

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146755.5 - phase: 0
(485 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
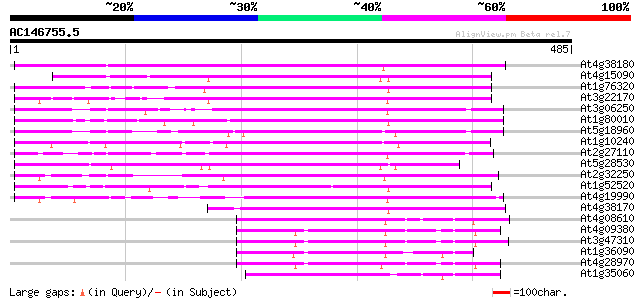
Score E
Sequences producing significant alignments: (bits) Value
At4g38180 unknown protein (At4g38180) 253 2e-67
At4g15090 unknown protein 206 3e-53
At1g76320 putative phytochrome A signaling protein 199 3e-51
At3g22170 far-red impaired response protein, putative 188 5e-48
At3g06250 unknown protein 182 4e-46
At1g80010 hypothetical protein 180 1e-45
At5g18960 FAR1 - like protein 177 2e-44
At1g10240 unknown protein 176 3e-44
At2g27110 Mutator-like transposase 174 1e-43
At5g28530 far-red impaired response protein (FAR1) - like 169 3e-42
At2g32250 Mutator-like transposase 167 1e-41
At1g52520 F6D8.26 165 5e-41
At4g19990 putative protein 159 4e-39
At4g38170 hypothetical protein 129 3e-30
At4g08610 predicted transposon protein 70 2e-12
At4g09380 putative protein 69 6e-12
At3g47310 putative protein 68 1e-11
At1g36090 hypothetical protein 68 1e-11
At4g28970 putative protein 67 2e-11
At1g35060 hypothetical protein 67 3e-11
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 253 bits (645), Expect = 2e-67
Identities = 139/432 (32%), Positives = 225/432 (51%), Gaps = 9/432 (2%)
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRNI-NGEVVQQTFLCHREGIREEKYINSTSRK 63
F E A FYN Y GF+ R S R+ +G ++Q+ F+C +EG R + R+
Sbjct: 79 FESEEAAKAFYNSYARRIGFSTRVSSSRRSRRDGAIIQRQFVCAKEGFRNMNEKRTKDRE 138
Query: 64 -REHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSEYDK 122
+ + ++R GC+A + V + S +W + F DHNH V L +HR++S K
Sbjct: 139 IKRPRTITRVGCKASLSVKMQ-DSGKWLVSGFVKDHNHELVPPDQVHCLRSHRQISGPAK 197
Query: 123 YQMNTMRQSGISTTRIHGYFASQAGGYQNVGYNRRDMYNEQRKRRMRW-NSDAEQAVNFL 181
++T++ +G+ RI + GG VG+ D N R R + + + +++L
Sbjct: 198 TLIDTLQAAGMGPRRIMSALIKEYGGISKVGFTEVDCRNYMRNNRQKSIEGEIQLLLDYL 257
Query: 182 KHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVI 241
+ M++ + F+ D S+ ++FW D + MD++ FGD + FD TY+ +Y P
Sbjct: 258 RQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDFTHFGDTVTFDTTYRSNRYRLPFAP 317
Query: 242 FSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAIRKV 301
F+GVNHH Q I+FG A I +ETE ++VWL ++ AM PVS+ TD D +R AI V
Sbjct: 318 FTGVNHHGQPILFGCAFIINETEASFVWLFNTWLAAMSAHPPVSITTDHDAVIRAAIMHV 377
Query: 302 FPEAHHRLCAWHLIRNATSN-----IKNLHFVSKFKDCLLGDVDVDVFQRKWEELVTEFG 356
FP A HR C WH+++ +K+ F S F C+ V+ F+R W L+ ++
Sbjct: 378 FPGARHRFCKWHILKKCQEKLSHVFLKHPSFESDFHKCVNLTESVEDFERCWFSLLDKYE 437
Query: 357 LEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVSALTNLHDFFQQF 416
L ++ W+ +Y R+ W + R FFA T R + ++S F Y++A TNL FF+ +
Sbjct: 438 LRDHEWLQAIYSDRRQWVPVYLRDTFFADMSLTHRSDSINSYFDGYINASTNLSQFFKLY 497
Query: 417 FRWLNYMRYREI 428
+ L +E+
Sbjct: 498 EKALESRLEKEV 509
>At4g15090 unknown protein
Length = 768
Score = 206 bits (523), Expect = 3e-53
Identities = 112/388 (28%), Positives = 199/388 (50%), Gaps = 14/388 (3%)
Query: 38 EVVQQTFLCHREGIREEKYINSTSRKREHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDD 97
+ + F C R G+ E + +S +R + C V+ D +W I F D
Sbjct: 32 DFIDAKFACSRYGVTPESESSGSSSRRSTVKKTDCKASMHVKRRPD---GKWIIHEFVKD 88
Query: 98 HNHSFVKE-KFERMLPAHRKMSEYDKYQMNTMRQSGISTTRIHGYFASQAGGYQNVG-YN 155
HNH + + + + K++E K ++ + T +++ + Q+GGY+N+G
Sbjct: 89 HNHELLPALAYHFRIQRNVKLAE--KNNIDILHAVSERTKKMYVEMSRQSGGYKNIGSLL 146
Query: 156 RRDMYNEQRKRRMRW--NSDAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVS 213
+ D+ ++ K R D++ + + K + ++ F+ ++ D L++LFW D S
Sbjct: 147 QTDVSSQVDKGRYLALEEGDSQVLLEYFKRIKKENPKFFYAIDLNEDQRLRNLFWADAKS 206
Query: 214 CMDYSIFGDVLAFDATYKKIKYNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKI 273
DY F DV++FD TY K PL +F GVNHH+Q ++ G A++ DE+ +T+VWL+K
Sbjct: 207 RDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQPMLLGCALVADESMETFVWLIKT 266
Query: 274 FVEAMGGKLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHLIRNAT---SNIKNLH--FV 328
++ AMGG+ P ++TD D + +A+ ++ P H WH++ S++ H F+
Sbjct: 267 WLRAMGGRAPKVILTDQDKFLMSAVSELLPNTRHCFALWHVLEKIPEYFSHVMKRHENFL 326
Query: 329 SKFKDCLLGDVDVDVFQRKWEELVTEFGLEENPWMLEMYQKRKMWAAAHFRGKFFAGFRT 388
KF C+ D F +W ++V++FGLE + W+L +++ R+ W F AG T
Sbjct: 327 LKFNKCIFRSWTDDEFDMRWWKMVSQFGLENDEWLLWLHEHRQKWVPTFMSDVFLAGMST 386
Query: 389 TSRCEGLHSEFGKYVSALTNLHDFFQQF 416
+ R E ++S F KY+ L +F +Q+
Sbjct: 387 SQRSESVNSFFDKYIHKKITLKEFLRQY 414
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 199 bits (506), Expect = 3e-51
Identities = 123/420 (29%), Positives = 195/420 (46%), Gaps = 20/420 (4%)
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRN-INGEVVQQTFLCHREGIREEKYINSTSRK 63
F E A++FY Y GF K R+ + E + F C R G +++ R
Sbjct: 3 FETHEDAYLFYKDYAKSVGFGTAKLSSRRSRASKEFIDAKFSCIRYGSKQQSDDAINPRA 62
Query: 64 REHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSEYDKY 123
+ GC+A + V +WY+ F +HNH + E+ +HR
Sbjct: 63 SP-----KIGCKASMHVKRRPDG-KWYVYSFVKEHNHDLLPEQ-AHYFRSHRNTELVKSN 115
Query: 124 QMNTMRQSGISTTRIHGYFASQAGGYQNVGYNRRDMYNEQRKRR--MRWNSDAEQAVNFL 181
R+ T Y ++ + M N+ K R + DAE + FL
Sbjct: 116 DSRLRRKKNTPLTD-----CKHLSAYHDLDFIDGYMRNQHDKGRRLVLDTGDAEILLEFL 170
Query: 182 KHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVI 241
M ++ F+ D L+++FW D DY F DV++F+ +Y KY PLV+
Sbjct: 171 MRMQEENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVSKYKVPLVL 230
Query: 242 FSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAIRKV 301
F GVNHH Q ++ G ++ D+T TYVWL++ ++ AMGG+ P ++TD + +++ AI V
Sbjct: 231 FVGVNHHVQPVLLGCGLLADDTVYTYVWLMQSWLVAMGGQKPKVMLTDQNNAIKAAIAAV 290
Query: 302 FPEAHHRLCAWHLIRNATSNIKNLH-----FVSKFKDCLLGDVDVDVFQRKWEELVTEFG 356
PE H C WH++ N+ F+ K C+ + F R+W +L+ +F
Sbjct: 291 LPETRHCYCLWHVLDQLPRNLDYWSMWQDTFMKKLFKCIYRSWSEEEFDRRWLKLIDKFH 350
Query: 357 LEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVSALTNLHDFFQQF 416
L + PWM +Y++RK WA RG FAG R E ++S F +YV T+L +F + +
Sbjct: 351 LRDVPWMRSLYEERKFWAPTFMRGITFAGLSMRCRSESVNSLFDRYVHPETSLKEFLEGY 410
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 188 bits (478), Expect = 5e-48
Identities = 124/433 (28%), Positives = 206/433 (46%), Gaps = 47/433 (10%)
Query: 5 FPDREVAFMFYNWYGCFHGF--AARKSRLIRNINGEVVQQTFLCHREGIREEKYINSTSR 62
F A+ FY Y GF A + SR + E + F C R G + E Y S +R
Sbjct: 76 FESHGEAYSFYQEYSRAMGFNTAIQNSRRSKTTR-EFIDAKFACSRYGTKRE-YDKSFNR 133
Query: 63 KREH------------KPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERM 110
R + ++ C+A + V +W I HSFV+E +
Sbjct: 134 PRARQSKQDPENMAGRRTCAKTDCKASMHVKRRPDG-KWVI--------HSFVREHNHEL 184
Query: 111 LPAHRKMSEYDKYQMNTMRQSGISTTRIHGYFASQAGGYQNVGYNRRDMYNEQRKRRMRW 170
LPA + +SE T +I+ A Q Y+ V + D + K R
Sbjct: 185 LPA-QAVSE--------------QTRKIYAAMAKQFAEYKTVISLKSDSKSSFEKGRTLS 229
Query: 171 --NSDAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDA 228
D + ++FL M S + F+ + D ++++FW D S +Y F DV++ D
Sbjct: 230 VETGDFKILLDFLSRMQSLNSNFFYAVDLGDDQRVKNVFWVDAKSRHNYGSFCDVVSLDT 289
Query: 229 TYKKIKYNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVIT 288
TY + KY PL IF GVN H Q ++ G A+I DE+ TY WL++ ++ A+GG+ P +IT
Sbjct: 290 TYVRNKYKMPLAIFVGVNQHYQYMVLGCALISDESAATYSWLMETWLRAIGGQAPKVLIT 349
Query: 289 DGDLSMRNAIRKVFPEAHHRLCAWHLIRNATSNIKNL-----HFVSKFKDCLLGDVDVDV 343
+ D+ M + + ++FP H L WH++ + N+ + +F+ KF+ C+ +
Sbjct: 350 ELDVVMNSIVPEIFPNTRHCLFLWHVLMKVSENLGQVVKQHDNFMPKFEKCIYKSGKDED 409
Query: 344 FQRKWEELVTEFGLEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYV 403
F RKW + + FGL+++ WM+ +Y+ RK WA + AG T+ R + +++ F KY+
Sbjct: 410 FARKWYKNLARFGLKDDQWMISLYEDRKKWAPTYMTDVLLAGMSTSQRADSINAFFDKYM 469
Query: 404 SALTNLHDFFQQF 416
T++ +F + +
Sbjct: 470 HKKTSVQEFVKVY 482
>At3g06250 unknown protein
Length = 764
Score = 182 bits (462), Expect = 4e-46
Identities = 113/431 (26%), Positives = 195/431 (45%), Gaps = 56/431 (12%)
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRN-INGEVVQQTFLCHREGIREEKYINSTSRK 63
F A FY Y GF R +L R+ ++G + + F+C +EG +
Sbjct: 196 FNSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITSRRFVCSKEGFQHP--------- 246
Query: 64 REHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRK-----MS 118
SR GC A +R+ S W + + DHNH K + +
Sbjct: 247 ------SRMGCGAYMRIKRQ-DSGGWIVDRLNKDHNHDLEPGKKNAGMKKITDDVTGGLD 299
Query: 119 EYDKYQMNTMRQSGISTTRIHGYFASQAGGYQNVGYNRRDMYNEQRKRRMRWNSDAEQAV 178
D ++N + + IS+TR + +G ++ Y +
Sbjct: 300 SVDLIELNDL-SNHISSTREN-----------TIG---KEWY--------------PVLL 330
Query: 179 NFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTP 238
++ + ++D F+ + ++GS +FW D S S FGD + FD +Y+K Y+ P
Sbjct: 331 DYFQSKQAEDMGFFYAIELDSNGSCMSIFWADSRSRFACSQFGDAVVFDTSYRKGDYSVP 390
Query: 239 LVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAI 298
F G NHH Q ++ G A++ DE+++ + WL + ++ AM G+ P S++ D DL ++ A+
Sbjct: 391 FATFIGFNHHRQPVLLGGALVADESKEAFSWLFQTWLRAMSGRRPRSMVADQDLPIQQAV 450
Query: 299 RKVFPEAHHRLCAWHLIRNATSNIKNL--HFVSKFKDCLLGDVDVDVFQRKWEELVTEFG 356
+VFP HHR AW + N+++ F +++ CL F W LV ++G
Sbjct: 451 AQVFPGTHHRFSAWQIRSKERENLRSFPNEFKYEYEKCLYQSQTTVEFDTMWSSLVNKYG 510
Query: 357 LEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVSALTNLHDFFQQF 416
L +N W+ E+Y+KR+ W A+ R FF G + +G +++LT+L +F ++
Sbjct: 511 LRDNMWLREIYEKREKWVPAYLRASFFGGIHVDGTFDPF---YGTSLNSLTSLREFISRY 567
Query: 417 FRWLNYMRYRE 427
+ L R E
Sbjct: 568 EQGLEQRREEE 578
Score = 38.9 bits (89), Expect = 0.006
Identities = 28/97 (28%), Positives = 41/97 (41%), Gaps = 17/97 (17%)
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRN-INGEVVQQTFLCHREGIREEKYINSTSRK 63
F E A +YN Y GF R +L R+ +G V + F+C +EG +
Sbjct: 34 FDTAEEARDYYNSYATRTGFKVRTGQLYRSRTDGTVSSRRFVCSKEGFQLN--------- 84
Query: 64 REHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNH 100
SR GC A +RV + +W + +HNH
Sbjct: 85 ------SRTGCPAFIRVQ-RRDTGKWVLDQIQKEHNH 114
>At1g80010 hypothetical protein
Length = 696
Score = 180 bits (457), Expect = 1e-45
Identities = 128/440 (29%), Positives = 206/440 (46%), Gaps = 27/440 (6%)
Query: 5 FPDREVAFMFYNWYGCFHGFAAR-KSRLIRNINGEVVQQTFLCHREGIREEKYINSTSRK 63
F + A+ FYN Y GFA R KS + + E C+ +G + K ++ SR+
Sbjct: 72 FESYDDAYSFYNSYARELGFAIRVKSSWTKRNSKEKRGAVLCCNCQGFKLLK--DAHSRR 129
Query: 64 REHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSEYDKY 123
+E +R GCQA +R+ + + RW + DHNHSF ++ +H+K S
Sbjct: 130 KE----TRTGCQAMIRLRL-IHFDRWKVDQVKLDHNHSFDPQRAHNS-KSHKKSSSSASP 183
Query: 124 QMNTMRQSG----ISTTRIHGYFASQAGGYQNVGYNRR-------DMYNEQRKRRMRWNS 172
T + + T +++ A + D + R+ +R
Sbjct: 184 ATKTNPEPPPHVQVRTIKLYRTLALDTPPALGTSLSSGETSDLSLDHFQSSRRLELRGGF 243
Query: 173 DAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKK 232
A Q F +SS + + + DGSL+++FW D + YS FGDVL FD T
Sbjct: 244 RALQDFFFQIQLSSPN--FLYLMDLADDGSLRNVFWIDARARAAYSHFGDVLLFDTTCLS 301
Query: 233 IKYNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDL 292
Y PLV F G+NHH +I+ G ++ D++ +TYVWL + ++ M G+ P IT+
Sbjct: 302 NAYELPLVAFVGINHHGDTILLGCGLLADQSFETYVWLFRAWLTCMLGRPPQIFITEQCK 361
Query: 293 SMRNAIRKVFPEAHHRLCAWHLIRNATSNIKNLH----FVSKFKDCLLGDVDVDVFQRKW 348
+MR A+ +VFP AHHRL H++ N ++ L F + G + V+ F+ W
Sbjct: 362 AMRTAVSEVFPRAHHRLSLTHVLHNICQSVVQLQDSDLFPMALNRVVYGCLKVEEFETAW 421
Query: 349 EELVTEFGLEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCE-GLHSEFGKYVSALT 407
EE++ FG+ N + +M+Q R++WA + + F AG T F YV T
Sbjct: 422 EEMIIRFGMTNNETIRDMFQDRELWAPVYLKDTFLAGALTFPLGNVAAPFIFSGYVHENT 481
Query: 408 NLHDFFQQFFRWLNYMRYRE 427
+L +F + + +L+ RE
Sbjct: 482 SLREFLEGYESFLDKKYTRE 501
>At5g18960 FAR1 - like protein
Length = 788
Score = 177 bits (448), Expect = 2e-44
Identities = 120/434 (27%), Positives = 193/434 (43%), Gaps = 59/434 (13%)
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRN-INGEVVQQTFLCHREGIREEKYINSTSRK 63
F A FY Y GF R +L R+ ++G + + F+C REG +
Sbjct: 217 FGSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITSRRFVCSREGFQHP--------- 267
Query: 64 REHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSEYDKY 123
SR GC A +R+ S W + + DHNH K
Sbjct: 268 ------SRMGCGAYMRIKRQ-DSGGWIVDRLNKDHNHDLEPGK----------------- 303
Query: 124 QMNTMRQSGISTTRIHGYFASQAGGYQNVGYNRRDMYNEQRKRRMRWNSDAEQAVNFL-K 182
+G+ G GG +V + + ++ R N ++ L
Sbjct: 304 ----KNDAGMKKIPDDG-----TGGLDSVDLIELNDFGNNHIKKTRENRIGKEWYPLLLD 354
Query: 183 HMSSK--DDMMFWRHTVHAD---GSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNT 237
+ S+ +DM F+ + V D GS +FW D + S FGD + FD +Y+K Y+
Sbjct: 355 YFQSRQTEDMGFF-YAVELDVNNGSCMSIFWADSRARFACSQFGDSVVFDTSYRKGSYSV 413
Query: 238 PLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNA 297
P G NHH Q ++ G A++ DE+++ ++WL + ++ AM G+ P S++ D DL ++ A
Sbjct: 414 PFATIIGFNHHRQPVLLGCAMVADESKEAFLWLFQTWLRAMSGRRPRSIVADQDLPIQQA 473
Query: 298 IRKVFPEAHHRLCAWHLIRNATSNIKNLHFVSKFK----DCLLGDVDVDVFQRKWEELVT 353
+ +VFP AHHR AW + N+ + F S+FK C+ + F W L+
Sbjct: 474 LVQVFPGAHHRYSAWQIREKERENL--IPFPSEFKYEYEKCIYQTQTIVEFDSVWSALIN 531
Query: 354 EFGLEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVSALTNLHDFF 413
++GL ++ W+ E+Y++R+ W A+ R FFAG E FG + ALT L +F
Sbjct: 532 KYGLRDDVWLREIYEQRENWVPAYLRASFFAGIPINGTIEPF---FGASLDALTPLREFI 588
Query: 414 QQFFRWLNYMRYRE 427
++ + L R E
Sbjct: 589 SRYEQALEQRREEE 602
Score = 40.8 bits (94), Expect = 0.002
Identities = 29/97 (29%), Positives = 41/97 (41%), Gaps = 17/97 (17%)
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRN-INGEVVQQTFLCHREGIREEKYINSTSRK 63
F E A FYN Y GF R +L R+ +G V + F+C +EG +
Sbjct: 49 FDTAEEAREFYNAYAARTGFKVRTGQLYRSRTDGTVSSRRFVCSKEGFQLN--------- 99
Query: 64 REHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNH 100
SR GC A +RV + +W + +HNH
Sbjct: 100 ------SRTGCTAFIRVQ-RRDTGKWVLDQIQKEHNH 129
>At1g10240 unknown protein
Length = 680
Score = 176 bits (446), Expect = 3e-44
Identities = 114/427 (26%), Positives = 204/427 (47%), Gaps = 22/427 (5%)
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRN--INGEVVQQTFLCHREGIREEKYINSTSR 62
F + A+ FY+ + GF+ R+ R + + ++ F+CHR G K ++
Sbjct: 54 FLTHDTAYEFYSTFAKRCGFSIRRHRTEGKDGVGKGLTRRYFVCHRAGNTPIKTLSEGKP 113
Query: 63 KREHKPLSRCGCQAKVRVH--IDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSEY 120
+R + SRCGCQA +R+ ++ S W + F + HNH ++ R LPA+R +S+
Sbjct: 114 QRNRRS-SRCGCQAYLRISKLTELGSTEWRVTGFANHHNHELLEPNQVRFLPAYRSISDA 172
Query: 121 DKYQMNTMRQSGISTTRIHGYFASQA---GGYQNVGYNRRDMYNEQRKRRMRWNSDAEQA 177
DK ++ ++GIS ++ + G+ + + +D+ N + + D +
Sbjct: 173 DKSRILMFSKTGISVQQMMRLLELEKCVEPGF--LPFTEKDVRNLLQSFKKLDPED--EN 228
Query: 178 VNFLKHMSS---KDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIK 234
++FL+ S KD + T+ A+ L+++ W S Y +FGD + FD T++
Sbjct: 229 IDFLRMCQSIKEKDPNFKFEFTLDANDKLENIAWSYASSIQSYELFGDAVVFDTTHRLSA 288
Query: 235 YNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSM 294
PL I+ GVN++ FG ++ DE ++ W L+ F M GK P +++TD ++ +
Sbjct: 289 VEMPLGIWVGVNNYGVPCFFGCVLLRDENLRSWSWALQAFTGFMNGKAPQTILTDHNMCL 348
Query: 295 RNAIRKVFPEAHHRLCAWHLIRNATSNIKNLHFVSKFKDC------LLGDVDVDVFQRKW 348
+ AI P H LC W ++ S N ++ D L V+ F+ W
Sbjct: 349 KEAIAGEMPATKHALCIWMVVGKFPSWF-NAGLGERYNDWKAEFYRLYHLESVEEFELGW 407
Query: 349 EELVTEFGLEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVSALTN 408
++V FGL N + +Y R +W+ + R F AG T R + +++ +++SA T
Sbjct: 408 RDMVNSFGLHTNRHINNLYASRSLWSLPYLRSHFLAGMTLTGRSKAINAFIQRFLSAQTR 467
Query: 409 LHDFFQQ 415
L F +Q
Sbjct: 468 LAHFVEQ 474
>At2g27110 Mutator-like transposase
Length = 851
Score = 174 bits (440), Expect = 1e-43
Identities = 112/419 (26%), Positives = 192/419 (45%), Gaps = 34/419 (8%)
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRNINGEVVQQTFLCHREGIREEKYINSTSRKR 64
F + A FY+ Y GF S+L+ +G V + F+C S+S KR
Sbjct: 55 FNSEKEAKSFYDEYSRQLGFT---SKLLPRTDGSVSVREFVC------------SSSSKR 99
Query: 65 EHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSEYDKYQ 124
+ LS C A VR+ + ++W + F +H H L R + +K
Sbjct: 100 SKRRLSE-SCDAMVRIELQ-GHEKWVVTKFVKEHTHGLASSNMLHCLRPRRHFANSEK-- 155
Query: 125 MNTMRQSGISTTRIHGYFASQAGGYQNVGYNRRDMYNEQRKRRMRWNSDAEQAVNFLKHM 184
+ Q G++ Y + A G M ++ R DA + + K M
Sbjct: 156 --SSYQEGVNVPSGMMYVSMDANSR---GARNASMATNTKRTIGR---DAHNLLEYFKRM 207
Query: 185 SSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVIFSG 244
+++ F+ + D + ++FW D S + Y+ FGD + D Y+ ++ P F+G
Sbjct: 208 QAENPGFFYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTLDTRYRCNQFRVPFAPFTG 267
Query: 245 VNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAIRKVFPE 304
VNHH Q+I+FG A+I DE++ +++WL K F+ AM + PVS++TD D +++ A +VFP
Sbjct: 268 VNHHGQAILFGCALILDESDTSFIWLFKTFLTAMRDQPPVSLVTDQDRAIQIAAGQVFPG 327
Query: 305 AHHRLCAWHLIRNATSNIKNL-----HFVSKFKDCLLGDVDVDVFQRKWEELVTEFGLEE 359
A H + W ++R + ++ F + +C+ ++ F+ W ++ ++ L
Sbjct: 328 ARHCINKWDVLREGQEKLAHVCLAYPSFQVELYNCINFTETIEEFESSWSSVIDKYDLGR 387
Query: 360 NPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVSALTNLHDFFQQFFR 418
+ W+ +Y R W +FR FFA + G S F YV+ T L FF+ + R
Sbjct: 388 HEWLNSLYNARAQWVPVYFRDSFFAAVFPSQGYSG--SFFDGYVNQQTTLPMFFRLYER 444
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 169 bits (428), Expect = 3e-42
Identities = 107/405 (26%), Positives = 202/405 (49%), Gaps = 22/405 (5%)
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRNINGEVVQQTFLCHREGIREEKYINSTSRKR 64
F + AF +Y+ + GF+ RK+R + N V ++ F+C+R G + + + R
Sbjct: 76 FTTDDEAFEYYSTFARKSGFSIRKARSTESQNLGVYRRDFVCYRSGFNQPRKKANVEHPR 135
Query: 65 EHKPLSRCGCQAKVRVHIDVSS--QRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSEYDK 122
E K + RCGC K+ + +V WY+ F + HNH +++ R+LPA+RK+ + D+
Sbjct: 136 ERKSV-RCGCDGKLYLTKEVVDGVSHWYVSQFSNVHNHELLEDDQVRLLPAYRKIQQSDQ 194
Query: 123 YQMNTMRQSGISTTRIHGYFASQAGGYQN-VGYNRRDMYNEQR--KRRMRWN-------- 171
++ + ++G RI + G + + +D+ N R K+ ++ N
Sbjct: 195 ERILLLSKAGFPVNRIVKLLELEKGVVSGQLPFIEKDVRNFVRACKKSVQENDAFMTEKR 254
Query: 172 -SDAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATY 230
SD + + K ++ +D + T + ++++ W G S YS+FGDV+ FD +Y
Sbjct: 255 ESDTLELLECCKGLAERDMDFVYDCTSDENQKVENIAWAYGDSVRGYSLFGDVVVFDTSY 314
Query: 231 KKIKYNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDG 290
+ + Y L +F G++++ ++++ G ++ DE+ ++ W L+ FV M G+ P +++TD
Sbjct: 315 RSVPYGLLLGVFFGIDNNGKAMLLGCVLLQDESCRSFTWALQTFVRFMRGRHPQTILTDI 374
Query: 291 DLSMRNAIRKVFPEAHHRLCAWHLIRNAT---SNIKNLHFVSKFK---DCLLGDVDVDVF 344
D +++AI + P +H + H++ S H+ +F+ D L +VD F
Sbjct: 375 DTGLKDAIGREMPNTNHVVFMSHIVSKLASWFSQTLGSHY-EEFRAGFDMLCRAGNVDEF 433
Query: 345 QRKWEELVTEFGLEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTT 389
+++W+ LVT FGL + +Y R W R F A T+
Sbjct: 434 EQQWDLLVTRFGLVPDRHAALLYSCRASWLPCCIREHFVAQTMTS 478
>At2g32250 Mutator-like transposase
Length = 684
Score = 167 bits (423), Expect = 1e-41
Identities = 108/431 (25%), Positives = 187/431 (43%), Gaps = 64/431 (14%)
Query: 5 FPDREVAFMFYNWYGCFHGF-----AARKSRLIRNINGEVVQQTFLCHREGIREEKYINS 59
F +E A+ FY Y GF A+R+S+ +G+ + C R G + EK
Sbjct: 44 FESKEAAYYFYREYARSVGFGITIKASRRSKR----SGKFIDVKIACSRFGTKREKATAI 99
Query: 60 TSRKREHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSE 119
R + GC+A + + ++W I F +HNH + F
Sbjct: 100 NPRS-----CPKTGCKAGLHMKRK-EDEKWVIYNFVKEHNHEICPDDF------------ 141
Query: 120 YDKYQMNTMRQSGISTTRIHGYFASQAGGYQNVGYNRRDMYNEQRKRRMRWNSDAEQAVN 179
Y +V + K+ ++ + E
Sbjct: 142 -----------------------------YVSVRGKNKPAGALAIKKGLQLALEEEDLKL 172
Query: 180 FLKH---MSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYN 236
L+H M K F+ +D ++++FW D + DY F DV+ FD Y + Y
Sbjct: 173 LLEHFMEMQDKQPGFFYAVDFDSDKRVRNVFWLDAKAKHDYCSFSDVVLFDTFYVRNGYR 232
Query: 237 TPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRN 296
P F GV+HH Q ++ G A+IG+ +E TY WL + +++A+GG+ P +ITD D + +
Sbjct: 233 IPFAPFIGVSHHRQYVLLGCALIGEVSESTYSWLFRTWLKAVGGQAPGVMITDQDKLLSD 292
Query: 297 AIRKVFPEAHHRLCAWHLIRNATSNI-----KNLHFVSKFKDCLLGDVDVDVFQRKWEEL 351
+ +VFP+ H C W ++ + + ++ F+ F +C+ + F+R+W +
Sbjct: 293 IVVEVFPDVRHIFCLWSVLSKISEMLNPFVSQDDGFMESFGNCVASSWTDEHFERRWSNM 352
Query: 352 VTEFGLEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVSALTNLHD 411
+ +F L EN W+ +++ RK W +F G AG R + S F KY+++ D
Sbjct: 353 IGKFELNENEWVQLLFRDRKKWVPHYFHGICLAGLSGPERSGSIASHFDKYMNSEATFKD 412
Query: 412 FFQQFFRWLNY 422
FF+ + ++L Y
Sbjct: 413 FFELYMKFLQY 423
>At1g52520 F6D8.26
Length = 703
Score = 165 bits (418), Expect = 5e-41
Identities = 114/419 (27%), Positives = 190/419 (45%), Gaps = 23/419 (5%)
Query: 5 FPDREVAFMFYNWYGCFHGFAAR-KSRLIRNINGEVVQQTFLCHREGIREEKYINSTSRK 63
F + A+ +YN Y GF R K+ + + E C +G K IN +R
Sbjct: 91 FESYDDAYNYYNCYASEVGFRVRVKNSWFKRRSKEKYGAVLCCSSQGF---KRINDVNRV 147
Query: 64 REHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSE--YD 121
R+ +R GC A +R+ V S+RW + DHNH + ++ + + +S D
Sbjct: 148 RKE---TRTGCPAMIRMR-QVDSKRWRVVEVTLDHNHLLGCKLYKSVKRKRKCVSSPVSD 203
Query: 122 KYQMNTMRQSGISTTRIHGYFASQAGGYQNVGYNRRDMYNEQRKRRMRWNSDAEQAVNFL 181
+ R + ++ +QN D+ N +R D+ N+
Sbjct: 204 AKTIKLYRACVVDNGSNVNPNSTLNKKFQN-STGSPDLLNLKR-------GDSAAIYNYF 255
Query: 182 KHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVI 241
M + F+ V+ +G L+++FW D S + S FGDV+ D++Y K+ PLV
Sbjct: 256 CRMQLTNPNFFYLMDVNDEGQLRNVFWADAFSKVSCSYFGDVIFIDSSYISGKFEIPLVT 315
Query: 242 FSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAIRKV 301
F+GVNHH ++ + + ET ++Y WLLK+++ M + P +++TD + AI +V
Sbjct: 316 FTGVNHHGKTTLLSCGFLAGETMESYHWLLKVWLSVM-KRSPQTIVTDRCKPLEAAISQV 374
Query: 302 FPEAHHRLCAWHLIRNATSNIKNLH----FVSKFKDCLLGDVDVDVFQRKWEELVTEFGL 357
FP +H R H++R + LH F + + V F+ W +V FG+
Sbjct: 375 FPRSHQRFSLTHIMRKIPEKLGGLHNYDAVRKAFTKAVYETLKVVEFEAAWGFMVHNFGV 434
Query: 358 EENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVSALTNLHDFFQQF 416
EN W+ +Y++R WA + + FFAG E L F +YV T L +F ++
Sbjct: 435 IENEWLRSLYEERAKWAPVYLKDTFFAGIAAAHPGETLKPFFERYVHKQTPLKEFLDKY 493
>At4g19990 putative protein
Length = 672
Score = 159 bits (401), Expect = 4e-39
Identities = 111/445 (24%), Positives = 193/445 (42%), Gaps = 78/445 (17%)
Query: 5 FPDREVAFMFYNWYGCFHGF-----AARKSRLIRNINGEVVQQTFLCHREGIREE----- 54
F +E AF FY Y GF A+R+SR+ G+ + F+C R G ++E
Sbjct: 27 FESKEEAFEFYKEYANSVGFTTIIKASRRSRM----TGKFIDAKFVCTRYGSKKEDIDTG 82
Query: 55 ------KYINSTSRKREHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFE 108
+ R R ++ S+ C+A + V RW ++ +HNH E F
Sbjct: 83 LGTDGFNIPQARKRGRINRSSSKTDCKAFLHVKRRQDG-RWVVRSLVKEHNH----EIFT 137
Query: 109 RMLPAHRKMSEYDKYQMNTMRQSGISTTRIHGYFASQAGGYQNVGYNRRDMYNEQRKRRM 168
+ R++S K + +++G + K R
Sbjct: 138 GQADSLRELSGRRKLE------------KLNGAIVKEV------------------KSRK 167
Query: 169 RWNSDAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDA 228
+ D E+ +NF M S L+++FW D DY+ F DV++ D
Sbjct: 168 LEDGDVERLLNFFTDMQS----------------LRNIFWVDAKGRFDYTCFSDVVSIDT 211
Query: 229 TYKKIKYNTPLVIFSGVNHHNQSIIFG-SAIIGDETEDTYVWLLKIFVEAMGGKLPVSVI 287
T+ K +Y PLV F+GVNHH Q ++ G ++ DE++ +VWL + +++AM G P ++
Sbjct: 212 TFIKNEYKLPLVAFTGVNHHGQFLLLGFGLLLTDESKSGFVWLFRAWLKAMHGCRPRVIL 271
Query: 288 TDGDLSMRNAIRKVFPEAHHRLCAWHLIRNATSNIKNL-----HFVSKFKDCLLGDVDVD 342
T D ++ A+ +VFP + H W + + ++ V + D + G +
Sbjct: 272 TKHDQMLKEAVLEVFPSSRHCFYMWDTLGQMPEKLGHVIRLEKKLVDEINDAIYGSCQSE 331
Query: 343 VFQRKWEELVTEFGLEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKY 402
F++ W E+V F + +N W+ +Y+ R+ W + + AG T R + ++S KY
Sbjct: 332 DFEKNWWEVVDRFHMRDNVWLQSLYEDREYWVPVYMKDVSLAGMCTAQRSDSVNSGLDKY 391
Query: 403 VSALTNLHDFFQQFFRWLNYMRYRE 427
+ T F +Q+ + + RY E
Sbjct: 392 IQRKTTFKAFLEQYKKMIQ-ERYEE 415
>At4g38170 hypothetical protein
Length = 531
Score = 129 bits (325), Expect = 3e-30
Identities = 68/263 (25%), Positives = 127/263 (47%), Gaps = 10/263 (3%)
Query: 172 SDAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYK 231
S E +N+LK ++ + + ++FW D ++Y+ FGD L FD TY+
Sbjct: 2 SRVEHVLNYLKRRQLENPGFLYA----IEDDCGNVFWADPTCRLNYTYFGDTLVFDTTYR 57
Query: 232 KIK-YNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDG 290
+ K Y P F+G NHH Q ++FG A+I +E+E ++ WL + +++AM P S+ +
Sbjct: 58 RGKRYQVPFAAFTGFNHHGQPVLFGCALILNESESSFAWLFQTWLQAMSAPPPPSITVEP 117
Query: 291 DLSMRNAIRKVFPEAHHRLCAWHLIRNATSNIKNLH-----FVSKFKDCLLGDVDVDVFQ 345
D ++ A+ +VF + R + + ++ F S+F +C+ F+
Sbjct: 118 DRLIQVAVSRVFSQTRLRFSQPLIFEETEEKLAHVFQAHPTFESEFINCVTETETAAEFE 177
Query: 346 RKWEELVTEFGLEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVSA 405
W+ +V + +E+N W+ +Y R+ W R F+ T L+S F +V A
Sbjct: 178 ASWDSIVRRYYMEDNDWLQSIYNARQQWVRVFIRDTFYGELSTNEGSSILNSFFQGFVDA 237
Query: 406 LTNLHDFFQQFFRWLNYMRYREI 428
T + +Q+ + ++ R +E+
Sbjct: 238 STTMQMLIKQYEKAIDSWREKEL 260
>At4g08610 predicted transposon protein
Length = 907
Score = 70.5 bits (171), Expect = 2e-12
Identities = 57/240 (23%), Positives = 99/240 (40%), Gaps = 8/240 (3%)
Query: 197 VHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVIFSGVNHHNQSIIFGS 256
V A ++LF GV + + V+ DAT+ K Y LV + + ++ + I S
Sbjct: 386 VDAQQKFKYLFVSLGVCIEGFKVMRKVVIVDATFLKTVYGDMLVFATAQDPNHHNYIIAS 445
Query: 257 AIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHLIR 316
A+I E + ++ W + +L + ++D S+ +I VFP A H C WHL +
Sbjct: 446 AVIDRENDASWSWFFNKLKTVIPDELGLVFVSDRHQSIIKSIMHVFPNARHGHCVWHLSQ 505
Query: 317 NATSNIK--NLHFVSKFKDCLLGDVDVDVFQRKWEELVTEFGLEENPWMLEMYQKRKMWA 374
N +K + F C + F R++ F L+++ + WA
Sbjct: 506 NVKVRVKTEKEEAAANFIACAHVYTQFE-FTREYARFRRRF--PNAGTYLDIWAPVENWA 562
Query: 375 AAHFRGKFFAGFRTTSRCEGLHSEF--GKYVSALTNLHDFFQQFFRWLNYMRYREIGVPE 432
+F G + T++ CE L+S F + L L ++ W N R + + E
Sbjct: 563 MCYFEGDRY-NIDTSNVCESLNSTFESARKYFLLPLLDAIIEKISEWFNKHRKKSVKYSE 621
>At4g09380 putative protein
Length = 960
Score = 68.9 bits (167), Expect = 6e-12
Identities = 55/234 (23%), Positives = 101/234 (42%), Gaps = 13/234 (5%)
Query: 197 VHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVIFSGV--NHHNQSIIF 254
+ + ++LF G + V+ DAT+ K Y LVI + NHH+ + F
Sbjct: 384 LEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGMLVIATAQDPNHHHYPLAF 443
Query: 255 GSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHL 314
G II E + +++W L+ + I+D S++ A++ V+P A H C WHL
Sbjct: 444 G--IIDSEKDVSWIWFLENLKTVYSDVPGLVFISDRHQSIKKAVKTVYPNALHAACIWHL 501
Query: 315 IRNATSNIK--NLHFVSKFKDCLLGDVDVDVFQRKWEELVTEFGLEENPWMLEMYQKRKM 372
+N +K KF+DC + + F++++ + + + + ++K
Sbjct: 502 CQNMRDRVKIDKDGAAVKFRDCAHAYTESE-FEKEFGHFTSLWPKAADFLVKVGFEK--- 557
Query: 373 WAAAHFRGKFFAGFRTTSRCEGLHSEFGK--YVSALTNLHDFFQQFFRWLNYMR 424
W+ HF+G + T++ E ++ F K L + +F W N R
Sbjct: 558 WSRCHFKGDKY-NIDTSNSAESINGVFKKARKYHLLPMIDVMISKFSEWFNEHR 610
>At3g47310 putative protein
Length = 735
Score = 68.2 bits (165), Expect = 1e-11
Identities = 55/241 (22%), Positives = 101/241 (41%), Gaps = 13/241 (5%)
Query: 197 VHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVIFSGV--NHHNQSIIF 254
+ + ++LF G + V+ DAT+ K Y LVI + NHH+ + F
Sbjct: 333 LEGEKKFKYLFIALGACIEGFRTMRKVIVVDATHLKTVYGGMLVIATAQDPNHHHYPLAF 392
Query: 255 GSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHL 314
G II E + +++W L+ + I+D S++ ++ V+P A H C WHL
Sbjct: 393 G--IIDSENDVSWIWFLEKLKTVYSDVPGLVFISDRHQSIKKVVKTVYPNALHAACIWHL 450
Query: 315 IRNATSNIK--NLHFVSKFKDCLLGDVDVDVFQRKWEELVTEFGLEENPWMLEMYQKRKM 372
+N +K KF+DC + + F++++ + + + + ++K
Sbjct: 451 CQNMRDRVKIDKDGAAVKFRDCAHAYTESE-FEKEFGHFTSLWPKAADFLVKVGFEK--- 506
Query: 373 WAAAHFRGKFFAGFRTTSRCEGLHSEFGK--YVSALTNLHDFFQQFFRWLNYMRYREIGV 430
W+ HF+G + T++ E ++ F K L + +F W N R
Sbjct: 507 WSRCHFKGDKY-NIDTSNSAESINGVFKKARKYHLLPMIDVMISKFSEWFNEHRQDSSSC 565
Query: 431 P 431
P
Sbjct: 566 P 566
>At1g36090 hypothetical protein
Length = 645
Score = 68.2 bits (165), Expect = 1e-11
Identities = 54/213 (25%), Positives = 90/213 (41%), Gaps = 19/213 (8%)
Query: 197 VHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVIFSG--VNHHNQSIIF 254
+ + ++LF G + V+ DAT+ K Y LVI + NHH+ + F
Sbjct: 344 LEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGMLVIATAHDPNHHHYPLAF 403
Query: 255 GSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHL 314
G II E + +++W L+ + I+D S++ A++ V+P A H C WHL
Sbjct: 404 G--IIDSEKDVSWIWFLEKLKTVYSDVPRLVFISDRHQSIKKAVKTVYPNALHAACIWHL 461
Query: 315 IRNATSNIK--NLHFVSKFKDCLLGDVDVDVFQRKWEELVTEFGLEENPWMLEMYQKRKM 372
+N +K KF+DC + E EFG + W + K+
Sbjct: 462 CQNMRDRVKIDKDGAAVKFRDCAHAYTE--------SEFEKEFGHFTSLWPKAVDFLVKV 513
Query: 373 ----WAAAHFRGKFFAGFRTTSRCEGLHSEFGK 401
W+ HF+G + T++ E ++ F K
Sbjct: 514 GFEKWSRCHFKGDKY-NIDTSNSAESINGVFKK 545
>At4g28970 putative protein
Length = 914
Score = 67.0 bits (162), Expect = 2e-11
Identities = 55/234 (23%), Positives = 100/234 (42%), Gaps = 13/234 (5%)
Query: 197 VHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVIFSGV--NHHNQSIIF 254
+ + ++LF G + V+ DAT+ K Y LVI + NHH+ + F
Sbjct: 384 LEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGMLVIATAQDPNHHHYPLAF 443
Query: 255 GSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHL 314
G II E + +++W L+ + I+D S++ A++ V+P A H C WHL
Sbjct: 444 G--IIDSEKDVSWIWFLEKLKTVYSDVPGLVFISDRHQSIKKAVKTVYPNALHAACIWHL 501
Query: 315 IRNATS--NIKNLHFVSKFKDCLLGDVDVDVFQRKWEELVTEFGLEENPWMLEMYQKRKM 372
+N I KF+DC + + F++++ + + + + ++K
Sbjct: 502 CQNMRDRVTIDKDGAAVKFRDCAHAYTESE-FEKEFGHFTSLWPKAADFLVKVGFEK--- 557
Query: 373 WAAAHFRGKFFAGFRTTSRCEGLHSEFGK--YVSALTNLHDFFQQFFRWLNYMR 424
W+ HF+G + T++ E ++ F K L + +F W N R
Sbjct: 558 WSRCHFKGDKY-NIDTSNSAESINGVFKKARKYHLLPMIDVMISKFSEWFNEHR 610
>At1g35060 hypothetical protein
Length = 873
Score = 66.6 bits (161), Expect = 3e-11
Identities = 55/224 (24%), Positives = 94/224 (41%), Gaps = 11/224 (4%)
Query: 205 HLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVIFSGVNHHNQSIIFGSAIIGDETE 264
++F G S + VL D T+ K KY L+ SG + + Q A++ E +
Sbjct: 507 YMFLAFGASIQGFKHLRRVLVVDGTHLKGKYKGVLLTASGQDANFQVYPLAFAVVDSEND 566
Query: 265 DTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHLIRNATSNIKN 324
D + W + +++++D S++ ++KVFP+AHH C HL RN + KN
Sbjct: 567 DAWTWFFTKLERIIADNNTLTILSDRHESIKVGVKKVFPQAHHGACIIHLCRNIQARFKN 626
Query: 325 LHFVSKFKDCLLGDVDVDVFQRKWEELVTEFGLEENPWMLEMYQKRKM--WAAAHFRGKF 382
L+ + + K++ L + NP ++ M W +F G+
Sbjct: 627 RGLTQ-----LVKNAGYEFTSGKFKTLYNQIN-AINPLCIKYLHDVGMAHWTRLYFPGQR 680
Query: 383 FAGFRTTSRCEGLHSEFGKYVSA-LTNLHDFFQQFF-RWLNYMR 424
F T++ E L+ K S+ + L F + RW N R
Sbjct: 681 F-NLMTSNIAETLNKALFKGRSSHIVELLRFIRSMLTRWFNAHR 723
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.139 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,726,303
Number of Sequences: 26719
Number of extensions: 444551
Number of successful extensions: 1490
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 72
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 1341
Number of HSP's gapped (non-prelim): 117
length of query: 485
length of database: 11,318,596
effective HSP length: 103
effective length of query: 382
effective length of database: 8,566,539
effective search space: 3272417898
effective search space used: 3272417898
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146755.5


