

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.6 + phase: 0
(346 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
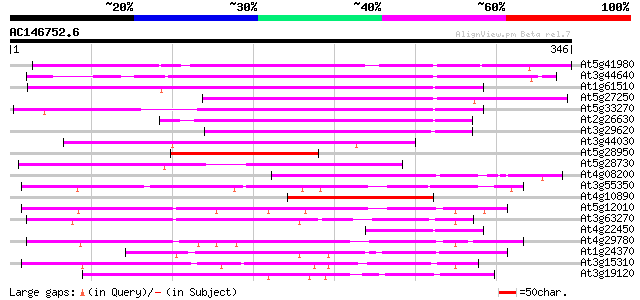
Score E
Sequences producing significant alignments: (bits) Value
At5g41980 unknown protein 228 4e-60
At3g44640 putative protein 223 9e-59
At1g61510 hypothetical protein 194 6e-50
At5g27250 putative protein 184 8e-47
At5g33270 putative protein 157 6e-39
At2g26630 En/Spm-like transposon protein 153 2e-37
At3g29620 hypothetical protein 130 8e-31
At3g44030 putative protein 129 3e-30
At5g28950 putative protein 114 1e-25
At5g28730 putative protein 100 2e-21
At4g08200 putative protein 86 3e-17
At3g55350 unknown protein 82 6e-16
At4g10890 putative protein 71 8e-13
At5g12010 putative protein 65 7e-11
At3g63270 unknown protein 65 7e-11
At4g22450 hypothetical protein 60 1e-09
At4g29780 unknown protein 60 2e-09
At1g24370 hypothetical protein 50 1e-06
At3g15310 unknown protein 49 4e-06
At3g19120 unknown protein 45 6e-05
>At5g41980 unknown protein
Length = 374
Score = 228 bits (581), Expect = 4e-60
Identities = 134/344 (38%), Positives = 187/344 (53%), Gaps = 29/344 (8%)
Query: 15 RMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGETIS 74
RM F KLCD+L+ G LR T +E Q+A L+I+ HN + R V F SGETIS
Sbjct: 47 RMDKPVFYKLCDLLQTRGLLRHTNRIKIEAQLAIFLFIIGHNLRTRAVQELFCYSGETIS 106
Query: 75 RHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVKVSAKD 134
RH + VL A++ + + F QP+ ++ LE PYFKDCVG +D HI V V +
Sbjct: 107 RHFNNVLNAVIAISKDFF-QPNSNSDTLENDD-----PYFKDCVGVVDSFHIPVMVGVDE 160
Query: 135 APRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDKLKIPEGKYY 194
+R TQNVLAA +FDL+F YVLAGWEGSASD +++ ALTR +KL++P+GKYY
Sbjct: 161 QGPFRNGNGLLTQNVLAASSFDLRFNYVLAGWEGSASDQQVLNAALTRRNKLQVPQGKYY 220
Query: 195 LVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGVLKKR 254
+VD + G I PY GV + +E KE+FN RH L AI R FG LK+R
Sbjct: 221 IVDNKYPNLPGFIAPYHGVSTNSRE--------EAKEMFNERHKLLHRAIHRTFGALKER 272
Query: 255 FEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDAELANQNVSHDNHEA 314
F IL ++ P Y ++ Q ++ A C LHNY+ +P +DL+ + E D A
Sbjct: 273 FPILLSA--PPYPLQTQVKLVIAACALHNYVRLEKP-DDLVFRMFEEETLAEAGEDREVA 329
Query: 315 SRSDM------------DEFAQGGIIKNGAAHQMWSNYKNNGQT 346
+ +E +++ A ++W++Y N T
Sbjct: 330 LEEEQVEIVGQEHGFRPEEVEDSLRLRDEIASELWNHYVQNMST 373
>At3g44640 putative protein
Length = 377
Score = 223 bits (569), Expect = 9e-59
Identities = 126/336 (37%), Positives = 184/336 (54%), Gaps = 45/336 (13%)
Query: 11 RNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSG 70
R + RM P+ F KLCD+ IL +N E+ +++
Sbjct: 72 RQLYRMEPEVFHKLCDV------------------------ILLYNGHISEIKLFYKHK- 106
Query: 71 ETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVKV 130
H+ LKA+ + ++ +P+ +P +I ST FYPYFKDC+GAIDGTHI V
Sbjct: 107 ------FHKALKALAFVAPNWMAKPE-VRVPAKIRESTSFYPYFKDCIGAIDGTHIFAMV 159
Query: 131 SAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTRE-DKLKIP 189
DA +R RK + +QNVLAAC FDL+F Y+L+GW+GSA DS+++ +ALTR ++L++P
Sbjct: 160 PTCDAASFRNRKGFISQNVLAACNFDLQFIYILSGWKGSAHDSKVLNDALTRNTNRLQVP 219
Query: 190 EGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSAR-NPPLNYKELFNLRHASLRNAIERAF 248
EGK+YLVD G+ + P+R RYHL++F + P ELFNLRHASLRN IER F
Sbjct: 220 EGKFYLVDCGYANRRKFLAPFRRTRYHLQDFRGQGRDPKTQNELFNLRHASLRNVIERIF 279
Query: 249 GVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDAELANQNVS 308
G+ K RF I ++ P Y K Q ++ AC LHN+L +++ E N + +
Sbjct: 280 GIFKSRFLIFKSA--PPYPFKTQTELVLACAGLHNFLHQECRSDEFPPETSINEENSDDN 337
Query: 309 HDNHEASRSDMD-------EFAQGGIIKNGAAHQMW 337
+N+ D+ E+A ++ +H MW
Sbjct: 338 EENNYEENGDVGILESQQREYANNW--RDTISHNMW 371
>At1g61510 hypothetical protein
Length = 608
Score = 194 bits (493), Expect = 6e-50
Identities = 107/284 (37%), Positives = 159/284 (55%), Gaps = 4/284 (1%)
Query: 12 NIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGE 71
+I+RM TF LC +L + L + +EE VA + ++ + R + ++ S E
Sbjct: 274 DILRMHQSTFRTLCKILSEQYKLEESCNIYLEESVAMFIEMVAQDLTVRVIAERYQHSLE 333
Query: 72 TISRHLHQVLKAILELEEKFI--VQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVK 129
T+ R L +VL A+L+L + + D + I + + R+ PYF DC+GA+DGTH+ V+
Sbjct: 334 TVKRKLDEVLSALLKLAADIVKPTRDDVTGISPFLVNDKRYMPYFIDCIGALDGTHVSVR 393
Query: 130 VSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDKLKIP 189
+ D RYRGRK T N+LA C F +KF G G A D++++ T E P
Sbjct: 394 PPSGDVERYRGRKSEATMNILAVCNFSMKFNIAYVGVPGRAHDTKVLTYCATHEASFPHP 453
Query: 190 E-GKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAF 248
GKYYLVD+G+ SG + P+R RYHL+ F+ PP N +ELFN RH+SLR+ IER F
Sbjct: 454 PAGKYYLVDSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLRSVIERTF 513
Query: 249 GVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNE 292
GV K ++ IL P Y +K I+ + LHNY+ ++ +
Sbjct: 514 GVWKAKWRILDRK-HPKYEVKKWIKIVTSTMALHNYIRDSQQED 556
>At5g27250 putative protein
Length = 348
Score = 184 bits (466), Expect = 8e-47
Identities = 104/232 (44%), Positives = 134/232 (56%), Gaps = 9/232 (3%)
Query: 120 AIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNA 179
AIDGTHI V + YR RK +QNVLAAC FDL+F YVL+GWEGSA DS+++++A
Sbjct: 116 AIDGTHINAMVQGPEKASYRNRKGVISQNVLAACNFDLEFIYVLSGWEGSAHDSKVLQDA 175
Query: 180 LTRE-DKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSAR-NPPLNYKELFNLRH 237
LTR ++L++PEGKYYL D GF + P R RYHL++F P N ELFNLRH
Sbjct: 176 LTRRTNRLQVPEGKYYLADCGFPNRRNFLAPLRSTRYHLQDFRGEGRDPTNQNELFNLRH 235
Query: 238 ASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYL----MSAEPNED 293
ASLRN IER FG+ K RF I ++ P + K Q I+ +C LHN+L S E + D
Sbjct: 236 ASLRNVIERIFGIFKSRFLIFKSA--PPFSFKTQAEIVLSCAALHNFLRQKCRSDEFSSD 293
Query: 294 LIAEVDAELANQNVSHDNHEAS-RSDMDEFAQGGIIKNGAAHQMWSNYKNNG 344
E D + ANQN + E + + E + A MW + N G
Sbjct: 294 EEDETDVDNANQNSEENGGEENVETQEQEREHARHWRATIAADMWRDATNIG 345
>At5g33270 putative protein
Length = 343
Score = 157 bits (398), Expect = 6e-39
Identities = 97/295 (32%), Positives = 148/295 (49%), Gaps = 40/295 (13%)
Query: 3 RLKNSETSRNIIRMGPQT----FLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAK 58
+LK + ++N+ G QT F ++ ++ + L + +EE VA + ++ +
Sbjct: 32 KLKGNIRNQNVNDAGRQTIIDRFYEVDGRVQEQYKLEESCNIYLEESVAMFIEMVAQDLT 91
Query: 59 NREVNFWFRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCV 118
R + ++ S ET+ R L +VL +C+
Sbjct: 92 VRVIAERYQHSLETVKRKLDEVL----------------------------------NCI 117
Query: 119 GAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKN 178
GA+DGTH+ V+ + D RYRGRK T N+LA C F +KFTY G G A D++++
Sbjct: 118 GALDGTHVSVRPPSGDVERYRGRKSEATMNILALCNFSMKFTYAYVGVPGRAHDTKVLTY 177
Query: 179 ALTREDKLKIPE-GKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRH 237
T E P GKYYLVD+G+ SG + P+R RYHL+ F+ PP N +ELFN RH
Sbjct: 178 CATHEASFPHPPAGKYYLVDSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRH 237
Query: 238 ASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNE 292
+SLR+ IER FGV K ++ IL Y +K I+ + LHNY+ ++ +
Sbjct: 238 SSLRSVIERTFGVWKAKWRILDRK-HLKYEVKKWIKIVTSTMALHNYIRDSQQED 291
>At2g26630 En/Spm-like transposon protein
Length = 292
Score = 153 bits (386), Expect = 2e-37
Identities = 80/194 (41%), Positives = 114/194 (58%), Gaps = 10/194 (5%)
Query: 93 VQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAA 152
+Q D +PL I+ F CVGA+DGTH+ V+ ++ A +Y+GRK PT NVLA
Sbjct: 28 LQLDPDELPLLIT--------FLYCVGALDGTHVPVRPPSQTAKKYKGRKLEPTMNVLAI 79
Query: 153 CTFDLKFTYVLAGWEGSASDSRIIKNALTREDKLKIP-EGKYYLVDAGFMLTSGLITPYR 211
C FD+KF Y G G A D++++ T E P GKYYLVD+G+ +G + P+R
Sbjct: 80 CNFDMKFIYAYVGVPGRAHDTKVLNYCATNEPYFSHPPNGKYYLVDSGYPTRTGYLGPHR 139
Query: 212 GVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQ 271
+RYHL +F PP+ +ELFN +H+ LR+ IER FGV K ++ I+ P YG+
Sbjct: 140 RMRYHLGQFGRGGPPVTARELFNRKHSGLRSVIERTFGVWKAKWRIVDRK-HPKYGLAKW 198
Query: 272 KLIIFACCILHNYL 285
I+ A LHN++
Sbjct: 199 IKIVTATMALHNFI 212
>At3g29620 hypothetical protein
Length = 222
Score = 130 bits (328), Expect = 8e-31
Identities = 68/166 (40%), Positives = 97/166 (57%), Gaps = 3/166 (1%)
Query: 121 IDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNAL 180
+DGTH+ VS +D RY RK + N+LA C FD+ FTY+ G GSA D++++ A+
Sbjct: 1 MDGTHVPAMVSGRDHQRYWNRKSICSMNILAVCNFDMLFTYIYVGVFGSAHDTKVLSLAM 60
Query: 181 TREDKLKIPE-GKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHAS 239
+ P GKYYLVD+G+ L G + P+R YH +F + PP N+KE FN RH+
Sbjct: 61 EGDPNFPHPHIGKYYLVDSGYALRRGYLGPFRQTWYHHNQFQNQAPPNNHKEKFNWRHSL 120
Query: 240 LRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYL 285
LR IER FGV K ++ I+ + Y I + I+ A LHN++
Sbjct: 121 LRCVIERTFGVWKGKWRIMQD--RAWYNIVTTRKIMVATMALHNFV 164
>At3g44030 putative protein
Length = 775
Score = 129 bits (323), Expect = 3e-30
Identities = 81/226 (35%), Positives = 115/226 (50%), Gaps = 9/226 (3%)
Query: 34 LRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGETISRHLHQVLKAILELEEKFIV 93
L PT S++E VA L I HN +V F ++ ET+ R +VL L +I
Sbjct: 550 LLPTLNISIKESVAMFLRICGHNEVQIDVGLRFGQNQETVQRKFREVLTETELLACDYIR 609
Query: 94 QPDGST---IPLEISSSTRFYPYFKDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVL 150
P IP + R++PYF VGA+DGTH+ VKV + Y R + + N++
Sbjct: 610 TPTRQELYRIPERLQVDRRYWPYFSGFVGAMDGTHVCVKVKPELQGMYWNRHDNASLNIM 669
Query: 151 AACTFDLKFTYVLAGWEGSASDSRIIKNALTREDKLKIPEG-KYYLVDAGFMLTSGLITP 209
A C ++ FTY+ G GS D+ +++ A + + +P KYYLVD+ + G +
Sbjct: 670 AICDLNMLFTYIWNGVPGSCHDTVVLQIAQQSDSEFPLPPSEKYYLVDSCYPNKQGFLAL 729
Query: 210 YRG-----VRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGV 250
YR VRYH+ +F PP N ELFN H SLR+ IER GV
Sbjct: 730 YRSSRNRVVRYHMSQFYPGPPPRNKHELFNQCHTSLRSVIERTVGV 775
>At5g28950 putative protein
Length = 148
Score = 114 bits (284), Expect = 1e-25
Identities = 54/92 (58%), Positives = 69/92 (74%), Gaps = 1/92 (1%)
Query: 100 IPLEISSSTRFYPYFKDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKF 159
+P +I STR YPYFKDCVGAID THI VS K P +R RK +QN+LAAC FD++F
Sbjct: 8 VPRKIRESTRLYPYFKDCVGAIDDTHIFAMVSQKKMPSFRNRKGDISQNMLAACNFDVEF 67
Query: 160 TYVLAGWEGSASDSRIIKNALTR-EDKLKIPE 190
YVL+GWEGSA DS+++ +ALTR ++L +PE
Sbjct: 68 MYVLSGWEGSAHDSKVLNDALTRNSNRLPVPE 99
>At5g28730 putative protein
Length = 296
Score = 99.8 bits (247), Expect = 2e-21
Identities = 68/242 (28%), Positives = 111/242 (45%), Gaps = 29/242 (11%)
Query: 6 NSETSRNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFW 65
N + + +IRM + F +LC++L + GL+ + S++E VA L I N R++
Sbjct: 20 NEVSCQTLIRMSSEAFTQLCEILHGKYGLQSSTNISLDESVAIFLIICASNDTQRDIALR 79
Query: 66 FRRSGETISRHLHQVLKAILELEEKFIVQ---PDGSTIPLEISSSTRFYPYFKDCVGAID 122
F + ETI R H VLKA+ L ++I + I + TR++P+ D +G
Sbjct: 80 FGHAQETIWRKFHDVLKAMERLAVEYIRPRKVEELRAISNRLQDDTRYWPFLMDLLGIA- 138
Query: 123 GTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTR 182
+ NVLA C D+ FTY G GS D+R++ A++
Sbjct: 139 -----------------------SFNVLAICDLDMLFTYCFVGMAGSTHDARVLSAAISD 175
Query: 183 EDKLKI-PEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARN-PPLNYKELFNLRHASL 240
+ + P+ KYYLVD+G+ G + PYR ++ + N +N E N++
Sbjct: 176 DPLFHVPPDSKYYLVDSGYANKRGYLAPYRREHREAQDIISNNFLTVNLFETHNIKDYDF 235
Query: 241 RN 242
N
Sbjct: 236 DN 237
>At4g08200 putative protein
Length = 202
Score = 85.9 bits (211), Expect = 3e-17
Identities = 59/191 (30%), Positives = 92/191 (47%), Gaps = 20/191 (10%)
Query: 162 VLAGWEGSASDSRIIKNALTREDKLKIPE-GKYYLVDAGFMLTSGLITPYRGVRYHLKEF 220
+ G G A D++++ A+ + P KYYLVD+G+ L G + YR +YH F
Sbjct: 21 IYVGVLGYAHDAKVLALAMQGDPNFSHPPISKYYLVDSGYGLHRGYLISYRQSQYHPSHF 80
Query: 221 SARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCI 280
+ PP NYKE FN H+SLR ER F V K +++I+ N Y ++ K ++
Sbjct: 81 QNQAPPNNYKEKFNRLHSSLRLVTERTFRVWKGKWKIMHNRAR--YDVRTTKKLVVETMA 138
Query: 281 LHNYLMSAEPNEDLIAEVDAELANQNVSHDNHEASRSDMDEFAQGGI----------IKN 330
LHN++ + I + D E AN DNH+ S + E G I++
Sbjct: 139 LHNFVRKSN-----ILDPDFE-ANWE-QDDNHQPSLKEEVEVQDDGQIFDSRQYMEGIRD 191
Query: 331 GAAHQMWSNYK 341
A +W+N++
Sbjct: 192 DIAMNLWNNHR 202
>At3g55350 unknown protein
Length = 406
Score = 81.6 bits (200), Expect = 6e-16
Identities = 85/336 (25%), Positives = 145/336 (42%), Gaps = 51/336 (15%)
Query: 8 ETSRNIIRMGPQTFLKLCDMLEREGGLRPTRWS-------SVEEQVAKSLYILTHNAKNR 60
+T ++ ++ +TF +C +++ + +P +S S+ ++VA +L L
Sbjct: 70 KTFESVFKISRKTFDYICSLVKADFTAKPANFSDSNGNPLSLNDRVAVALRRLGSGESLS 129
Query: 61 EVNFWFRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGA 120
+ F + T+S+ + +++ +EE+ I + EI S +C GA
Sbjct: 130 VIGETFGMNQSTVSQITWRFVES---MEERAIHHLSWPSKLDEIKSKFEKISGLPNCCGA 186
Query: 121 IDGTHIRVKVSAKDAPR---YRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIK 177
ID THI + + A + G K + + + A D++F V+AGW GS +D ++K
Sbjct: 187 IDITHIVMNLPAVEPSNKVWLDGEKNF-SMTLQAVVDPDMRFLDVIAGWPGSLNDDVVLK 245
Query: 178 NA----------LTREDKLKIPE----GKYYLVDAGFMLTSGLITPYRGVRYHLKEFSAR 223
N+ +KL + E +Y + D+GF L L+TPY+G L +
Sbjct: 246 NSGFYKLVEKGKRLNGEKLPLSERTELREYIVGDSGFPLLPWLLTPYQGKPTSLPQTE-- 303
Query: 224 NPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHN 283
FN RH+ A + A LK R+ I+ N IIF CC+LHN
Sbjct: 304 ---------FNKRHSEATKAAQMALSKLKDRWRII-NGVMWMPDRNRLPRIIFVCCLLHN 353
Query: 284 YLMSAEPNEDLIAEVDAELANQNVS--HDNHEASRS 317
++ E D L +Q +S HD + RS
Sbjct: 354 IIIDME---------DQTLDDQPLSQQHDMNYRQRS 380
>At4g10890 putative protein
Length = 380
Score = 71.2 bits (173), Expect = 8e-13
Identities = 34/91 (37%), Positives = 55/91 (60%), Gaps = 1/91 (1%)
Query: 172 DSRIIKNALTREDKLKIPEG-KYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYK 230
D++++K E P KYYLV++ + T+G + P+R + YHL +F PP+ +
Sbjct: 73 DTKVLKYCARNESFSPHPSNRKYYLVNSVYPTTTGYLGPHRRILYHLGQFGRGGPPVTVQ 132
Query: 231 ELFNLRHASLRNAIERAFGVLKKRFEILSNS 261
ELFN +H LR+ I+R FGV K ++ IL ++
Sbjct: 133 ELFNRKHLDLRSVIDRTFGVWKAKWRILDHT 163
Score = 27.7 bits (60), Expect = 9.6
Identities = 18/60 (30%), Positives = 30/60 (50%)
Query: 27 MLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGETISRHLHQVLKAILE 86
ML R GL+ +EE VA L + N R++ +++S + R + VL A+L+
Sbjct: 1 MLTRRYGLQELENVYLEESVAMFLEEVDKNRTVRDIVARYQQSLNVVKRKIDDVLSALLK 60
>At5g12010 putative protein
Length = 502
Score = 64.7 bits (156), Expect = 7e-11
Identities = 75/318 (23%), Positives = 133/318 (41%), Gaps = 34/318 (10%)
Query: 8 ETSRNIIRMGPQTFLKLCDMLEREGGLRPTRWSS---VEEQVAKSLYILTHNAKNREVNF 64
E + RM TF +CD L T + V ++VA ++ L R V+
Sbjct: 173 EDFKKAFRMSKSTFELICDELNSAVAKEDTALRNAIPVRQRVAVCIWRLATGEPLRLVSK 232
Query: 65 WFRRSGETISRHLHQVLKAILE-LEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDG 123
F T + + +V KAI + L K++ PD ++ I + VG++
Sbjct: 233 KFGLGISTCHKLVLEVCKAIKDVLMPKYLQWPDDESLR-NIRERFESVSGIPNVVGSMYT 291
Query: 124 THI-----RVKVSAKDAPRYRGRKEYPTQNVLAACTFDLK--FTYVLAGWEGSASDSRII 176
THI ++ V++ R+ R + + ++ + K FT + GW GS D +++
Sbjct: 292 THIPIIAPKISVASYFNKRHTERNQKTSYSITIQAVVNPKGVFTDLCIGWPGSMPDDKVL 351
Query: 177 KNALT--REDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFN 234
+ +L R + + +G + G L ++ PY + + FN
Sbjct: 352 EKSLLYQRANNGGLLKGMWVAGGPGHPLLDWVLVPYTQQNLTWTQHA-----------FN 400
Query: 235 LRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKL--IIFACCILHNYLMSAEPN- 291
+ + ++ + AFG LK R+ L TE +K Q L ++ ACC+LHN E
Sbjct: 401 EKMSEVQGVAKEAFGRLKGRWACLQKRTE----VKLQDLPTVLGACCVLHNICEMREEKM 456
Query: 292 --EDLIAEVDAELANQNV 307
E ++ +D E+ +NV
Sbjct: 457 EPELMVEVIDDEVLPENV 474
>At3g63270 unknown protein
Length = 396
Score = 64.7 bits (156), Expect = 7e-11
Identities = 75/303 (24%), Positives = 124/303 (40%), Gaps = 44/303 (14%)
Query: 11 RNIIRMGPQTFLKLCDMLEREGGLRPT--------RWSSVEEQVAKSLYILTHNAKNREV 62
++ R TF +C ++ + RP R SVE+QVA +L L V
Sbjct: 66 KHFFRASKTTFSYICSLVREDLISRPPSGLINIEGRLLSVEKQVAIALRRLASGDSQVSV 125
Query: 63 NFWFRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAID 122
F T+S+ + ++A+ E + + PD I EI S +C GAID
Sbjct: 126 GAAFGVGQSTVSQVTWRFIEALEERAKHHLRWPDSDRIE-EIKSKFEEMYGLPNCCGAID 184
Query: 123 GTHIRVKVSAKDAPR-YRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIK---- 177
THI + + A A + +++ + + +++F ++ GW G + S+++K
Sbjct: 185 TTHIIMTLPAVQASDDWCDQEKNYSMFLQGVFDHEMRFLNMVTGWPGGMTVSKLLKFSGF 244
Query: 178 ------------NALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNP 225
N T +I E Y + + L LITP+ +
Sbjct: 245 FKLCENAQILDGNPKTLSQGAQIRE--YVVGGISYPLLPWLITPHDS-----------DH 291
Query: 226 PLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKL--IIFACCILHN 283
P + FN RH +R+ AF LK + ILS + + +KL II CC+LHN
Sbjct: 292 PSDSMVAFNERHEKVRSVAATAFQQLKGSWRILS---KVMWRPDRRKLPSIILVCCLLHN 348
Query: 284 YLM 286
++
Sbjct: 349 III 351
>At4g22450 hypothetical protein
Length = 457
Score = 60.5 bits (145), Expect = 1e-09
Identities = 30/73 (41%), Positives = 43/73 (58%), Gaps = 1/73 (1%)
Query: 220 FSARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACC 279
F+ PP N +ELFN RH+SLR+ IER FGV K ++ IL P Y +K I+ +
Sbjct: 334 FNRGGPPTNSRELFNRRHSSLRSVIERTFGVWKAKWRILDRK-HPKYEVKKWIKIVTSTM 392
Query: 280 ILHNYLMSAEPNE 292
LHNY+ ++ +
Sbjct: 393 ALHNYIRDSQQED 405
>At4g29780 unknown protein
Length = 540
Score = 59.7 bits (143), Expect = 2e-09
Identities = 84/324 (25%), Positives = 141/324 (42%), Gaps = 37/324 (11%)
Query: 11 RNIIRMGPQTFLKLCDMLEREGGLRPTRWSSV---EEQVAKSLYILTHNAKNREVNFWFR 67
R RM TF +C+ L+ + T ++V ++ L A R V+ F
Sbjct: 214 RREFRMSKSTFNLICEELDTTVTKKNTMLRDAIPAPKRVGVCVWRLATGAPLRHVSERFG 273
Query: 68 RSGETISRHLHQVLKAILE-LEEKFIVQPDGSTIPLEISSSTRFYPYFK--DCVGAIDGT 124
T + + +V +AI + L K+++ P S I S+ +F K + VG+I T
Sbjct: 274 LGISTCHKLVIEVCRAIYDVLMPKYLLWPSDSEIN---STKAKFESVHKIPNVVGSIYTT 330
Query: 125 HI-----RVKVSAKDAPRY--RGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRII- 176
HI +V V+A R+ R +K + V D FT V G GS +D +I+
Sbjct: 331 HIPIIAPKVHVAAYFNKRHTERNQKTSYSITVQGVVNADGIFTDVCIGNPGSLTDDQILE 390
Query: 177 KNALTREDKLK-IPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNL 235
K++L+R+ + + + + ++GF LT L+ PY R + FN
Sbjct: 391 KSSLSRQRAARGMLRDSWIVGNSGFPLTDYLLVPY-----------TRQNLTWTQHAFNE 439
Query: 236 RHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKL--IIFACCILHNYLMSAEPNED 293
++ AF LK R+ L TE +K Q L ++ ACC+LHN + E+
Sbjct: 440 SIGEIQGIATAAFERLKGRWACLQKRTE----VKLQDLPYVLGACCVLHN--ICEMRKEE 493
Query: 294 LIAEVDAELANQNVSHDNHEASRS 317
++ E+ E+ + +N+ S S
Sbjct: 494 MLPELKFEVFDDVAVPENNIRSAS 517
>At1g24370 hypothetical protein
Length = 413
Score = 50.4 bits (119), Expect = 1e-06
Identities = 57/246 (23%), Positives = 106/246 (42%), Gaps = 20/246 (8%)
Query: 72 TISRHLHQVLKAILELEE-KFIVQPDGSTIP--LEISSSTRFYPYFKDCVGAIDGTHIRV 128
T L + +AI+E+ +++ PD + + L I S F +G++D H +
Sbjct: 27 TALESLKRFCRAIVEVFACRYLRSPDANDVARLLHIGESRGF----PRMLGSLDCMHWKW 82
Query: 129 K-VSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIK--NALTREDK 185
K +Y GR PT + A +DL + G GS +D +++ + +
Sbjct: 83 KNCPTAWGGQYAGRSRSPTIILEAVADYDLWIWHAYFGLPGSNNDINVLEASHLFANLAE 142
Query: 186 LKIPEGKYYL----VDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLR 241
P Y + + + L G+ + + + + R P K+LF ++ + R
Sbjct: 143 GTAPPASYVINEKPYNMSYYLADGIYPKWSTLVQTIHD--PRGPK---KKLFAMKQEACR 197
Query: 242 NAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDAE 301
+ERAFGVL+ RF I++ + + I+ +C I+HN ++ E + D E E
Sbjct: 198 KDVERAFGVLQSRFAIVAGPSR-LWNKTVLHDIMTSCIIMHNMIIEDERDIDAPIEERVE 256
Query: 302 LANQNV 307
+ + V
Sbjct: 257 VPIEEV 262
>At3g15310 unknown protein
Length = 415
Score = 48.9 bits (115), Expect = 4e-06
Identities = 73/301 (24%), Positives = 123/301 (40%), Gaps = 31/301 (10%)
Query: 8 ETSRNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVE-------EQVAKSLYILTHNAKNR 60
+T R RM FL++ D L E R + ++ ++ +L + +
Sbjct: 73 QTFRRRFRMKKPLFLRIVDRLSSELMFFQHRRDATGRFGHSPIQKCTAAIRLLAYGYASD 132
Query: 61 EVNFWFRRSGETISRHLHQVLKAILEL-EEKFIVQPDGSTIPLEIS-SSTRFYPYFKDCV 118
V+ + R T L K I+ ++++ P + + ++ R +P +
Sbjct: 133 AVDEYLRMGETTAMSCLENFTKGIISFFGDEYLRAPTATNLRRLLNIGKIRGFP---GMI 189
Query: 119 GAIDGTHIRVK--VSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRII 176
G++D H K +A RG + PT + A + DL +V G G+ +D I+
Sbjct: 190 GSLDCMHWEWKNCPTAWKGQYTRGSGK-PTIVLEAIASQDLWIWHVFFGPPGTLNDINIL 248
Query: 177 KNALTREDKL--KIPEGKYYL----VDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYK 230
+ +D L + P KY + + LT G+ + S R P
Sbjct: 249 DRSPIFDDILQGRAPNVKYKVNGREYHLAYYLTDGIYPKWATFIQ-----SIRLPQNRKA 303
Query: 231 ELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKL--IIFACCILHNYLMSA 288
LF + R +ERAFGVL+ RF I+ N PA +K+ I+ AC ILHN ++
Sbjct: 304 TLFATHQEADRKDVERAFGVLQARFHIIKN---PALVWDKEKIGNIMKACIILHNMIVED 360
Query: 289 E 289
E
Sbjct: 361 E 361
>At3g19120 unknown protein
Length = 446
Score = 45.1 bits (105), Expect = 6e-05
Identities = 68/271 (25%), Positives = 109/271 (40%), Gaps = 30/271 (11%)
Query: 46 VAKSLYILTHNAKNREVNFWFRRSGETISRHLHQVLKAIL-ELEEKFIVQPDGSTIPLEI 104
VA L L H + + + IS+ + V + + +L +FI P G +E
Sbjct: 152 VAMVLSRLAHGCSAKTLASRYSLDPYLISKITNMVTRLLATKLYPEFIKIPVGKRRLIET 211
Query: 105 SSSTRFYPYFKDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLK--FTYV 162
+ + GAID T ++++ K PR +Y VL D K F V
Sbjct: 212 TQGFEELTSLPNICGAIDSTPVKLRRRTKLNPRNIYGCKYGYDAVLLQVVADHKKIFWDV 271
Query: 163 LAGWEGSASDSRIIKNALTRE---------DKLKIPEGKY---YLV-DAGFMLTSGLITP 209
G DS +++L + +K+ G + Y+V D + L S L+TP
Sbjct: 272 CVKAPGGEDDSSHFRDSLLYKRLTSGDIVWEKVINIRGHHVRPYIVGDWCYPLLSFLMTP 331
Query: 210 YRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIK 269
+ + PP N LF+ R+ + A G+LK R++IL + G+
Sbjct: 332 F-------SPNGSGTPPEN---LFDGMLMKGRSVVVEAIGLLKARWKILQSLN---VGVN 378
Query: 270 AQKLIIFACCILHNYLMSA-EPNEDLIAEVD 299
I ACC+LHN A EP ++ + D
Sbjct: 379 HAPQTIVACCVLHNLCQIAREPEPEIWKDPD 409
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,742,027
Number of Sequences: 26719
Number of extensions: 319086
Number of successful extensions: 797
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 730
Number of HSP's gapped (non-prelim): 44
length of query: 346
length of database: 11,318,596
effective HSP length: 100
effective length of query: 246
effective length of database: 8,646,696
effective search space: 2127087216
effective search space used: 2127087216
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146752.6


