

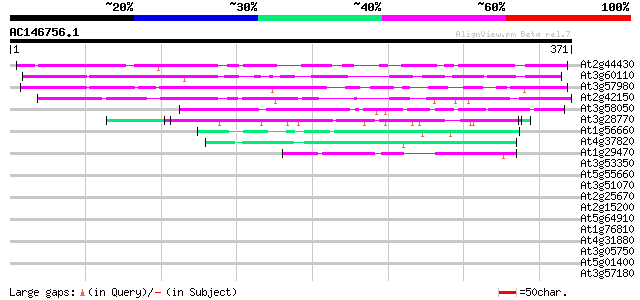
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146756.1 - phase: 0
(371 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g44430 unknown protein 189 3e-48
At3g60110 unknown protein 144 6e-35
At3g57980 unknown protein 134 8e-32
At2g42150 unknown protein 131 7e-31
At3g58050 putative protein 52 5e-07
At3g28770 hypothetical protein 48 8e-06
At1g56660 hypothetical protein 47 2e-05
At4g37820 unknown protein 45 8e-05
At1g29470 unknown protein 45 8e-05
At3g53350 unknown protein 41 0.001
At5g55660 putative protein 41 0.001
At3g51070 putative protein 40 0.002
At2g25670 unknown protein 40 0.002
At2g15200 putative protein on transposon FARE2.7 (cds2) 40 0.002
At5g64910 unknown protein 39 0.003
At1g76810 translation initiation factor IF-2 like protein 39 0.003
At4g31880 unknown protein 39 0.006
At3g05750 hypothetical protein 38 0.008
At5g01400 putative protein 38 0.010
At3g57180 unknown protein 38 0.010
>At2g44430 unknown protein
Length = 646
Score = 189 bits (479), Expect = 3e-48
Identities = 134/372 (36%), Positives = 195/372 (52%), Gaps = 77/372 (20%)
Query: 5 ENHQNQNKNPNQDLQGWGTWEELLLASAVDRHGFKDWDTIAMEVQSRTNRTSLLATAHHC 64
E+ N N +Q Q WGTWEELLLA AV RHGF DWD++A EV+SR++ + LLA+A+ C
Sbjct: 35 EDEDNSNTKKSQT-QAWGTWEELLLACAVKRHGFGDWDSVATEVRSRSSLSHLLASANDC 93
Query: 65 EQKFHDLNRRFKDDVPPPQQNGDVSAVTAEDSD-------HVPWLDKLRKQRVAELRRDV 117
K+ DL RRF + Q+ DV+A E+ + ++PWL++LR RVAELRR+V
Sbjct: 94 RHKYRDLKRRFHE-----QEKTDVTATVEEEEEEEERVGNNIPWLEQLRNLRVAELRREV 148
Query: 118 QLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAI 177
+ D SILSLQL+VKKLE+E+ E E +PDL E + +E G + + A+
Sbjct: 149 ERYDCSILSLQLKVKKLEEER-----EVGEEKPDL--ENERKEERSENDGSESEHREKAV 201
Query: 178 RRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGS 237
+A EESDR+N+S+NESNST + GE + G
Sbjct: 202 ---------------SAAEESDRENRSMNESNSTAT------AGEEERVCG--------- 231
Query: 238 VQVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEG 297
+EP QT E ++G+ N + V ++ E E+ SV+R G
Sbjct: 232 -------DEPSQT--------REDDSGNDKNPDPDPVNKDATAAEE----EEGSVSR--G 270
Query: 298 GGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKSEPLFGVLEMIKRHQK 357
S E+ S + + +RR++ +GE E+ KS+PL +L++I+ H +
Sbjct: 271 SEASHSDELGESGTSESKWKRKRRKQGGAGEIRSAES------KSQPLISLLDLIRSHPR 324
Query: 358 FSLFERRLEKNQ 369
SLFERRL +
Sbjct: 325 GSLFERRLRSQE 336
>At3g60110 unknown protein
Length = 644
Score = 144 bits (364), Expect = 6e-35
Identities = 117/360 (32%), Positives = 171/360 (47%), Gaps = 76/360 (21%)
Query: 9 NQNKNPNQDLQGWGTWEELLLASAVDRHGFKDWDTIAMEVQSRTNRTSLLATAHHCEQKF 68
+ N N NQ Q WGTWEEL+L AV RH F DWD++A EVQ+R +R+SL+ +A +C K+
Sbjct: 40 DDNSNLNQIKQVWGTWEELVLTCAVKRHAFSDWDSVAKEVQAR-SRSSLIVSAVNCRLKY 98
Query: 69 HDLNRRFKDDVPPPQQNGDVSAVTAEDSDHVPWLDKLRKQRVAELR---RDVQLSDVSIL 125
DL RRF+D V +N + +A ++ + WL++LR +A+ DV +
Sbjct: 99 QDLKRRFQDSVDVGDENTEAAANEEDEVGEISWLEQLRSLHMADSAVRFNDVTTRYCTEK 158
Query: 126 SLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTT 185
SLQL+VKKLE+EK + + + +PDL +N++T +PV R++ TT
Sbjct: 159 SLQLKVKKLEEEK---DGDDGDNKPDL---------KNDET-----KPV----RVNRETT 197
Query: 186 NTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLN 245
+D+ DN+S+NESNST S +D
Sbjct: 198 ESDR----------DDNRSMNESNSTASVDKIADHDRLD--------------------- 226
Query: 246 EPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSE 305
G K E S N D V P EER V RSE + E E
Sbjct: 227 -----GDKMVKANENSRNPDPD-----PVNKAETPEEEERTVS----KRSEMSNSGELDE 272
Query: 306 VQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKSEPLFGVLEMIKRHQKFSLFERRL 365
+S+ L K R+ Q+ R G K + KS+PL ++++I+ H + S+FE RL
Sbjct: 273 SGTSNCLGK-RKGQKYRSGGGGGGVKSAGD-----KSQPLIDIIKLIRSHPRGSVFESRL 326
>At3g57980 unknown protein
Length = 650
Score = 134 bits (337), Expect = 8e-32
Identities = 118/372 (31%), Positives = 174/372 (46%), Gaps = 64/372 (17%)
Query: 8 QNQNKNPNQDLQGWGTWEELLLASAVDRHGFKDWDTIAMEVQSRTNRTSLLATAHHCEQK 67
+++N + + Q W T EELLLA AV RHG WD++A EV + N T TA C K
Sbjct: 3 KSENDKNSPEKQTWSTMEELLLACAVHRHGTDSWDSVASEVHKQ-NSTFRTLTAIDCRHK 61
Query: 68 FHDLNRRFKDDVPPPQQNGDVSAVTAEDSDHVPWLDKLRKQRVAELRRDVQLSDVSILSL 127
++DL RRF ++ P + D + AE S VPWL++LRK RV ELRR+V+ D+SI SL
Sbjct: 62 YNDLKRRFSRNLVSPG-SADEETLAAEISS-VPWLEELRKLRVDELRREVERYDLSISSL 119
Query: 128 QLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDE----PVPAIRRLDES 183
QL+VK LEDE+ K + + +A + E +G + E P P
Sbjct: 120 QLKVKTLEDEREKSLKTENSDLDRIAETKENHTESGNNSGVPVTELKNSPDPNDNSPGTG 179
Query: 184 TTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSG 243
+ NT++ + A + N+ E N E + SG G +S
Sbjct: 180 SENTNRAVKIAEPVDEEPNRIGGEDND-----------EKPAREDSGRG------SCESV 222
Query: 244 LNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRES 303
E D+ KR E N+ E V +E K E+D T+E+
Sbjct: 223 AKESDRAEPKR-----EGNDSP-----------ELVESMDESKGEED---------TKET 257
Query: 304 SEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAV------VKSEPLFGVLEMIKRHQK 357
S+ QSS+S RKE +D + +N+D ++ V+S+PL +E+++ H
Sbjct: 258 SDGQSSASFP--------RKETVDQD-QPDNKDQSLTVNKIFVESQPLSDFIEILQSHPI 308
Query: 358 FSLFERRLEKNQ 369
S F RRLE +
Sbjct: 309 GSHFSRRLETQE 320
>At2g42150 unknown protein
Length = 631
Score = 131 bits (329), Expect = 7e-31
Identities = 115/365 (31%), Positives = 174/365 (47%), Gaps = 88/365 (24%)
Query: 19 QGWGTWEELLLASAVDRHGFKDWDTIAMEVQSRT-NRTSLLATAHHCEQKFHDLNRRFKD 77
Q W TWEELLLA AV RHG + W++++ E+Q + N SL A+A C K+ DL RF
Sbjct: 14 QTWSTWEELLLACAVHRHGTESWNSVSAEIQKLSPNLCSLTASA--CRHKYFDLKSRFTQ 71
Query: 78 DVPPPQQNGDVSAVTAEDSDHVPWLDKLRKQRVAELRRDVQLSDVSILSLQLQVKKLEDE 137
++P P+ ++S PWL++LRK RV ELRR+V+ D+SI +LQ +VK+LE+E
Sbjct: 72 ELPVPESVAEISTA--------PWLEELRKLRVDELRREVEQYDLSISTLQSKVKQLEEE 123
Query: 138 KAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPV--PAIRRLDESTTNTDKLLPAAG 195
+ E +PD E E +K D EPV P ++ ++E+ + K +
Sbjct: 124 R-----EMSFIKPD--TETENLDLERKKERSDSGEPVPNPPVQLMNETISPDPKEI--GS 174
Query: 196 EESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRK 255
E ++R+ + GSG G + K
Sbjct: 175 ENTEREEEMA----------------------GSG--------------------GGESK 192
Query: 256 SVEEESNNGSYDNNEAKAVTCESV---PPSEERKVEDDSVT---RSEGGGTRE---SSEV 306
E+S GS CESV P + +VE SVT SE G +R +S+V
Sbjct: 193 LAGEDSCRGS----------CESVEKEPTTNSERVEPVSVTELIESEDGASRGEEITSDV 242
Query: 307 QSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKSEPLFGVLEMIKRHQKFSLFERRLE 366
QSS+SL + ++ +++ S AK V+S+PL +E++ H S F RRLE
Sbjct: 243 QSSASLPRKGTSEPDKEDQSPTSAK-----DFTVESQPLISFVEILLSHPCGSHFSRRLE 297
Query: 367 KNQVL 371
+ + +
Sbjct: 298 RQETI 302
>At3g58050 putative protein
Length = 1209
Score = 52.0 bits (123), Expect = 5e-07
Identities = 68/265 (25%), Positives = 116/265 (43%), Gaps = 29/265 (10%)
Query: 113 LRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDE 172
L + + ++ I++L+ QVK LE+E+ ++ EE+ E + E +L + E+ + E
Sbjct: 511 LEQHLHVACKEIITLEKQVKLLEEEEKEKREEEERKEKKRSKEREKKLRKKER----LKE 566
Query: 173 PVPAIRRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPG 232
+ + ++ D LL ++ EE D N +N+ S ++TG D+ S P
Sbjct: 567 KDKGKEKKNPECSDKDMLLNSSREEEDLPNLYDETNNTINSEESEIETGYADL---SPP- 622
Query: 233 HGSGSVQVK---SGLNEP-------DQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPS 282
GS VQ + G P D+ R K +E+E N + N+ K V +
Sbjct: 623 -GSPDVQERQCLDGCPSPRAENHYCDRPDRDIKDLEDE--NVYFTNDHQKPVHQNA---R 676
Query: 283 EERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKS 342
++V+ D+ R R S+ S S S+ R + R EV N + V S
Sbjct: 677 YWKEVQSDNALR--WSDKRRYSDNASFVSRSEA-RYRNDRLEVPSRGFNGSNRQLRVNAS 733
Query: 343 EPLFGVLEMIKRHQKFSLFERRLEK 367
+ G L IK H+KF + R+ +
Sbjct: 734 KT--GGLNGIKSHEKFQCCDNRISE 756
>At3g28770 hypothetical protein
Length = 2081
Score = 48.1 bits (113), Expect = 8e-06
Identities = 51/246 (20%), Positives = 105/246 (41%), Gaps = 28/246 (11%)
Query: 107 KQRVAELRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKT 166
KQ A++ D S+ + + + E++K K NE +V + + + + EK
Sbjct: 729 KQEEAQIYGGESKDDKSVEAKGKKKESKENKKTKTNENRVRNKEENVQGNKKESEKVEKG 788
Query: 167 GGDIDEPVPAIRRLDE----STTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGE 222
+ ++ D ST N D+ +GE++ D + + S ++ + + G
Sbjct: 789 EKKESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKEDKEESKDYQSVEAK-EKNENGG 847
Query: 223 VDVKLGSGPGH----GSGSVQVKSGLNEPDQTGRKRKSVEEESNNGS-----YDNN---E 270
VD +G+ SV+VK+ N+ + +KR+ V+ + + + NN +
Sbjct: 848 VDTNVGNKEDSKDLKDDRSVEVKA--NKEESMKKKREEVQRNDKSSTKEVRDFANNMDID 905
Query: 271 AKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDA 330
+ + ESV ++ K E G E+ + ++SS K + ++++KE +
Sbjct: 906 VQKGSGESVKYKKDEKKE---------GNKEENKDTINTSSKQKGKDKKKKKKESKNSNM 956
Query: 331 KMENED 336
K + ED
Sbjct: 957 KKKEED 962
Score = 45.4 bits (106), Expect = 5e-05
Identities = 57/295 (19%), Positives = 107/295 (35%), Gaps = 47/295 (15%)
Query: 65 EQKFHDLNRRFKDDVPPPQQNGDVSAVTAEDSDHVPWLDKLRKQRVAELRRDVQLSDVSI 124
++K H+ N+ K + ++ + + + + ++KL Q + + D S
Sbjct: 1083 DKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKS- 1141
Query: 125 LSLQLQVKKLEDEKAKENEEKVET-EPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDES 183
++L K+ + ++ KENEEK ET E + + S + + + EK
Sbjct: 1142 QHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEK------------------ 1183
Query: 184 TTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSG 243
+S +D Q E S LK E D K + + K
Sbjct: 1184 -------------KSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKE 1230
Query: 244 LNEP--------DQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRS 295
N+P Q+G K++S+E ES +S E ++ DS S
Sbjct: 1231 KNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADS 1290
Query: 296 EGGGTRESSE------VQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKSEP 344
+S E +Q+ S + R + RK+ + + ++ K++P
Sbjct: 1291 HSDSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKP 1345
Score = 43.9 bits (102), Expect = 1e-04
Identities = 51/262 (19%), Positives = 107/262 (40%), Gaps = 34/262 (12%)
Query: 103 DKLRKQRVAELRRDVQLSDVSILSLQLQVKKLEDE----------KAKENEEKVETEPDL 152
DK R+++ +E R+ + + S L+ KK E+E K+K+ E+K E E +
Sbjct: 1035 DKKREEKDSEERKSKKEKEES---RDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNK 1091
Query: 153 AVSGEGRLPENEK----TGGDIDEPVPAIRRLDESTTNTDK----------LLPAAGEES 198
++ E E +K +E + +L++ +N K + +ES
Sbjct: 1092 SMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKES 1151
Query: 199 DRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVE 258
D+ + NE S ++ K+ + +V + + + E ++ K+ +
Sbjct: 1152 DKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEED 1211
Query: 259 EESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRES--SEVQSSSSLSKTR 316
+ +N + K E P +++K T + GG +ES SE + + + K++
Sbjct: 1212 RKKQTSVEENKKQKETKKEKNKPKDDKK-----NTTKQSGGKKESMESESKEAENQQKSQ 1266
Query: 317 RTQRRRKEVSGEDAKMENEDVA 338
T + + S + M+ + A
Sbjct: 1267 ATTQADSDESKNEILMQADSQA 1288
Score = 34.3 bits (77), Expect = 0.11
Identities = 44/238 (18%), Positives = 92/238 (38%), Gaps = 28/238 (11%)
Query: 133 KLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLP 192
KL++E K+N+EK E+E + + E + E +K+ + +E+ K
Sbjct: 988 KLKEEN-KDNKEKKESEDSASKNREKKEYEEKKS-----------KTKEEAKKEKKKSQD 1035
Query: 193 AAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGR 252
EE D + + + A K E + H KS E +
Sbjct: 1036 KKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENH-------KSKKKEDKKEHE 1088
Query: 253 KRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSL 312
KS+++E + +E + + K+ED + + +E + S
Sbjct: 1089 DNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKK-----KEDKNEKKKSQH 1143
Query: 313 SKTRRTQRRRKEVSGEDAKMENEDVAVVKSEPLFGVLEMIKRHQKFSLFERRLEKNQV 370
K + + +KE + K E +++ KS+ E+ K+ +K S +++ ++ ++
Sbjct: 1144 VKLVKKESDKKEKKENEEKSETKEIESSKSQK----NEVDKKEKKSSKDQQKKKEKEM 1197
Score = 30.4 bits (67), Expect = 1.6
Identities = 49/255 (19%), Positives = 91/255 (35%), Gaps = 62/255 (24%)
Query: 135 EDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAA 194
ED++ ++ + VE + G N++ D+ + R E N ++ +
Sbjct: 826 EDKEESKDYQSVEAKEKNENGGVDTNVGNKEDSKDLKDD-----RSVEVKANKEESMKKK 880
Query: 195 GEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPD------ 248
EE R N+ +ST D ++DV+ GSG + K G E +
Sbjct: 881 REEVQR-----NDKSSTKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKEENKDTINT 935
Query: 249 ---QTGRKRKSVEEESNNGS----------YDNNEAKAV--------------------- 274
Q G+ +K ++ES N + Y NNE K
Sbjct: 936 SSKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKD 995
Query: 275 ------TCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGE 328
+ +S + E+K ++ ++++ +E + Q K ++ +KE
Sbjct: 996 NKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKE---- 1051
Query: 329 DAKMENEDVAVVKSE 343
K E+ D+ K E
Sbjct: 1052 --KEESRDLKAKKKE 1064
>At1g56660 hypothetical protein
Length = 522
Score = 46.6 bits (109), Expect = 2e-05
Identities = 47/219 (21%), Positives = 88/219 (39%), Gaps = 33/219 (15%)
Query: 125 LSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDEST 184
+ ++++ K +E KAK++EE SG+ + + +K G ++D S
Sbjct: 33 VEMEVKAKSIEKVKAKKDEES---------SGKSKKDKEKKKGKNVD-----------SE 72
Query: 185 TNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGL 244
DK ++ +D + V++ + G L+ E DVK+ + K
Sbjct: 73 VKEDK-----DDDKKKDGKMVSKKHEEG--HGDLEVKESDVKVEEHEKEHKKGKEKKHEE 125
Query: 245 NEPDQTGRKRKSVEEESNNGSYDNNEA--KAVTCESVPPSEERKVEDD----SVTRSEGG 298
E ++ G+K+K+ +E+ +G + N+ K E V +E E+D +
Sbjct: 126 LEEEKEGKKKKNKKEKDESGPEEKNKKADKEKKHEDVSQEKEELEEEDGKKNKKKEKDES 185
Query: 299 GTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDV 337
GT E + K K+V G+ K E D+
Sbjct: 186 GTEEKKKKPKKEKKQKEESKSNEDKKVKGKKEKGEKGDL 224
Score = 37.0 bits (84), Expect = 0.017
Identities = 48/232 (20%), Positives = 92/232 (38%), Gaps = 29/232 (12%)
Query: 131 VKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDE----PVPAIRRLDESTTN 186
++K ++EK KE++E T+ ++ + + EK +E P + DEST
Sbjct: 224 LEKEDEEKKKEHDE---TDQEMKEKDSKKNKKKEKDESCAEEKKKKPDKEKKEKDESTEK 280
Query: 187 TDKLLPAA---GEESDRDNQS--VNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQV- 240
DK L GE+ +++++ E ++T D + K V
Sbjct: 281 EDKKLKGKKGKGEKPEKEDEGKKTKEHDATEQEMDDEAADHKEGKKKKNKDKAKKKETVI 340
Query: 241 -----KSGLNEPDQTG----RKRKSVEEESNNGSYDNNEAKAVT--CESVPPSEERKVED 289
K ++ D G +K K E++S G D E K E+ S + K+E+
Sbjct: 341 DEVCEKETKDKDDDEGETKQKKNKKKEKKSEKGEKDVKEDKKKENPLETEVMSRDIKLEE 400
Query: 290 DSVTRSEGGGTRESSEV-----QSSSSLSKTRRTQRRRKEVSGEDAKMENED 336
+ E T E + +S K ++ +++ K+ ++ KM ++
Sbjct: 401 PEAEKKEEDDTEEKKKSKVEGGESEEGKKKKKKDKKKNKKKDTKEPKMTEDE 452
Score = 34.7 bits (78), Expect = 0.086
Identities = 52/256 (20%), Positives = 97/256 (37%), Gaps = 47/256 (18%)
Query: 87 DVSAVTAEDSDHVPWLDKLRKQRVAELRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKV 146
+V + ED D DK + ++ + + D+ + ++V++ E E K E+K
Sbjct: 68 NVDSEVKEDKDD----DKKKDGKMVSKKHEEGHGDLEVKESDVKVEEHEKEHKKGKEKKH 123
Query: 147 ETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQSVN 206
E +L EG+ +N+K + DE P +E DK ++ ++ V+
Sbjct: 124 E---ELEEEKEGKKKKNKK---EKDESGP-----EEKNKKADK---------EKKHEDVS 163
Query: 207 ESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSY 266
+ D K + + + +SG E + +K K +EES +
Sbjct: 164 QEKEELEEEDGKKNKKKE--------------KDESGTEEKKKKPKKEKKQKEESKSNED 209
Query: 267 DNNEAKAVTCES---VPPSEERKVEDDSV---TRSEGGGTRESSEVQSSSSLSKTRRTQR 320
+ K E EE+K E D + + + E S + K ++ +
Sbjct: 210 KKVKGKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKKEKDESCAEEKKKKPDK 269
Query: 321 RRKEVSGEDAKMENED 336
+KE +D E ED
Sbjct: 270 EKKE---KDESTEKED 282
>At4g37820 unknown protein
Length = 532
Score = 44.7 bits (104), Expect = 8e-05
Identities = 43/210 (20%), Positives = 83/210 (39%), Gaps = 14/210 (6%)
Query: 130 QVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDID-EPVPAIRRLDESTTNTD 188
Q K EDEK K + E++ V G+ ++ + D E P ++ +ES++ +
Sbjct: 292 QTKNEEDEKEKVQSSEEESK----VKESGKNEKDASSSQDESKEEKPERKKKEESSSQGE 347
Query: 189 KLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPD 248
G+E + + + +S+S + + S + ++K
Sbjct: 348 ------GKEEEPEKREKEDSSSQEESKEEEPENKEKEASSSQEENEIKETEIKEKEESSS 401
Query: 249 QTGRKRKSVEE---ESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSE 305
Q G + K E+ ES N+E K ES S +K ++ S+ ++S
Sbjct: 402 QEGNENKETEKKSSESQRKENTNSEKKIEQVESTDSSNTQKGDEQKTDESKRESGNDTSN 461
Query: 306 VQSSSSLSKTRRTQRRRKEVSGEDAKMENE 335
++ SKT ++ +GE + +NE
Sbjct: 462 KETEDDSSKTESEKKEENNRNGETEETQNE 491
Score = 40.0 bits (92), Expect = 0.002
Identities = 43/213 (20%), Positives = 78/213 (36%), Gaps = 4/213 (1%)
Query: 135 EDEKAKENEE---KVETEPDLAVSGEGRLP-ENEKTGGDIDEPVPAIRRLDESTTNTDKL 190
EDE +NE V E L + G E ++T ++D D + +
Sbjct: 204 EDESGPKNEVLEGSVIKEVSLNTTENGSDDGEQQETKSELDSKTGEKGFSDSNGELPETN 263
Query: 191 LPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQT 250
L + ++ +ES S+G +T + + S +SG NE D +
Sbjct: 264 LSTSNATETTESSGSDESGSSGKSTGYQQTKNEEDEKEKVQSSEEESKVKESGKNEKDAS 323
Query: 251 GRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSS 310
+ +S EE+ + + ++ E P E++ E + E SS
Sbjct: 324 SSQDESKEEKPERKKKEESSSQGEGKEEEPEKREKEDSSSQEESKEEEPENKEKEASSSQ 383
Query: 311 SLSKTRRTQRRRKEVSGEDAKMENEDVAVVKSE 343
++ + T+ + KE S EN++ SE
Sbjct: 384 EENEIKETEIKEKEESSSQEGNENKETEKKSSE 416
Score = 38.9 bits (89), Expect = 0.005
Identities = 44/210 (20%), Positives = 88/210 (40%), Gaps = 29/210 (13%)
Query: 137 EKAKENEEKVETEPDL-AVSGE-------GRLPENEKTGGDIDEPVPAIRRLDES----- 183
E ++ E+ ET+ +L + +GE G LPE + + E + DES
Sbjct: 228 ENGSDDGEQQETKSELDSKTGEKGFSDSNGELPETNLSTSNATETTESSGS-DESGSSGK 286
Query: 184 ------TTNTD---KLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHG 234
T N + + + ++ EES NE +++ S+ ++ + K G
Sbjct: 287 STGYQQTKNEEDEKEKVQSSEEESKVKESGKNEKDASSSQDESKEEKPERKKKEESSSQG 346
Query: 235 SGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTR 294
G + EP++ ++ S +EES +N E +A + + +E ++++ +
Sbjct: 347 EGKEE------EPEKREKEDSSSQEESKEEEPENKEKEASSSQEENEIKETEIKEKEESS 400
Query: 295 SEGGGTRESSEVQSSSSLSKTRRTQRRRKE 324
S+ G + +E +SS S K ++ E
Sbjct: 401 SQEGNENKETEKKSSESQRKENTNSEKKIE 430
Score = 35.0 bits (79), Expect = 0.066
Identities = 30/107 (28%), Positives = 46/107 (42%), Gaps = 4/107 (3%)
Query: 248 DQTGRKRKSVEEES---NNGSYDNNEAKAVTCESVPPSEERK-VEDDSVTRSEGGGTRES 303
++T + VE+E N G D + K E V EE K VE+++ +EG G E
Sbjct: 74 EETKDVKDEVEDEEGSKNEGGGDVSTDKENGDEIVEREEEEKAVEENNEKEAEGTGNEEG 133
Query: 304 SEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKSEPLFGVLE 350
+E ++ K E+S E+A+ N SE + G E
Sbjct: 134 NEDSNNGESEKVVDESEGGNEISNEEAREINYKGDDASSEVMHGTEE 180
>At1g29470 unknown protein
Length = 768
Score = 44.7 bits (104), Expect = 8e-05
Identities = 42/158 (26%), Positives = 67/158 (41%), Gaps = 28/158 (17%)
Query: 181 DESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPG-HGSGSVQ 239
DE T+ + E++D + E NS A + E D K G G +G G
Sbjct: 78 DEKNEETEVVTETNEEKTDPEKSG--EENSGEKTESAEERKEFDDKNGDGDRKNGDGEKD 135
Query: 240 VKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGG 299
+S E D+T +K K+ EES SEE K ED + T G
Sbjct: 136 TES---ESDETKQKEKTQLEES--------------------SEENKSEDSNGTEENAGE 172
Query: 300 TRESSEVQSSSSLSKTRRTQRRRKEV--SGEDAKMENE 335
+ E++E +S + +T + + K+V +G+ A++ E
Sbjct: 173 SEENTEKKSEENAGETEESTEKSKDVFPAGDQAEITKE 210
Score = 28.9 bits (63), Expect = 4.7
Identities = 22/93 (23%), Positives = 40/93 (42%), Gaps = 8/93 (8%)
Query: 247 PDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSV---TRSEGGGTRES 303
PD+ + + V E + + + + E +EERK DD R G G
Sbjct: 77 PDEKNEETEVVTETNEEKTDPEKSGEENSGEKTESAEERKEFDDKNGDGDRKNGDG---- 132
Query: 304 SEVQSSSSLSKTRRTQRRRKEVSGEDAKMENED 336
E + S +T++ ++ + E S E+ K E+ +
Sbjct: 133 -EKDTESESDETKQKEKTQLEESSEENKSEDSN 164
>At3g53350 unknown protein
Length = 396
Score = 41.2 bits (95), Expect = 0.001
Identities = 43/205 (20%), Positives = 84/205 (40%), Gaps = 22/205 (10%)
Query: 158 GRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGEES-DRDNQSVNESNSTGSRFD 216
GR+PE E T + E +++ E ++ L A EE+ D +Q ++ + S SR +
Sbjct: 71 GRIPELESTISQLQEE---LKKAKEELNRSEALKREAQEEAEDAKHQLMDINASEDSRIE 127
Query: 217 AL--------KTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDN 268
L KT + +++ HG S + S +NE + K E E Y+
Sbjct: 128 ELRKLSQERDKTWQSELEAMQRQ-HGMDSTALSSAINEVQKLKSKLFESESELEQSKYEV 186
Query: 269 NEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGE 328
+ + R++E++ V + + E E++ + +LS+ TQ + + E
Sbjct: 187 RSLEKLV---------RQLEEERVNSRDSSSSMEVEELKEAMNLSRQEITQLKSAVEAAE 237
Query: 329 DAKMENEDVAVVKSEPLFGVLEMIK 353
E + ++ + E +K
Sbjct: 238 TRYQEEYIQSTLQIRSAYEQTEAVK 262
>At5g55660 putative protein
Length = 759
Score = 40.8 bits (94), Expect = 0.001
Identities = 40/187 (21%), Positives = 75/187 (39%), Gaps = 6/187 (3%)
Query: 138 KAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGEE 197
K KE EK+ E L + + T D + L++ TD L+ +
Sbjct: 404 KVKEKFEKINKEKLLEFCDLFDISVAKATTKKEDIVTKLVEFLEKPHATTDVLVNEKEKG 463
Query: 198 SDR------DNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTG 251
R + + S+S S KT E H + + +E ++
Sbjct: 464 VKRKRTPKKSSPAAGSSSSKRSAKSQKKTEEATRTNKKSVAHSDDESEEEKEDDEEEEKE 523
Query: 252 RKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSS 311
++ + EEE+ NG D +E +A + E + E + T+ + G+R SS+ + S+
Sbjct: 524 QEVEEEEEENENGIPDKSEDEAPQLSESEENVESEEESEEETKKKKRGSRTSSDKKESAG 583
Query: 312 LSKTRRT 318
S++++T
Sbjct: 584 KSRSKKT 590
>At3g51070 putative protein
Length = 895
Score = 40.4 bits (93), Expect = 0.002
Identities = 49/239 (20%), Positives = 95/239 (39%), Gaps = 17/239 (7%)
Query: 116 DVQLSDVSILSL---QLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDE 172
+ + SDVS S + + K E ++ + E+ PD AV E E K+ + E
Sbjct: 58 ETERSDVSASSNGNDEPEPTKQESDEQQAFEDNPGKLPDDAVKSED---EQRKSAKEKSE 114
Query: 173 PVPAIRRLDESTTNTD-KLLPAAGEESDRDNQSVNES-----NSTGSRFDALKTGEVDVK 226
+ + E+ N D K+ +++ ++NQ+V ES F+ + + D
Sbjct: 115 TTSSKTQTQETQQNNDDKISEEKEKDNGKENQTVQESEEGQMKKVVKEFEKEQKQQRDED 174
Query: 227 LGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNG---SYDNNEAKAVTCESVPPSE 283
G+ P G Q G +PD ++ E++SN + + + + SE
Sbjct: 175 AGTQPKGTQGQEQ-GQGKEQPDVEQGNKQGQEQDSNTDVTFTDATKQEQPMETGQGETSE 233
Query: 284 ERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKS 342
K E++ + G E+ + Q+ + + + K + E+ + E A +S
Sbjct: 234 TSKNEENGQPEEQNSGNEETGQ-QNEEKTTASEENGKGEKSMKDENGQQEEHTTAEEES 291
Score = 37.4 bits (85), Expect = 0.013
Identities = 49/214 (22%), Positives = 86/214 (39%), Gaps = 34/214 (15%)
Query: 131 VKKLEDEKAKENEEKVETEPDLAVS---GEGR----LPENEKTGGDIDEPVPAIRRLDES 183
VK+ E E+ ++ +E T+P G+G+ + + K G + D ++
Sbjct: 160 VKEFEKEQKQQRDEDAGTQPKGTQGQEQGQGKEQPDVEQGNKQGQEQDSNTDVT--FTDA 217
Query: 184 TTNTDKLLPAAGEESDRDNQSVN---ESNSTGSRFDALKTGEV-DVKLGSGPGHGSGSVQ 239
T + GE S+ N E ++G+ +TG+ + K + +G G
Sbjct: 218 TKQEQPMETGQGETSETSKNEENGQPEEQNSGNE----ETGQQNEEKTTASEENGKGEKS 273
Query: 240 VKSGLNEPDQTGRKRK--SVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEG 297
+K D+ G++ + + EEES N E E++ EERK E SE
Sbjct: 274 MK------DENGQQEEHTTAEEESGN----KEEESTSKDENMEQQEERKDEKKHEQGSEA 323
Query: 298 GG-----TRESSEVQSSSSLSKTRRTQRRRKEVS 326
G +ES+E Q S T ++++ S
Sbjct: 324 SGFGSGIPKESAESQKSWKSQATESKDEKQRQTS 357
>At2g25670 unknown protein
Length = 318
Score = 40.4 bits (93), Expect = 0.002
Identities = 41/202 (20%), Positives = 84/202 (41%), Gaps = 15/202 (7%)
Query: 134 LEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPA 193
+E+E+ +E E +V EP++ + E P E + + R + + LL
Sbjct: 123 VEEEQEQETEVQVHPEPEVKKAPEVPAPPKE-----AERQLSKKERKKKELAELEALLAD 177
Query: 194 AGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRK 253
G + ++N + ES G GE + K + G S + K + D+
Sbjct: 178 FG-VAPKENNGLEESQEAGQEKKEDVNGEGEKKENAAGGESKASKKKK----KKDKQKEV 232
Query: 254 RKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVE-DDSVTRSEGGGTRESSEVQSSSSL 312
++S E+++NN N +A S P EE ++ + + + ++S + +++
Sbjct: 233 KESQEQQANN----NADAVDEAAGSEPTEEESPIDVKERIKKLASMKKKKSGKEVDAAAK 288
Query: 313 SKTRRTQRRRKEVSGEDAKMEN 334
+ RRK+++ K +N
Sbjct: 289 AAAEEAAARRKKLAAAKKKEKN 310
>At2g15200 putative protein on transposon FARE2.7 (cds2)
Length = 650
Score = 40.4 bits (93), Expect = 0.002
Identities = 53/229 (23%), Positives = 81/229 (35%), Gaps = 36/229 (15%)
Query: 132 KKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLL 191
+K EDEK ++ EEK E E E L + E G + E ++ DE
Sbjct: 421 QKEEDEKKEQEEEKQEEE-----GKEEELEKVEYRGDEGKEKQEIPKQGDEEMEGE---- 471
Query: 192 PAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTG 251
EE + E R D K + K G G Q + G E ++
Sbjct: 472 ----EEKQEEEGKEEEEEKVEYRGDEGKEKQEIPKQGDEEMEGEEEKQEEEGKEEEEEKV 527
Query: 252 RKRKSVEEESNNGSYDNNEAKAVTC------------ESVP--------PSEERKVEDDS 291
K ++VEE + ++ EA + ES P P EE E D
Sbjct: 528 LKEENVEEHDEHDETEDQEAYVILSDDEDNGTTPTEKESQPQKEETTEVPKEENVEEHDK 587
Query: 292 VTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGE---DAKMENEDV 337
+E + + T + +R+KE + E + K ++EDV
Sbjct: 588 HDETEDQEAYVILSDDEDNGTAPTEKESQRQKEETTEVPRETKKDDEDV 636
>At5g64910 unknown protein
Length = 487
Score = 39.3 bits (90), Expect = 0.003
Identities = 35/155 (22%), Positives = 58/155 (36%), Gaps = 4/155 (2%)
Query: 194 AGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRK 253
A + +++ +V E+ +T +T E + K+ S G + ++ N+ ++ +
Sbjct: 23 ASSSASKNDDAVVEATTTQETQPTQETEETEDKVESPAPEEEGKNEEEANENQEEEAAKV 82
Query: 254 RKSVEEESNN---GSYDNNEAKAVTCESVPPSEERKVE-DDSVTRSEGGGTRESSEVQSS 309
EE N D E K EE V+ D+S ++ E SSE Q
Sbjct: 83 ESKAAEEGGNEEEAKEDKEEEKEEAAREDKEEEEEAVKPDESASQKEEAKGASSSEPQLR 142
Query: 310 SSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKSEP 344
K K+VS AK + SEP
Sbjct: 143 RGKRKRGTKTEAEKKVSTPRAKKRAKTTKAQASEP 177
>At1g76810 translation initiation factor IF-2 like protein
Length = 1280
Score = 39.3 bits (90), Expect = 0.003
Identities = 51/252 (20%), Positives = 95/252 (37%), Gaps = 25/252 (9%)
Query: 139 AKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGEES 198
++++E +E E V+ G+ ++K GG + V DE+ T+ K +
Sbjct: 224 SRDDENTIEDEESPEVTFSGKKKSSKKKGGSVLASVGDDSVADETKTSDTKNVEVVETGK 283
Query: 199 DRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPG----HGSGSVQVKSGLNEP------- 247
+ + N+S T + L ++ LG P S V+ K+ EP
Sbjct: 284 SKKKKKNNKSGRTVQEEEDL--DKLLAALGETPAAERPASSTPVEEKAAQPEPVAPVENA 341
Query: 248 -DQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVE--DDSVTRSEGGGTRESS 304
++ G + + ++ E KA + S E K E ++SVT +++
Sbjct: 342 GEKEGEEETAAAKKKKKKKEKEKEKKAAAAAAATSSVEVKEEKQEESVTEPLQPKKKDAK 401
Query: 305 EVQSSSSLSK---------TRRTQRRRKEVSGEDAKMENEDVAVVKSEPLFGVLEMIKRH 355
+ + K RR + ++ E+ K+ E+ + E L E KR
Sbjct: 402 GKAAEKKIPKHVREMQEALARRQEAEERKKKEEEEKLRKEEEERRRQEELEAQAEEAKRK 461
Query: 356 QKFSLFERRLEK 367
+K E+ L K
Sbjct: 462 RKEKEKEKLLRK 473
Score = 36.6 bits (83), Expect = 0.023
Identities = 44/246 (17%), Positives = 103/246 (40%), Gaps = 18/246 (7%)
Query: 103 DKLRKQRVAELRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGR-LP 161
++L Q R+ + +L +L+ K L ++ E +++ + L +G G +
Sbjct: 449 EELEAQAEEAKRKRKEKEKEKLLRKKLEGKLLTAKQKTEAQKREAFKNQLLAAGGGLPVA 508
Query: 162 ENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTG 221
+N+ P+ A ++ D + E ++NQ+ + T TG
Sbjct: 509 DNDGDATSSKRPIYANKKKSSRQKGIDTSVQGEDEVEPKENQA--DEQDTLGEVGLTDTG 566
Query: 222 EVDV--KLGSGPGHGSGSVQVKSGLNEPDQTGR---KRKSVEEESNNGSYDNNEAKAVTC 276
+VD+ + + G V ++G+ E D+ K + + G +D+ E
Sbjct: 567 KVDLIELVNTDENSGPADVAQENGVEEDDEEDEWDAKSWGTVDLNLKGDFDDEE------ 620
Query: 277 ESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENED 336
E P +++++ D+++++ G + V+++ + RT+R + +DA + +
Sbjct: 621 EEAQPVVKKELK-DAISKAHDSGKPLIAAVKATPEVEDATRTKRATR---AKDASKKGKG 676
Query: 337 VAVVKS 342
+A +S
Sbjct: 677 LAPSES 682
Score = 30.4 bits (67), Expect = 1.6
Identities = 40/205 (19%), Positives = 78/205 (37%), Gaps = 16/205 (7%)
Query: 132 KKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLL 191
KK + +K KE E+K S E + EK + EP+ ++ D +K +
Sbjct: 354 KKKKKKKEKEKEKKAAAAAAATSSVEVK---EEKQEESVTEPLQP-KKKDAKGKAAEKKI 409
Query: 192 PAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTG 251
P E + E+ + + K E + KL ++++ E
Sbjct: 410 PKHVRE-------MQEALARRQEAEERKKKEEEEKLRKEEEERRRQEELEAQAEEA---- 458
Query: 252 RKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSS 311
KRK E+E E K +T + +++R+ + + + GG ++ ++SS
Sbjct: 459 -KRKRKEKEKEKLLRKKLEGKLLTAKQKTEAQKREAFKNQLLAAGGGLPVADNDGDATSS 517
Query: 312 LSKTRRTQRRRKEVSGEDAKMENED 336
+++ G D ++ ED
Sbjct: 518 KRPIYANKKKSSRQKGIDTSVQGED 542
>At4g31880 unknown protein
Length = 873
Score = 38.5 bits (88), Expect = 0.006
Identities = 39/166 (23%), Positives = 71/166 (42%), Gaps = 15/166 (9%)
Query: 180 LDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQ 239
LDES + D+ E +D+ Q E ST KTG+ S GSG+
Sbjct: 660 LDESELSQDE------EAADQTGQE--EDASTVPLTKKAKTGKQSKMDNSSAKKGSGA-- 709
Query: 240 VKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDD-SVTRSEGG 298
G ++ T + S + + + + ++K + E SEE E++ T + G
Sbjct: 710 ---GSSKAKATPASKSSKTSQDDKTASKSKDSKEASREEEASSEEESEEEEPPKTVGKSG 766
Query: 299 GTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKSEP 344
+R ++ S S K++ + ++++E S +++ VKS P
Sbjct: 767 SSRSKKDISSVSKSGKSKASSKKKEEPSKATTSSKSKS-GPVKSVP 811
Score = 30.8 bits (68), Expect = 1.2
Identities = 48/213 (22%), Positives = 85/213 (39%), Gaps = 30/213 (14%)
Query: 118 QLSDVSILSLQLQVK-----KLEDEKAKENE----EKVETEPDLAVSGEGRLPENEKTGG 168
Q D S + L + K K+++ AK+ K + P S + +++KT
Sbjct: 677 QEEDASTVPLTKKAKTGKQSKMDNSSAKKGSGAGSSKAKATP---ASKSSKTSQDDKTAS 733
Query: 169 DIDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFD---ALKTGE--V 223
+ A R + S+ E + ++V +S S+ S+ D K+G+
Sbjct: 734 KSKDSKEASREEEASSEEES--------EEEEPPKTVGKSGSSRSKKDISSVSKSGKSKA 785
Query: 224 DVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSE 283
K P + S + KSG K K+ + ++ +GS +KA S SE
Sbjct: 786 SSKKKEEPSKATTSSKSKSG--PVKSVPAKSKTGKGKAKSGSASTPASKAKESASESESE 843
Query: 284 ERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTR 316
E E + T+++ G ++ S QS S + R
Sbjct: 844 ETPKEPEPATKAKSGKSQGS---QSKSGKKRKR 873
Score = 30.0 bits (66), Expect = 2.1
Identities = 47/230 (20%), Positives = 94/230 (40%), Gaps = 33/230 (14%)
Query: 76 KDDVPPPQQNGDVSAVTAEDSDHVPWLDKLRKQRVAELRRDVQLSDVSI-LSLQLQVKKL 134
KD + P + V+A T+ +++ + L + + +V ++ L Q KK
Sbjct: 393 KDVLTSPPVDSSVTAATSSENEKNKSVQILPSKTSGDETANVSSPSMAEELPEQSVPKKT 452
Query: 135 EDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAA 194
++K KE+ + E +P +++ E ++ E E T+ ++ +
Sbjct: 453 ANQKKKESSTE-EVKPSASIATE-----------EVSE---------EPNTSEPQVTKKS 491
Query: 195 GEE---SDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTG 251
G++ S + +V S + S K E V GS + Q + E +
Sbjct: 492 GKKVASSSKTKPTVPPSKKSTSETKVAKQSEKKVV-------GSDNAQESTKPKEEKKKP 544
Query: 252 RKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTR 301
+ K+++EES + S +NE AV+ + S+ +K +V S T+
Sbjct: 545 GRGKAIDEESLHTSSGDNEKPAVSSGKL-ASKSKKEAKQTVEESPNSNTK 593
>At3g05750 hypothetical protein
Length = 798
Score = 38.1 bits (87), Expect = 0.008
Identities = 25/104 (24%), Positives = 40/104 (38%)
Query: 240 VKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGG 299
VKSGL E + RK + V + + ++ T + G
Sbjct: 345 VKSGLKESSASTRKTVDKPNNQKQNQFAETSVSNQRGRKVMKKVNKVLVENGTTTKKPGF 404
Query: 300 TRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKSE 343
T S++ +SSSLS+ + R +K +G N D + K E
Sbjct: 405 TATSAKKSTSSSLSRKKNLSRSKKPANGVQEAGVNSDKRIKKGE 448
>At5g01400 putative protein
Length = 1467
Score = 37.7 bits (86), Expect = 0.010
Identities = 38/218 (17%), Positives = 83/218 (37%), Gaps = 16/218 (7%)
Query: 130 QVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDK 189
QV+ E + ++E +++ +EP + + + + +D P+ +S ++
Sbjct: 1253 QVQANETQTSQEQQQQQASEPQQ--TSQSQQVSVPLSHSQVDHQEPSQVVASQSQSSPIG 1310
Query: 190 LLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQ 249
+ +A +S S + S+ + TG ++ +G ++ N P
Sbjct: 1311 TVQSAMSQSQNSPIDTGRSEMSQSQNSPIDTGRSEMSQSQNSPIDTGRSEMSQSQNSPID 1370
Query: 250 TGRKRKSVEEES----------NNGSYDNNEAKAVTCESVP-PSEERKVEDDSVTRSEGG 298
TGR S + S G D ++ V+ S P P+ + D +
Sbjct: 1371 TGRSEMSESQSSPIGQSQSSPIGTGQSDMSQTPQVSDSSAPEPTSHTRTSDPQASSQT-- 1428
Query: 299 GTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENED 336
R+ E ++ S+ T+ + + S E+ + E E+
Sbjct: 1429 -LRDDDEKIDDTATSENEVTEIEKSKESSEEEEEEEEE 1465
>At3g57180 unknown protein
Length = 637
Score = 37.7 bits (86), Expect = 0.010
Identities = 30/111 (27%), Positives = 47/111 (42%), Gaps = 15/111 (13%)
Query: 245 NEPDQTG--RKRKSVEEESNNGSY-DNNEAKAVTCESVPPSEERKVEDDSV--------- 292
N+PD G +KRK + + +N+E EE + EDD +
Sbjct: 108 NDPDLPGYYQKRKVIANNLEGDEHVENDELAGFEMVDDDADEEEEGEDDEMDDEIKNAIE 167
Query: 293 ---TRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVV 340
+ SE G ES E + K R ++ RK+++ E+AK +N D V
Sbjct: 168 GSNSESESGFEWESDEWEEKKEEKKKRVSKTERKKIAREEAKKDNYDDVTV 218
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.306 0.126 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,782,835
Number of Sequences: 26719
Number of extensions: 405936
Number of successful extensions: 2077
Number of sequences better than 10.0: 251
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 207
Number of HSP's that attempted gapping in prelim test: 1800
Number of HSP's gapped (non-prelim): 430
length of query: 371
length of database: 11,318,596
effective HSP length: 101
effective length of query: 270
effective length of database: 8,619,977
effective search space: 2327393790
effective search space used: 2327393790
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146756.1


