjct: 332 SKTNSGNGGTNDGASGIGSNDGSTGTNPGAGGGTDSNIEGTENNVGGKETNPGASGIGN- 390
Query: 166 IKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGG 225
S G G+S GT T K N G + G
Sbjct: 391 -------------SDGSTGTSPEGTESNADGTKTNTGGKESNTGSESNTNSSPQKLEAQG 437
Query: 226 GSG 228
G+G
Sbjct: 438 GNG 440
>At5g19090 unknown protein
Length = 587
Score = 50.1 bits (118), Expect = 1e-06
Identities = 45/164 (27%), Positives = 61/164 (36%), Gaps = 28/164 (17%)
Query: 87 ETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNK 146
E D+ PP K KP+ G N P+N P +
Sbjct: 246 EFDDHPPPPNKMKPMMGG-------------GNMIMPNNMMPNM------------MMPN 280
Query: 147 AEGVSDKKKNNGAAGFLN-RIK-KNESVEVLRSGSGGG-GGSSGSSKGGGGGGTNTKEQK 203
A+ + + KN G G +I+ K V G GGG GG G GGGG N +
Sbjct: 281 AQQMLNAHKNGGGPGPAGGKIEGKGMPFPVQMGGGGGGPGGKKGGPGGGGGNMGNQNQGG 340
Query: 204 KVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGR 247
N GKG G + GGG +G+K ++ N N G+
Sbjct: 341 GGKNGGKGGGGHPLDGKMGGGGGGPNGNKGGGGVQMNGGPNGGK 384
Score = 35.4 bits (80), Expect = 0.035
Identities = 23/70 (32%), Positives = 32/70 (44%), Gaps = 8/70 (11%)
Query: 128 PRTGSSANKPAETPKLKNKAEGVS-DKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSS 186
P+ GS+ N+ P L N+ + + D G G N+ ++ GGGGG
Sbjct: 77 PKGGSNNNQ--NQPNLANQFKAMQIDHGGKGGGGGGGGPANNNKGQKI-----GGGGGGG 129
Query: 187 GSSKGGGGGG 196
G GGGGGG
Sbjct: 130 GGGGGGGGGG 139
Score = 29.3 bits (64), Expect = 2.5
Identities = 18/47 (38%), Positives = 19/47 (40%), Gaps = 9/47 (19%)
Query: 184 GSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSG 230
G G GGGG N K QK + G G G GGG G G
Sbjct: 102 GGKGGGGGGGGPANNNKGQK--IGGGGGGGG-------GGGGGGGGG 139
Score = 29.3 bits (64), Expect = 2.5
Identities = 21/87 (24%), Positives = 31/87 (35%), Gaps = 21/87 (24%)
Query: 122 PPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGG 181
P S S P G+ P++ G N ++ + + + G GG
Sbjct: 419 PQSMSMPMGGAMGGPMGSLPQM------------GGGPGPMSNNMQAVQGLPAMGPGGGG 466
Query: 182 GGGSS---------GSSKGGGGGGTNT 199
GGG S G G GGGG ++
Sbjct: 467 GGGPSAEAPPGYFQGQVSGNGGGGQDS 493
Score = 28.9 bits (63), Expect = 3.2
Identities = 24/85 (28%), Positives = 29/85 (33%), Gaps = 19/85 (22%)
Query: 156 NNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSK------------GGGGGGTNTKEQK 203
N G G ++ N + G GGGGG G GGGGGG +
Sbjct: 368 NKGGGG----VQMNGGPNGGKKGGGGGGGGGGGPMSGGLPPGFRPMGGGGGGGGGPQSMS 423
Query: 204 KVVNNGKGEKGKERASRYNDGGGSG 228
+ G G S GGG G
Sbjct: 424 MPMG---GAMGGPMGSLPQMGGGPG 445
>At5g03380 farnesylated protein - like
Length = 392
Score = 50.1 bits (118), Expect = 1e-06
Identities = 57/210 (27%), Positives = 85/210 (40%), Gaps = 30/210 (14%)
Query: 69 KKERPSSDVKPTLKPSYS--ETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNS 126
KKE P S KPS + E E+ P A EKP ++ + AS PP S
Sbjct: 96 KKETPPSSGGAEKKPSPAAEEKPAEKKPAAVEKPGEKKEEKKKEEG-EKKASPPPPPKES 154
Query: 127 TPRTGSSANKPAETPKLK---NKAEGVS----DKKKN----------NGAAGFLNRIKKN 169
T + + K+K NK +GV+ D K+ +LN K
Sbjct: 155 TVVLKTKLHCEGCEHKIKRIVNKIKGVNSVAIDSAKDLVIVKGIIDVKQLTPYLNE-KLK 213
Query: 170 ESVEVLRSGSGGGGGSSGSSKGGGGGGTNTK---EQKKVVN------NGKGEKGKERASR 220
+VEV+ + G + ++ GG K E+K++ + +G GEK KE A
Sbjct: 214 RTVEVVPAKKDDGAPVAAAAAAPAGGEKKDKVAGEKKEIKDVGEKKVDGGGEKKKEVAVG 273
Query: 221 YNDGGGSGSGDKRNKNIESNSKRNVGRPPK 250
GGG G GD +++ + G PP+
Sbjct: 274 GGGGGGGGGGDGGAMDVKKSEYNGYGYPPQ 303
>At3g18610 unknown protein
Length = 636
Score = 50.1 bits (118), Expect = 1e-06
Identities = 56/252 (22%), Positives = 96/252 (37%), Gaps = 30/252 (11%)
Query: 43 KRSSIPVKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVT 102
K+ I VK ++ S + P++ K D K S E+ ++ P +KP
Sbjct: 227 KKEPIVVKKDSSDESSSDEETPVVKKKPTTVVKDAKAESSSSEEESSSDDEPTPAKKPTV 286
Query: 103 GARSRRSNKNLSSN--------ASNKKPP-------SNSTPRTGSSANKPAETPKLKNKA 147
++ + K+ SS+ + ++KPP S ++ + SS E+ K ++K
Sbjct: 287 VKNAKPAAKDSSSSEEDSDEEESDDEKPPTKKAKVSSKTSKQESSSDESSDESDKEESKD 346
Query: 148 EGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVN 207
E V+ KKK++ K N + GG SK G + + + +
Sbjct: 347 EKVTPKKKDSDVEMVDAEQKSNAKQPKTPTNQTQGG-----SKTLFAGNLSYQIARSDIE 401
Query: 208 NGKGEKGK---ERASRYNDGGGSGSG-------DKRNKNIESNSKRNVGRPPKKAAETVA 257
N E G+ R S ++DG G G ++ K +E N K +GR +
Sbjct: 402 NFFKEAGEVVDVRLSSFDDGSFKGYGHIEFASPEEAQKALEMNGKLLLGRDVRLDLANER 461
Query: 258 STKRGRESSASG 269
T R G
Sbjct: 462 GTPRNSNPGRKG 473
Score = 27.3 bits (59), Expect = 9.4
Identities = 52/226 (23%), Positives = 80/226 (35%), Gaps = 38/226 (16%)
Query: 70 KERPSS--DVKPTLKPSYSETDEE-EPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNS 126
KE PS D + + S +DEE P K + +P+ K +S+ S S
Sbjct: 79 KETPSKLKDESSSEEEDDSSSDEEIAPAKKRPEPI---------KKAKVESSSSDDDSTS 129
Query: 127 TPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESV-EVLRSGSGGGGGS 185
T +PA K K ++ D ++ +KK +V E + S
Sbjct: 130 DEETAPVKKQPAVLEKAKVESSSSDDDSSSDEET---VPVKKQPAVLEKAKIESSSSDDD 186
Query: 186 SGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGD----------KRNK 235
S S + K+Q V+ EK K +S +DG S K++
Sbjct: 187 SSSDE----ETVPMKKQTAVL-----EKAKAESSSSDDGSSSDEEPTPAKKEPIVVKKDS 237
Query: 236 NIESNSKRNVGRPPKKAAETVASTKRGRESS---ASGGKDKRPKKR 278
+ ES+S KK V K SS +S + P K+
Sbjct: 238 SDESSSDEETPVVKKKPTTVVKDAKAESSSSEEESSSDDEPTPAKK 283
>At3g18810 protein kinase, putative
Length = 700
Score = 48.9 bits (115), Expect = 3e-06
Identities = 46/198 (23%), Positives = 76/198 (38%), Gaps = 32/198 (16%)
Query: 87 ETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPP--------SNSTPRTGSSANKPA 138
++ E PP + + +++ + S++S+ PP SN +P+ SS ++
Sbjct: 5 QSPENSPPSPTPPSPSSSDNQQQSSPPPSDSSSPSPPAPPPPDDSSNGSPQPPSSDSQSP 64
Query: 139 ETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTN 198
+P+ N +G NNG N N G+ ++G++K G N
Sbjct: 65 PSPQGNNNNDG------NNGNNNNDNNNNNN--------GNNNNDNNNGNNKDNNNNGNN 110
Query: 199 TKEQKKVVNNGKGEKGKERASRYND----GGGSGSGDKRNKNIESNSKRNVGRPPKKAAE 254
NNG G ND G GS ++ NS RN PP+ A
Sbjct: 111 NNGNN---NNGNDNNGNNNNGNNNDNNNQNNGGGSNNRSPPPPSRNSDRNSPSPPRALAP 167
Query: 255 TVASTKRGRESSASGGKD 272
+S G S++SG +
Sbjct: 168 PRSS---GGGSNSSGNNE 182
Score = 33.1 bits (74), Expect = 0.17
Identities = 39/186 (20%), Positives = 64/186 (33%), Gaps = 18/186 (9%)
Query: 73 PSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSS----NASNKKPPS---- 124
P S P+ S ++ PP P A + + S ++ ++ PPS
Sbjct: 11 PPSPTPPSPSSSDNQQQSSPPPSDSSSPSPPAPPPPDDSSNGSPQPPSSDSQSPPSPQGN 70
Query: 125 -NSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGG 183
N+ G++ N N + + K+N G N N + + + G
Sbjct: 71 NNNDGNNGNNNNDNNNNNNGNNNNDNNNGNNKDNNNNGNNNNGNNNNGNDNNGNNNNGNN 130
Query: 184 GSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERA---SRYNDGGGSGSGDKRNKNIESN 240
+ + GGG N N+ + RA R + GG + SG N E N
Sbjct: 131 NDNNNQ-NNGGGSNNRSPPPPSRNSDRNSPSPPRALAPPRSSGGGSNSSG-----NNEPN 184
Query: 241 SKRNVG 246
+ VG
Sbjct: 185 TAAIVG 190
Score = 32.0 bits (71), Expect = 0.38
Identities = 32/159 (20%), Positives = 49/159 (30%), Gaps = 17/159 (10%)
Query: 122 PPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGG 181
PPS + P SS N+ +P SD + A N S + S S
Sbjct: 11 PPSPTPPSPSSSDNQQQSSPP-------PSDSSSPSPPAPPPPDDSSNGSPQPPSSDSQS 63
Query: 182 GGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNS 241
G++ G G N + NN G ND + D N +N
Sbjct: 64 PPSPQGNNNNDGNNGNNNNDNN---NNNNGNNN-------NDNNNGNNKDNNNNGNNNNG 113
Query: 242 KRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRSK 280
N G + +++ G ++ P S+
Sbjct: 114 NNNNGNDNNGNNNNGNNNDNNNQNNGGGSNNRSPPPPSR 152
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 47.0 bits (110), Expect = 1e-05
Identities = 26/68 (38%), Positives = 36/68 (52%), Gaps = 7/68 (10%)
Query: 163 LNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYN 222
+N+ +K ES R+G GGGGG G GGGGGG + N G G+ + ++ +
Sbjct: 78 VNKTEKCES----RAGGGGGGGGGG---GGGGGGQGSGGGGGEGNGGNGKDNSHKRNKSS 130
Query: 223 DGGGSGSG 230
GGG G G
Sbjct: 131 GGGGGGGG 138
Score = 44.3 bits (103), Expect = 7e-05
Identities = 37/128 (28%), Positives = 46/128 (35%), Gaps = 19/128 (14%)
Query: 109 SNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKK 168
S N + S+ S ST + + A + NK E + G G
Sbjct: 43 SESNFVNATSSSVLDSKSTANISTVEFENALDDQNVNKTEKCESRAGGGGGGG------- 95
Query: 169 NESVEVLRSGSGGGGGSSGSSKGGG-GGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGS 227
G GGGGG GS GGG G G N K+ N G G GGGS
Sbjct: 96 --------GGGGGGGGGQGSGGGGGEGNGGNGKDNSHKRNKSSGGGG---GGGGGGGGGS 144
Query: 228 GSGDKRNK 235
G+G R +
Sbjct: 145 GNGSGRGR 152
Score = 41.6 bits (96), Expect = 5e-04
Identities = 29/81 (35%), Positives = 34/81 (41%), Gaps = 14/81 (17%)
Query: 155 KNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGS----SKGGGGGGTNTKEQKKVVNNGK 210
+ NG G N K+N+S G GGGGG SG+ +GGGGGG G
Sbjct: 113 EGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGRGRGGGGGG----------GGGG 162
Query: 211 GEKGKERASRYNDGGGSGSGD 231
G G GGG G D
Sbjct: 163 GGGGGGGGGGGGGGGGGGGSD 183
Score = 37.4 bits (85), Expect = 0.009
Identities = 28/88 (31%), Positives = 32/88 (35%), Gaps = 13/88 (14%)
Query: 144 KNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQK 203
+NK+ G G G N + G GGGGG G GGGGGG
Sbjct: 126 RNKSSGGGGGGGGGGGGGSGNGSGRGRGGGGGGGGGGGGGGGGGGGGGGGGGG------- 178
Query: 204 KVVNNGKGEKGKERASRYNDG-GGSGSG 230
G G GK + G GG G G
Sbjct: 179 -----GGGSDGKGGGWGFGFGWGGDGPG 201
>At1g65440
Length = 1703
Score = 46.6 bits (109), Expect = 1e-05
Identities = 39/125 (31%), Positives = 56/125 (44%), Gaps = 22/125 (17%)
Query: 115 SNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFL-----NRIKKN 169
S ++ KK ST GS + NK++G +G G + KK+
Sbjct: 1589 SESAGKKTGGGSTGGWGSESGG--------NKSDGAGSWGSGSGGGGSGGWGNDSGGKKS 1640
Query: 170 ESVEVLRSGSGGGGGSSGSSKGG------GGGGTNTKEQKKVVNNGKGEKGKERASRYND 223
SGSGGGG G+ GG GG G+ + +K ++G+G G E +SR +D
Sbjct: 1641 SEDGGFGSGSGGGGSDWGNESGGKKSSADGGWGSESGGKK---SDGEGGWGNEPSSRKSD 1697
Query: 224 GGGSG 228
GGG G
Sbjct: 1698 GGGGG 1702
Score = 35.4 bits (80), Expect = 0.035
Identities = 28/97 (28%), Positives = 39/97 (39%), Gaps = 9/97 (9%)
Query: 178 GSGGGGG----SSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKR 233
G+ GGGG S+G GGG G E ++G G G + GGGSG
Sbjct: 1580 GNSGGGGWGSESAGKKTGGGSTGGWGSESGGNKSDGAGSWGSG-----SGGGGSGGWGND 1634
Query: 234 NKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGG 270
+ +S+ G + G++SSA GG
Sbjct: 1635 SGGKKSSEDGGFGSGSGGGGSDWGNESGGKKSSADGG 1671
Score = 27.3 bits (59), Expect = 9.4
Identities = 22/82 (26%), Positives = 28/82 (33%), Gaps = 27/82 (32%)
Query: 179 SGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDG-------------- 224
SGG G S S+GG G ++ G R Y +G
Sbjct: 1495 SGGSGWGSSQSEGGWKGNSD-------------RSGSGRGGEYRNGGGRDGHPSGAPRPY 1541
Query: 225 GGSGSGDKRNKNIESNSKRNVG 246
GG G G R + + NS R G
Sbjct: 1542 GGRGRGRGRGRRDDMNSDRQDG 1563
>At1g15340 unknown protein
Length = 384
Score = 46.6 bits (109), Expect = 1e-05
Identities = 59/284 (20%), Positives = 108/284 (37%), Gaps = 36/284 (12%)
Query: 8 QSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKGDHKPIIN 67
+S+ I+QK + P PD K P+L +KR S K + K + +
Sbjct: 77 RSSRISQKVKATTPT-----PD-----KEPLL--KKRRSSLTKKDNKEAAEKNEEAAV-- 122
Query: 68 DKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNST 127
+ + DV K +E ++E+ + + + + + NK+
Sbjct: 123 ----KENMDVDKDGKTENAEAEKEKEKEGVTEIAEAEKENNEGEKTEAEKVNKEGEKTEA 178
Query: 128 PRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEV----LRSGSGGGG 183
+ G + AE K KAE N A + K ES+EV L +G G
Sbjct: 179 GKEGQTEIAEAEKEKEGEKAEA------ENKEAEVVR--DKKESMEVDTSELEKKAGSGE 230
Query: 184 GSSGSSKGGGGGGTNTKEQKKVVNNG------KGEKGKERASRYNDGGGSGSGDKRNKNI 237
G+ SK G T KE ++VV EK + + S + G + N+
Sbjct: 231 GAEEPSKVEGLKDTEMKEAQEVVTEADVEKKPAEEKTENKGSVTTEANGEQNVTLGEPNL 290
Query: 238 ESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRSKK 281
+++++ + G+ K+ E + +E+ +K+ + + K
Sbjct: 291 DADAEADKGKESKEYDEKTTEAEANKENDTQESDEKKTEAAANK 334
>At1g60640 unknown protein
Length = 340
Score = 46.2 bits (108), Expect = 2e-05
Identities = 50/198 (25%), Positives = 86/198 (43%), Gaps = 19/198 (9%)
Query: 87 ETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNK 146
E D+ E +++ RS+R K ++K +S + G N+ E P+ ++
Sbjct: 158 EVDDLETASQEKEKKGNRRSQRQGKR------SQKQEKDSLTKNGE--NEEVEDPETPSQ 209
Query: 147 AEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTK---EQK 203
+ + K N A L R +K E + G +SG+ G N++ ++K
Sbjct: 210 EKQI---KGNRRARRELRRSQKQEKDSSTKHGENEEVDNSGTPSQGKQIKENSRARRQRK 266
Query: 204 KVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGR 263
++ GKG K + D + S +K+ K S+S+R + R K+ E + STK G
Sbjct: 267 RLEKQGKGSLTKHGENEEVDNPETPSQEKQIKG-NSSSRRQLKRSEKQ--EKIPSTKEGE 323
Query: 264 ESSASGGKDKRPKKRSKK 281
AS + R K+R K
Sbjct: 324 NEDAS--QKSRKKRRQTK 339
>At4g34440 serine/threonine protein kinase - like
Length = 670
Score = 45.8 bits (107), Expect = 3e-05
Identities = 62/246 (25%), Positives = 86/246 (34%), Gaps = 42/246 (17%)
Query: 5 GQAQSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTF--SHKGDH 62
G SN + ++S PP P S P S S PP + S P P+ T +
Sbjct: 17 GTPPSNGTSPSNESSPPTPPSSPPPS-SISAPPPDISASFSPPPAPPTQETSPPTSPSSS 75
Query: 63 KPIINDKK----ERPSSDVK----PTLKPSYSETDEEEPPKAKEKPVTGA---RSRRSNK 111
P++ + E PS P P+ +T + P+ P GA R+R +
Sbjct: 76 PPVVANPSPQTPENPSPPAPEGSTPVTPPAPPQTPSNQSPERPTPPSPGANDDRNRTNGG 135
Query: 112 NLSSNASNKKPPS--NSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNG----------- 158
N + + S PPS N T G S + P +N G SD N
Sbjct: 136 NNNRDGSTPSPPSSGNRTSGDGGSPSPPRSISPPQN--SGDSDSSSGNHPQANIGLIIGV 193
Query: 159 --AAGFL-----------NRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKV 205
AG L NR KK +S +V G+ G GG T + V
Sbjct: 194 LVGAGLLLLLAVCICICCNRKKKKKSPQVNHMHYYNNNPYGGAPSGNGGYYKGTPQDHVV 253
Query: 206 VNNGKG 211
G+G
Sbjct: 254 NMAGQG 259
>At1g31280 unknown protein
Length = 1013
Score = 45.8 bits (107), Expect = 3e-05
Identities = 30/74 (40%), Positives = 36/74 (48%), Gaps = 6/74 (8%)
Query: 176 RSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNK 235
R G G G G G GGGGGG EQ + G GE+G+ R S GG G G +
Sbjct: 7 RGGRGDGRGRGGRGYGGGGGGG---EQGRDRGYGGGEQGRGRGSE-RGGGNRGQGRGEQQ 62
Query: 236 NIESNSKRNVGRPP 249
+ S S+R G PP
Sbjct: 63 DFRSQSQR--GPPP 74
>At3g46740 chloroplast import-associated channel protein homolog
Length = 818
Score = 45.4 bits (106), Expect = 3e-05
Identities = 41/123 (33%), Positives = 56/123 (45%), Gaps = 14/123 (11%)
Query: 84 SYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPR------TGSSANKP 137
++S + P T SRR K LS ++S S +PR + S N+
Sbjct: 3 AFSVNGQLIPTATSSTASTSLSSRR--KFLSPSSSRLPRISTQSPRVPSIKCSKSLPNRD 60
Query: 138 AETPK----LKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGG 193
ET LKN A+ ++ ++ A+ FL RI SV L G GGG G+ G GGG
Sbjct: 61 TETSSKDSLLKNLAKPLAVASVSSAASFFLFRISNLPSV--LTGGGGGGDGNFGGFGGGG 118
Query: 194 GGG 196
GGG
Sbjct: 119 GGG 121
>At3g17110 hypothetical protein
Length = 175
Score = 45.1 bits (105), Expect = 4e-05
Identities = 25/54 (46%), Positives = 25/54 (46%), Gaps = 10/54 (18%)
Query: 177 SGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSG 230
SGSGGG G G GGGGGG G G G S Y G GSGSG
Sbjct: 41 SGSGGGNGGGGGGGGGGGGG----------GGGGGGDGSGSGSGYGSGYGSGSG 84
Score = 40.0 bits (92), Expect = 0.001
Identities = 24/58 (41%), Positives = 28/58 (47%), Gaps = 11/58 (18%)
Query: 177 SGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRN 234
SG+G GGG G GGGGGG ++ NG G S Y +G GSG G N
Sbjct: 124 SGAGRGGGGGGGGGGGGGGGGGSR------GNGSG-----YGSGYGEGYGSGYGGGDN 170
Score = 39.7 bits (91), Expect = 0.002
Identities = 21/51 (41%), Positives = 23/51 (44%), Gaps = 1/51 (1%)
Query: 180 GGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSG 230
GGGGG G GGGGGG + +G G G N GGG G G
Sbjct: 49 GGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYG-SGSGYGGGNNGGGGGGGG 98
Score = 39.7 bits (91), Expect = 0.002
Identities = 21/54 (38%), Positives = 23/54 (41%), Gaps = 1/54 (1%)
Query: 178 GSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKG-EKGKERASRYNDGGGSGSG 230
G+ GGGG G GGGGGG +G G G R GGG G G
Sbjct: 88 GNNGGGGGGGGRGGGGGGGNGGSGNGSGYGSGSGYGSGAGRGGGGGGGGGGGGG 141
Score = 38.1 bits (87), Expect = 0.005
Identities = 25/60 (41%), Positives = 26/60 (42%), Gaps = 3/60 (5%)
Query: 177 SGSGGGGGS---SGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKR 233
SGSG G GS SGS GGG G + G G G S Y G G GSG R
Sbjct: 69 SGSGSGYGSGYGSGSGYGGGNNGGGGGGGGRGGGGGGGNGGSGNGSGYGSGSGYGSGAGR 128
Score = 37.4 bits (85), Expect = 0.009
Identities = 20/55 (36%), Positives = 24/55 (43%), Gaps = 6/55 (10%)
Query: 178 GSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDK 232
G GGGGG G GGG G + +G G G N+GGG G G +
Sbjct: 51 GGGGGGGGGGGGGGGGDGSGSGSGYGSGYGSGSGYGGG------NNGGGGGGGGR 99
Score = 37.4 bits (85), Expect = 0.009
Identities = 23/58 (39%), Positives = 25/58 (42%), Gaps = 4/58 (6%)
Query: 177 SGSGGGGGSSGSSKG----GGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSG 230
SGSG GGG++G G GGGGG G G A R GGG G G
Sbjct: 81 SGSGYGGGNNGGGGGGGGRGGGGGGGNGGSGNGSGYGSGSGYGSGAGRGGGGGGGGGG 138
Score = 35.8 bits (81), Expect = 0.026
Identities = 23/60 (38%), Positives = 25/60 (41%), Gaps = 7/60 (11%)
Query: 178 GSGGGGGSSGSSKGGG---GGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRN 234
G GGGGG G G G G G + NNG G G R GGG G+G N
Sbjct: 57 GGGGGGGGGGDGSGSGSGYGSGYGSGSGYGGGNNGGGGGGGGRGG----GGGGGNGGSGN 112
Score = 31.2 bits (69), Expect = 0.65
Identities = 19/57 (33%), Positives = 22/57 (38%), Gaps = 9/57 (15%)
Query: 178 GSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRN 234
G GGG G SG+ G G G + G G G GGG G G + N
Sbjct: 102 GGGGGNGGSGNGSGYGSGSGYGSGAGRGGGGGGGGGG---------GGGGGGGSRGN 149
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.299 0.122 0.330
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,640,099
Number of Sequences: 26719
Number of extensions: 422602
Number of successful extensions: 9241
Number of sequences better than 10.0: 664
Number of HSP's better than 10.0 without gapping: 306
Number of HSP's successfully gapped in prelim test: 369
Number of HSP's that attempted gapping in prelim test: 2766
Number of HSP's gapped (non-prelim): 2433
length of query: 281
length of database: 11,318,596
effective HSP length: 98
effective length of query: 183
effective length of database: 8,700,134
effective search space: 1592124522
effective search space used: 1592124522
T: 11
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146756.6


