

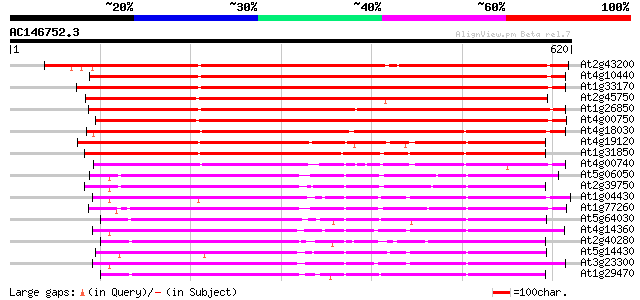
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.3 - phase: 0 /pseudo
(620 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g43200 hypothetical protein 652 0.0
At4g10440 putative protein 608 e-174
At1g33170 unknown protein 607 e-174
At2g45750 hypothetical protein 602 e-172
At1g26850 unknown protein 593 e-170
At4g00750 unknown protein 591 e-169
At4g18030 unknown protein 536 e-152
At4g19120 ERD3 protein (ERD3) 491 e-139
At1g31850 unknown protein 486 e-137
At4g00740 unknown protein 441 e-124
At5g06050 ankyrin-like protein 432 e-121
At2g39750 unknown protein 430 e-120
At1g04430 unknown protein 421 e-118
At1g77260 unknown protein 415 e-116
At5g64030 ankyrin-like protein 412 e-115
At4g14360 412 e-115
At2g40280 unknown protein 409 e-114
At5g14430 unknown protein 408 e-114
At3g23300 ankyrin repeat protein 405 e-113
At1g29470 unknown protein 390 e-108
>At2g43200 hypothetical protein
Length = 611
Score = 652 bits (1682), Expect = 0.0
Identities = 320/604 (52%), Positives = 416/604 (67%), Gaps = 33/604 (5%)
Query: 39 KRLILLHYHFNMFSKYLNIFFITTFFIF---------LYIITSSFFTS------------ 77
KRL L F +FS Y + I T I L++ + S + S
Sbjct: 14 KRLFLFFTPFLLFSLYYILTTIKTITISSQDRHHPPQLHVPSISHYYSLPETSENRSSPP 73
Query: 78 PLTLHSPITTKIS---HFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQ-NNERL 133
PL L P ++ S +F C N+TNY PC DP +++ + ++R+ERHCP E+
Sbjct: 74 PLLLPPPPSSSSSLSSYFPLCPKNFTNYLPCHDPSTARQYSIERHYRRERHCPDIAQEKF 133
Query: 134 TCLIPKPIGYKNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFP 193
CL+PKP GYK PFPWP+S+ AWF NVPF +L E KK+QNW+ L GDRFVFPGGGTSFP
Sbjct: 134 RCLVPKPTGYKTPFPWPESRKYAWFRNVPFKRLAELKKTQNWVRLEGDRFVFPGGGTSFP 193
Query: 194 DGVKGYVDDLKKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQV 253
GVK YVD + +LP L SG IRTVLD+GCGVASFGA L++Y ILTMSIAP D H+AQV
Sbjct: 194 GGVKDYVDVILSVLP--LASGSIRTVLDIGCGVASFGAFLLNYKILTMSIAPRDIHEAQV 251
Query: 254 MFALERGLPAMLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFW 313
FALERGLPAMLGV ST++L +PS+SFD+ HCSRCLV W + DGLYL E+DR+LRP G+W
Sbjct: 252 QFALERGLPAMLGVLSTYKLPYPSRSFDMVHCSRCLVNWTSYDGLYLMEVDRVLRPEGYW 311
Query: 314 VLSGPPINWRVNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKC 373
VLSGPP+ RV +K + + L+ + L ++ ++CWEK+AE + IW+KP NH++C
Sbjct: 312 VLSGPPVASRVKFKNQKRDSKELQNQMEKLNDVFRRLCWEKIAESYPVVIWRKPSNHLQC 371
Query: 374 MQKLNTLSSPKFCNSSDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPR 433
++L L P C+SSD DA WY +M CI PLP+V D ++ VL+ WP RLN PR
Sbjct: 372 RKRLKALKFPGLCSSSDPDAAWYKEMEPCITPLPDVNDTNKT---VLKNWPERLN-HVPR 427
Query: 434 LRKENHDVFSLKTYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKY 493
++ + ++ + D +W++RV YY+ K LS+GKYRNV+DMNAG GGFAAAL+KY
Sbjct: 428 MKTGSIQGTTIAGFKADTNLWQRRVLYYDTKFKFLSNGKYRNVIDMNAGLGGFAAALIKY 487
Query: 494 PVWVMNVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCD 553
P+WVMNVVPFD K N LG++Y+RGLIGTYM+WCE STYPRTYDLIHA +FS+Y+DKCD
Sbjct: 488 PMWVMNVVPFDLKPNTLGVVYDRGLIGTYMNWCEALSTYPRTYDLIHANGVFSLYLDKCD 547
Query: 554 ITDIVIEMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIM 613
I DI++EM RILRPEG VIIRD DV++KVK IT++MRW G + +D + H +++
Sbjct: 548 IVDILLEMQRILRPEGAVIIRDRFDVLVKVKAITNQMRWNG--TMYPEDNSVFDHGTILI 605
Query: 614 VLNN 617
V N+
Sbjct: 606 VDNS 609
>At4g10440 putative protein
Length = 633
Score = 608 bits (1567), Expect = e-174
Identities = 270/526 (51%), Positives = 374/526 (70%), Gaps = 5/526 (0%)
Query: 89 ISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFP 148
I +F+ C + + Y PCED +R ++F + +ERHCP +E L CLIP P YK PF
Sbjct: 90 IKYFEPCELSLSEYTPCEDRQRGRRFDRNMMKYRERHCPVKDELLYCLIPPPPNYKIPFK 149
Query: 149 WPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLP 208
WP+S+D AW+ N+P +L K QNWI + GDRF FPGGGT FP G Y+DD+ +L+P
Sbjct: 150 WPQSRDYAWYDNIPHKELSVEKAVQNWIQVEGDRFRFPGGGTMFPRGADAYIDDIARLIP 209
Query: 209 VNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVF 268
L G IRT +D GCGVASFGA L+ DI+ +S AP D H+AQV FALERG+PA++G+
Sbjct: 210 --LTDGGIRTAIDTGCGVASFGAYLLKRDIMAVSFAPRDTHEAQVQFALERGVPAIIGIM 267
Query: 269 STHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKA 328
+ RL +P+++FD+AHCSRCL+PW NDGLYL E+DR+LRPGG+W+LSGPPINW+ ++
Sbjct: 268 GSRRLPYPARAFDLAHCSRCLIPWFKNDGLYLMEVDRVLRPGGYWILSGPPINWKQYWRG 327
Query: 329 WQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNS 388
W+ L+KEQ+++E++A +CW+KV E G ++IWQKP+NHI+C + SP C+S
Sbjct: 328 WERTEEDLKKEQDSIEDVAKSLCWKKVTEKGDLSIWQKPLNHIECKKLKQNNKSPPICSS 387
Query: 389 SDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTYS 448
++D+ WY + CI PLPE + D+ AGG LE WP R PPR+ + + + +
Sbjct: 388 DNADSAWYKDLETCITPLPETNNPDDSAGGALEDWPDRAFAVPPRIIRGTIPEMNAEKFR 447
Query: 449 EDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSN 508
EDN +WK+R+++Y+ ++ LS G++RN+MDMNA GGFAA+++KYP WVMNVVP DA+
Sbjct: 448 EDNEVWKERIAHYKKIVPELSHGRFRNIMDMNAFLGGFAASMLKYPSWVMNVVPVDAEKQ 507
Query: 509 NLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPE 568
LG+IYERGLIGTY DWCE FSTYPRTYD+IHA LFS+Y +CD+T I++EM RILRPE
Sbjct: 508 TLGVIYERGLIGTYQDWCEGFSTYPRTYDMIHAGGLFSLYEHRCDLTLILLEMDRILRPE 567
Query: 569 GTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
GTV++RD+ + + KV++I M+W+ + D + +PE I+V
Sbjct: 568 GTVVLRDNVETLNKVEKIVKGMKWKS---QIVDHEKGPFNPEKILV 610
>At1g33170 unknown protein
Length = 709
Score = 607 bits (1565), Expect = e-174
Identities = 276/542 (50%), Positives = 377/542 (68%), Gaps = 7/542 (1%)
Query: 75 FTSPLTLHSPITTK-ISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERL 133
F S L IT + + +F+ C + + Y PCED +R ++F + +ERHCP +E L
Sbjct: 92 FESHHKLELKITNQTVKYFEPCDMSLSEYTPCEDRERGRRFDRNMMKYRERHCPSKDELL 151
Query: 134 TCLIPKPIGYKNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFP 193
CLIP P YK PF WP+S+D AW+ N+P +L K QNWI + G+RF FPGGGT FP
Sbjct: 152 YCLIPPPPNYKIPFKWPQSRDYAWYDNIPHKELSIEKAIQNWIQVEGERFRFPGGGTMFP 211
Query: 194 DGVKGYVDDLKKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQV 253
G Y+DD+ +L+P L G IRT +D GCGVASFGA L+ DI+ MS AP D H+AQV
Sbjct: 212 RGADAYIDDIARLIP--LTDGAIRTAIDTGCGVASFGAYLLKRDIVAMSFAPRDTHEAQV 269
Query: 254 MFALERGLPAMLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFW 313
FALERG+PA++G+ + RL +P+++FD+AHCSRCL+PW NDGLYL E+DR+LRPGG+W
Sbjct: 270 QFALERGVPAIIGIMGSRRLPYPARAFDLAHCSRCLIPWFQNDGLYLTEVDRVLRPGGYW 329
Query: 314 VLSGPPINWRVNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKC 373
+LSGPPINW+ +K W+ L++EQ+++E+ A +CW+KV E G ++IWQKPINH++C
Sbjct: 330 ILSGPPINWKKYWKGWERSQEDLKQEQDSIEDAARSLCWKKVTEKGDLSIWQKPINHVEC 389
Query: 374 MQKLNTLSSPKFCNSSD-SDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPP 432
+ +P C+ SD D WY + +C+ PLPE DE AGG LE WP R PP
Sbjct: 390 NKLKRVHKTPPLCSKSDLPDFAWYKDLESCVTPLPEANSSDEFAGGALEDWPNRAFAVPP 449
Query: 433 RLRKENHDVFSLKTYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVK 492
R+ + + + EDN +WK+R+SYY+ ++ LS G++RN+MDMNA GGFAAA++K
Sbjct: 450 RIIGGTIPDINAEKFREDNEVWKERISYYKQIMPELSRGRFRNIMDMNAYLGGFAAAMMK 509
Query: 493 YPVWVMNVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKC 552
YP WVMNVVP DA+ LG+I+ERG IGTY DWCE FSTYPRTYDLIHA LFS+Y ++C
Sbjct: 510 YPSWVMNVVPVDAEKQTLGVIFERGFIGTYQDWCEGFSTYPRTYDLIHAGGLFSIYENRC 569
Query: 553 DITDIVIEMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMI 612
D+T I++EM RILRPEGTV+ RD+ +++ K++ IT+ MRW+ + D + +PE I
Sbjct: 570 DVTLILLEMDRILRPEGTVVFRDTVEMLTKIQSITNGMRWKSR---ILDHERGPFNPEKI 626
Query: 613 MV 614
++
Sbjct: 627 LL 628
>At2g45750 hypothetical protein
Length = 631
Score = 602 bits (1551), Expect = e-172
Identities = 277/516 (53%), Positives = 363/516 (69%), Gaps = 7/516 (1%)
Query: 84 PITTKISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGY 143
P+T F C+ + + PCED KR KF ++ ++RHCP+ E L C IP P GY
Sbjct: 79 PVTETAVSFPSCAAALSEHTPCEDAKRSLKFSRERLEYRQRHCPEREEILKCRIPAPYGY 138
Query: 144 KNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDL 203
K PF WP S+D AWF+NVP T+L KK+QNW+ DRF FPGGGT FP G Y+DD+
Sbjct: 139 KTPFRWPASRDVAWFANVPHTELTVEKKNQNWVRYENDRFWFPGGGTMFPRGADAYIDDI 198
Query: 204 KKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPA 263
+L ++L G IRT +D GCGVASFGA L+ +I TMS AP D H+AQV FALERG+PA
Sbjct: 199 GRL--IDLSDGSIRTAIDTGCGVASFGAYLLSRNITTMSFAPRDTHEAQVQFALERGVPA 256
Query: 264 MLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWR 323
M+G+ +T RL +PS++FD+AHCSRCL+PW NDG YL E+DR+LRPGG+W+LSGPPINW+
Sbjct: 257 MIGIMATIRLPYPSRAFDLAHCSRCLIPWGQNDGAYLMEVDRVLRPGGYWILSGPPINWQ 316
Query: 324 VNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSP 383
+K W+ L EQ +E++A +CW+KV + +AIWQKP NHI C + L +P
Sbjct: 317 KRWKGWERTMDDLNAEQTQIEQVARSLCWKKVVQRDDLAIWQKPFNHIDCKKTREVLKNP 376
Query: 384 KFC-NSSDSDAGWYTKMTACIFPLPEVKDIDE---IAGGVLEKWPIRLNDSPPRLRKENH 439
+FC + D D WYTKM +C+ PLPEV D ++ +AGG +EKWP RLN PPR+ K
Sbjct: 377 EFCRHDQDPDMAWYTKMDSCLTPLPEVDDAEDLKTVAGGKVEKWPARLNAIPPRVNKGAL 436
Query: 440 DVFSLKTYSEDNMIWKKRVSYYEVMLKSL-SSGKYRNVMDMNAGFGGFAAALVKYPVWVM 498
+ + + + E+ +WK+RVSYY+ + L +G+YRN++DMNA GGFAAAL PVWVM
Sbjct: 437 EEITPEAFLENTKLWKQRVSYYKKLDYQLGETGRYRNLVDMNAYLGGFAAALADDPVWVM 496
Query: 499 NVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIV 558
NVVP +AK N LG+IYERGLIGTY +WCE STYPRTYD IHA ++F++Y +C+ +I+
Sbjct: 497 NVVPVEAKLNTLGVIYERGLIGTYQNWCEAMSTYPRTYDFIHADSVFTLYQGQCEPEEIL 556
Query: 559 IEMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEG 594
+EM RILRP G VIIRD DV++KVKE+T + WEG
Sbjct: 557 LEMDRILRPGGGVIIRDDVDVLIKVKELTKGLEWEG 592
>At1g26850 unknown protein
Length = 616
Score = 593 bits (1529), Expect = e-170
Identities = 278/527 (52%), Positives = 359/527 (67%), Gaps = 6/527 (1%)
Query: 88 KISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPF 147
K+ F+ C YT+Y PC+D +R FP+ + +ERHC NE+L CLIP P GY PF
Sbjct: 82 KVKAFEPCDGRYTDYTPCQDQRRAMTFPRDSMIYRERHCAPENEKLHCLIPAPKGYVTPF 141
Query: 148 PWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLL 207
WPKS+D ++N P+ L K QNWI GD F FPGGGT FP G Y+D L ++
Sbjct: 142 SWPKSRDYVPYANAPYKALTVEKAIQNWIQYEGDVFRFPGGGTQFPQGADKYIDQLASVI 201
Query: 208 PVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGV 267
P +++G +RT LD GCGVAS+GA L ++ MS AP D H+AQV FALERG+PA++GV
Sbjct: 202 P--MENGTVRTALDTGCGVASWGAYLWSRNVRAMSFAPRDSHEAQVQFALERGVPAVIGV 259
Query: 268 FSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYK 327
T +L +P+++FD+AHCSRCL+PW ANDG+YL E+DR+LRPGG+W+LSGPPINW+VNYK
Sbjct: 260 LGTIKLPYPTRAFDMAHCSRCLIPWGANDGMYLMEVDRVLRPGGYWILSGPPINWKVNYK 319
Query: 328 AWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCN 387
AWQ L++EQ +EE A +CWEK E G+IAIWQK +N C + + + FC
Sbjct: 320 AWQRPKEDLQEEQRKIEEAAKLLCWEKKYEHGEIAIWQKRVNDEACRSRQDDPRA-NFCK 378
Query: 388 SSDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTY 447
+ D+D WY KM ACI P PE DE+AGG L+ +P RLN PPR+ + ++ Y
Sbjct: 379 TDDTDDVWYKKMEACITPYPETSSSDEVAGGELQAFPDRLNAVPPRISSGSISGVTVDAY 438
Query: 448 SEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKS 507
+DN WKK V Y+ + L +G+YRN+MDMNAGFGGFAAAL +WVMNVVP A+
Sbjct: 439 EDDNRQWKKHVKAYKRINSLLDTGRYRNIMDMNAGFGGFAAALESQKLWVMNVVPTIAEK 498
Query: 508 NNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRP 567
N LG++YERGLIG Y DWCE FSTYPRTYDLIHA LFS+Y +KC+ DI++EM RILRP
Sbjct: 499 NRLGVVYERGLIGIYHDWCEAFSTYPRTYDLIHANHLFSLYKNKCNADDILLEMDRILRP 558
Query: 568 EGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
EG VIIRD D ++KVK I MRW+ V D ++ PE +++
Sbjct: 559 EGAVIIRDDVDTLIKVKRIIAGMRWDAKLV---DHEDGPLVPEKVLI 602
>At4g00750 unknown protein
Length = 633
Score = 591 bits (1524), Expect = e-169
Identities = 272/524 (51%), Positives = 367/524 (69%), Gaps = 8/524 (1%)
Query: 95 CSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPKSKD 154
C ++ Y PCE R FP++ +ERHCP+ +E + C IP P GY PF WP+S+D
Sbjct: 99 CGVEFSEYTPCEFVNRSLNFPRERLIYRERHCPEKHEIVRCRIPAPYGYSLPFRWPESRD 158
Query: 155 NAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNLDSG 214
AWF+NVP T+L KK+QNW+ DRF+FPGGGT FP G Y+D++ +L +NL G
Sbjct: 159 VAWFANVPHTELTVEKKNQNWVRYEKDRFLFPGGGTMFPRGADAYIDEIGRL--INLKDG 216
Query: 215 RIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTHRLT 274
IRT +D GCGVASFGA LM +I+TMS AP D H+AQV FALERG+PA++GV ++ RL
Sbjct: 217 SIRTAIDTGCGVASFGAYLMSRNIVTMSFAPRDTHEAQVQFALERGVPAIIGVLASIRLP 276
Query: 275 FPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQTEPT 334
FP+++FD+AHCSRCL+PW +G YL E+DR+LRPGG+W+LSGPPINW+ ++K W+
Sbjct: 277 FPARAFDIAHCSRCLIPWGQYNGTYLIEVDRVLRPGGYWILSGPPINWQRHWKGWERTRD 336
Query: 335 VLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSSDSDAG 394
L EQ+ +E +A +CW K+ + +A+WQKP NH+ C + L P FC+ + + G
Sbjct: 337 DLNSEQSQIERVARSLCWRKLVQREDLAVWQKPTNHVHCKRNRIALGRPPFCHRTLPNQG 396
Query: 395 WYTKMTACIFPLPEV--KDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLKTYSEDNM 452
WYTK+ C+ PLPEV +I E+AGG L +WP RLN PPR++ + + + + +
Sbjct: 397 WYTKLETCLTPLPEVTGSEIKEVAGGQLARWPERLNALPPRIKSGSLEGITEDEFVSNTE 456
Query: 453 IWKKRVSYYEVMLKSLS-SGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSNNLG 511
W++RVSYY+ + L+ +G+YRN +DMNA GGFA+ALV PVWVMNVVP +A N LG
Sbjct: 457 KWQRRVSYYKKYDQQLAETGRYRNFLDMNAHLGGFASALVDDPVWVMNVVPVEASVNTLG 516
Query: 512 IIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPEGTV 571
+IYERGLIGTY +WCE STYPRTYD IHA ++FS+Y D+CD+ DI++EM RILRP+G+V
Sbjct: 517 VIYERGLIGTYQNWCEAMSTYPRTYDFIHADSVFSLYKDRCDMEDILLEMDRILRPKGSV 576
Query: 572 IIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMVL 615
IIRD DV+ KVK+ITD M+WEG + D +N E I+ L
Sbjct: 577 IIRDDIDVLTKVKKITDAMQWEGR---IGDHENGPLEREKILFL 617
>At4g18030 unknown protein
Length = 621
Score = 536 bits (1381), Expect = e-152
Identities = 259/538 (48%), Positives = 347/538 (64%), Gaps = 20/538 (3%)
Query: 86 TTKISH--------FQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLI 137
T KI H F+ C +Y PC++ R KFP++N +ERHCP +NE+L CL+
Sbjct: 71 TVKIPHKADPKPVSFKPCDVKLKDYTPCQEQDRAMKFPRENMIYRERHCPPDNEKLRCLV 130
Query: 138 PKPIGYKNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVK 197
P P GY PFPWPKS+D ++N PF L K QNW+ G+ F FPGGGT FP G
Sbjct: 131 PAPKGYMTPFPWPKSRDYVHYANAPFKSLTVEKAGQNWVQFQGNVFKFPGGGTMFPQGAD 190
Query: 198 GYVDDLKKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFAL 257
Y+++L ++P+ G +RT LD GCGVAS+GA ++ ++LTMS AP D H+AQV FAL
Sbjct: 191 AYIEELASVIPIK--DGSVRTALDTGCGVASWGAYMLKRNVLTMSFAPRDNHEAQVQFAL 248
Query: 258 ERGLPAMLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSG 317
ERG+PA++ V + L +P+++FD+A CSRCL+PW AN+G YL E+DR+LRPGG+WVLSG
Sbjct: 249 ERGVPAIIAVLGSILLPYPARAFDMAQCSRCLIPWTANEGTYLMEVDRVLRPGGYWVLSG 308
Query: 318 PPINWRVNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKL 377
PPINW+ +K W L EQ +E +A +CWEK E G IAI++K IN C +
Sbjct: 309 PPINWKTWHKTWNRTKAELNAEQKRIEGIAESLCWEKKYEKGDIAIFRKKINDRSCDR-- 366
Query: 378 NTLSSP-KFCNSSDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRK 436
S+P C D+D WY ++ C+ P P+V + +E+AGG L+K+P RL PP + K
Sbjct: 367 ---STPVDTCKRKDTDDVWYKEIETCVTPFPKVSNEEEVAGGKLKKFPERLFAVPPSISK 423
Query: 437 ENHDVFSLKTYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVW 496
+ ++Y ED +WKKRV+ Y+ + + + S +YRNVMDMNAG GGFAAAL W
Sbjct: 424 GLINGVDEESYQEDINLWKKRVTGYKRINRLIGSTRYRNVMDMNAGLGGFAAALESPKSW 483
Query: 497 VMNVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITD 556
VMNV+P N L ++YERGLIG Y DWCE FSTYPRTYD IHA +FS+Y C + D
Sbjct: 484 VMNVIP-TINKNTLSVVYERGLIGIYHDWCEGFSTYPRTYDFIHASGVFSLYQHSCKLED 542
Query: 557 IVIEMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
I++E RILRPEG VI RD DV+ V++I D MRW+ + D ++ PE I+V
Sbjct: 543 ILLETDRILRPEGIVIFRDEVDVLNDVRKIVDGMRWD---TKLMDHEDGPLVPEKILV 597
>At4g19120 ERD3 protein (ERD3)
Length = 600
Score = 491 bits (1265), Expect = e-139
Identities = 247/526 (46%), Positives = 335/526 (62%), Gaps = 22/526 (4%)
Query: 76 TSPLTLHSPITTKISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTC 135
+S L + + K F CS++Y +Y PC DP++ KK+ ERHCP +R C
Sbjct: 54 SSSLDVDDSLQVKSVSFSECSSDYQDYTPCTDPRKWKKYGTHRLTFMERHCPPVFDRKQC 113
Query: 136 LIPKPIGYKNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDG 195
L+P P GYK P WPKSKD W+ NVP+ + + K +QNW+ G++F+FPGGGT FP G
Sbjct: 114 LVPPPDGYKPPIRWPKSKDECWYRNVPYDWINKQKSNQNWLRKEGEKFIFPGGGTMFPHG 173
Query: 196 VKGYVDDLKKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMF 255
V YVD ++ L+P + G IRT +D GCGVAS+G L+D ILT+S+AP D H+AQV F
Sbjct: 174 VSAYVDLMQDLIP-EMKDGTIRTAIDTGCGVASWGGDLLDRGILTVSLAPRDNHEAQVQF 232
Query: 256 ALERGLPAMLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVL 315
ALERG+PA+LG+ ST RL FPS SFD+AHCSRCL+PW G+YL E+ RILRPGGFWVL
Sbjct: 233 ALERGIPAILGIISTQRLPFPSNSFDMAHCSRCLIPWTEFGGVYLLEVHRILRPGGFWVL 292
Query: 316 SGPPINWRVNYKAWQTEPTVLEKEQN--NLEELAMQMCWEKVAEGGQIAIWQKPINHIKC 373
SGPP+N+ +K W T T+ E+ N L+EL MC++ A+ IA+WQK +++ C
Sbjct: 293 SGPPVNYENRWKGWDT--TIEEQRSNYEKLQELLSSMCFKMYAKKDDIAVWQKSPDNL-C 349
Query: 374 MQKLNT---LSSPKFCNSSDSDAGWYTKMTAC-IFPLPEVKDIDEIAGGVLEKWPIRLND 429
KL+ PK +S + D+ WYT + C + P P++K D + KWP RL+
Sbjct: 350 YNKLSNDPDAYPPKCDDSLEPDSAWYTPLRPCVVVPSPKLKKTDLES---TPKWPERLHT 406
Query: 430 SPPRLRK---ENHDVFSLKTYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGF 486
+P R+ N +VF D+ WK R +Y+ +L ++ S K RNVMDMN +GG
Sbjct: 407 TPERISDVPGGNGNVF-----KHDDSKWKTRAKHYKKLLPAIGSDKIRNVMDMNTAYGGL 461
Query: 487 AAALVKYPVWVMNVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFS 546
AAALV P+WVMNVV A +N L ++++RGLIGTY DWCE FSTYPRTYDL+H LF+
Sbjct: 462 AAALVNDPLWVMNVVSSYA-ANTLPVVFDRGLIGTYHDWCEAFSTYPRTYDLLHVDGLFT 520
Query: 547 MYIDKCDITDIVIEMHRILRPEGTVIIRDSRDVILKVKEITDKMRW 592
+CD+ +++EM RILRP G IIR+S + + ++RW
Sbjct: 521 SESQRCDMKYVMLEMDRILRPSGYAIIRESSYFADSIASVAKELRW 566
>At1g31850 unknown protein
Length = 603
Score = 486 bits (1250), Expect = e-137
Identities = 239/514 (46%), Positives = 334/514 (64%), Gaps = 12/514 (2%)
Query: 83 SPITTKISHFQFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIG 142
+PI K F C + + +Y PC DPKR KK+ ERHCP E+ CLIP P G
Sbjct: 67 TPIQIKSVSFPECGSEFQDYTPCTDPKRWKKYGVHRLSFLERHCPPVYEKNECLIPPPDG 126
Query: 143 YKNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDD 202
YK P WPKS++ W+ NVP+ + + K +Q+W+ GD+F FPGGGT FP GV YVD
Sbjct: 127 YKPPIRWPKSREQCWYRNVPYDWINKQKSNQHWLKKEGDKFHFPGGGTMFPRGVSHYVDL 186
Query: 203 LKKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLP 262
++ L+P + G +RT +D GCGVAS+G L+D IL++S+AP D H+AQV FALERG+P
Sbjct: 187 MQDLIP-EMKDGTVRTAIDTGCGVASWGGDLLDRGILSLSLAPRDNHEAQVQFALERGIP 245
Query: 263 AMLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINW 322
A+LG+ ST RL FPS +FD+AHCSRCL+PW G+YL EI RI+RPGGFWVLSGPP+N+
Sbjct: 246 AILGIISTQRLPFPSNAFDMAHCSRCLIPWTEFGGIYLLEIHRIVRPGGFWVLSGPPVNY 305
Query: 323 RVNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKL--NTL 380
++ W T + + N L+ L MC++K A+ IA+WQK ++ C K+ N
Sbjct: 306 NRRWRGWNTTMEDQKSDYNKLQSLLTSMCFKKYAQKDDIAVWQK-LSDKSCYDKIAKNME 364
Query: 381 SSPKFCNSS-DSDAGWYTKMTACIF-PLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKEN 438
+ P C+ S + D+ WYT + C+ P P+VK + G + KWP RL+ +P R+ +
Sbjct: 365 AYPPKCDDSIEPDSAWYTPLRPCVVAPTPKVK---KSGLGSIPKWPERLHVAPERIGDVH 421
Query: 439 HDVFSLKTYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVM 498
S + D+ WK RV +Y+ +L +L + K RNVMDMN +GGF+AAL++ P+WVM
Sbjct: 422 GG--SANSLKHDDGKWKNRVKHYKKVLPALGTDKIRNVMDMNTVYGGFSAALIEDPIWVM 479
Query: 499 NVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIV 558
NVV +N+L ++++RGLIGTY DWCE FSTYPRTYDL+H +LF++ +C++ I+
Sbjct: 480 NVVS-SYSANSLPVVFDRGLIGTYHDWCEAFSTYPRTYDLLHLDSLFTLESHRCEMKYIL 538
Query: 559 IEMHRILRPEGTVIIRDSRDVILKVKEITDKMRW 592
+EM RILRP G VIIR+S + + + +RW
Sbjct: 539 LEMDRILRPSGYVIIRESSYFMDAITTLAKGIRW 572
>At4g00740 unknown protein
Length = 600
Score = 441 bits (1135), Expect = e-124
Identities = 233/533 (43%), Positives = 311/533 (57%), Gaps = 38/533 (7%)
Query: 93 QFCSTNYTNYCPCEDPKRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPKS 152
++C + PCEDP+R + ++ F +ERHCP E CLIP P GYK P PWP+S
Sbjct: 82 EYCPAEAVAHMPCEDPRRNSQLSREMNFYRERHCPLPEETPLCLIPPPSGYKIPVPWPES 141
Query: 153 KDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNLD 212
W +N+P+ K+ + K Q W+ G+ F FPGGGT FP G Y++ L + +P+N
Sbjct: 142 LHKIWHANMPYNKIADRKGHQGWMKREGEYFTFPGGGTMFPGGAGQYIEKLAQYIPLN-- 199
Query: 213 SGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTHR 272
G +RT LD+GCGVASFG +L+ IL +S AP D H +Q+ FALERG+PA + + T R
Sbjct: 200 GGTLRTALDMGCGVASFGGTLLSQGILALSFAPRDSHKSQIQFALERGVPAFVAMLGTRR 259
Query: 273 LTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQTE 332
L FP+ SFD+ HCSRCL+P+ A + Y E+DR+LRPGG+ V+SGPP+ W K W
Sbjct: 260 LPFPAYSFDLMHCSRCLIPFTAYNATYFIEVDRLLRPGGYLVISGPPVQWPKQDKEWA-- 317
Query: 333 PTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSS--D 390
+L+ +A +C+E +A G IW+KP+ C+ N + C+ S
Sbjct: 318 ---------DLQAVARALCYELIAVDGNTVIWKKPVGD-SCLPSQNEFGL-ELCDESVPP 366
Query: 391 SDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPR--LRKENHDVFSLKTYS 448
SDA WY K+ C+ VK E A G + KWP RL P R + K DVF
Sbjct: 367 SDA-WYFKLKRCVTRPSSVK--GEHALGTISKWPERLTKVPSRAIVMKNGLDVF-----E 418
Query: 449 EDNMIWKKRVSYYEVMLK-SLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKS 507
D W +RV+YY L L S RNVMDMNA FGGFAA L PVWVMNV+P K
Sbjct: 419 ADARRWARRVAYYRDSLNLKLKSPTVRNVMDMNAFFGGFAATLASDPVWVMNVIP-ARKP 477
Query: 508 NNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMY------IDKCDITDIVIEM 561
L +IY+RGLIG Y DWCEPFSTYPRTYD IH + S+ +C + D+++EM
Sbjct: 478 LTLDVIYDRGLIGVYHDWCEPFSTYPRTYDFIHVSGIESLIKRQDSSKSRCSLVDLMVEM 537
Query: 562 HRILRPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
RILRPEG V+IRDS +V+ KV + +RW + + + ES E I++
Sbjct: 538 DRILRPEGKVVIRDSPEVLDKVARMAHAVRWSSS---IHEKEPESHGREKILI 587
>At5g06050 ankyrin-like protein
Length = 682
Score = 432 bits (1112), Expect = e-121
Identities = 232/527 (44%), Positives = 312/527 (59%), Gaps = 28/527 (5%)
Query: 89 ISHFQFCSTNYTNYCPCEDP----KRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYK 144
+ F+ CS N T Y PC D KR + F ER+CP + L C +P P GY+
Sbjct: 146 VRKFEICSENMTEYIPCLDNVEAIKRLNSTARGERF--ERNCPNDGMGLNCTVPIPQGYR 203
Query: 145 NPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLK 204
+P PWP+S+D WF+NVP TKLVE K QNWI D+F FPGGGT F G Y+D +
Sbjct: 204 SPIPWPRSRDEVWFNNVPHTKLVEDKGGQNWIYKENDKFKFPGGGTQFIHGADQYLDQIS 263
Query: 205 KLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAM 264
+++P R VLD+GCGVASFGA LM ++LTMSIAP D H+ Q+ FALERG+PAM
Sbjct: 264 QMIPDISFGNHTRVVLDIGCGVASFGAYLMSRNVLTMSIAPKDVHENQIQFALERGVPAM 323
Query: 265 LGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRV 324
+ F+T RL +PS++FD+ HCSRC + W +DG+ L E++R+LR GG++V + P+
Sbjct: 324 VAAFTTRRLLYPSQAFDLVHCSRCRINWTRDDGILLLEVNRMLRAGGYFVWAAQPV---- 379
Query: 325 NYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPK 384
YK LE++ + L ++CW V + G IAIWQKP+N+ C SP
Sbjct: 380 -YK----HEKALEEQWEEMLNLTTRLCWVLVKKEGYIAIWQKPVNN-TCYLSRGAGVSPP 433
Query: 385 FCNSSDS-DAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFS 443
CNS D D WY + ACI + E G L WP RL P RL+ D +
Sbjct: 434 LCNSEDDPDNVWYVDLKACITRIEE-----NGYGANLAPWPARLLTPPDRLQTIQIDSYI 488
Query: 444 LK--TYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAAL--VKYPVWVMN 499
+ + ++ WK+ +S Y L G RNV+DM AGFGGFAAAL +K WV+N
Sbjct: 489 ARKELFVAESKYWKEIISNYVNALHWKQIG-LRNVLDMRAGFGGFAAALAELKVDCWVLN 547
Query: 500 VVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVI 559
V+P N L +IY+RGL+G DWCEPF TYPRTYDL+HA LFS+ +C++T +++
Sbjct: 548 VIPVSG-PNTLPVIYDRGLLGVMHDWCEPFDTYPRTYDLLHAAGLFSIERKRCNMTTMML 606
Query: 560 EMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNES 606
EM RILRP G V IRD+ +V +++EI + MRW A+ + S
Sbjct: 607 EMDRILRPGGRVYIRDTINVTSELQEIGNAMRWHTSLRETAEGPHSS 653
>At2g39750 unknown protein
Length = 694
Score = 430 bits (1106), Expect = e-120
Identities = 226/519 (43%), Positives = 309/519 (58%), Gaps = 28/519 (5%)
Query: 83 SPITTKISHFQFCSTNYTNYCPCEDP----KRQKKFPKKNYFRKERHCPQNNERLTCLIP 138
S +I F C + Y PC D K+ K + F ERHCP+ + L CL+P
Sbjct: 168 SKARVRIKKFGMCPESMREYIPCLDNTDVIKKLKSTERGERF--ERHCPEKGKGLNCLVP 225
Query: 139 KPIGYKNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKG 198
P GY+ P PWPKS+D WFSNVP T+LVE K QNWI+ ++F FPGGGT F G
Sbjct: 226 PPKGYRQPIPWPKSRDEVWFSNVPHTRLVEDKGGQNWISRDKNKFKFPGGGTQFIHGADQ 285
Query: 199 YVDDLKKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALE 258
Y+D + K++ IR +DVGCGVASFGA L+ D++TMS+AP D H+ Q+ FALE
Sbjct: 286 YLDQMSKMVSDITFGKHIRVAMDVGCGVASFGAYLLSRDVMTMSVAPKDVHENQIQFALE 345
Query: 259 RGLPAMLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGP 318
RG+PAM F+T RL +PS++FD+ HCSRC + W +DG+ L EI+R+LR GG++ +
Sbjct: 346 RGVPAMAAAFATRRLLYPSQAFDLIHCSRCRINWTRDDGILLLEINRMLRAGGYFAWAAQ 405
Query: 319 PINWRVNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLN 378
P+ ++ EP LE++ + L + +CW+ V + G +AIWQKP N+ C
Sbjct: 406 PV--------YKHEP-ALEEQWTEMLNLTISLCWKLVKKEGYVAIWQKPFNN-DCYLSRE 455
Query: 379 TLSSPKFCNSSDS-DAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKE 437
+ P C+ SD D WYT + CI +PE + GG + WP RL+ P RL+
Sbjct: 456 AGTKPPLCDESDDPDNVWYTNLKPCISRIPE-----KGYGGNVPLWPARLHTPPDRLQTI 510
Query: 438 NHDVFSLK--TYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAAL--VKY 493
D + + + ++ W + + Y LK K RNV+DM AGFGGFAAAL K
Sbjct: 511 KFDSYIARKELFKAESKYWNEIIGGYVRALK-WKKMKLRNVLDMRAGFGGFAAALNDHKL 569
Query: 494 PVWVMNVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCD 553
WV++VVP N L +IY+RGL+G DWCEPF TYPRTYD +HA LFS+ +C+
Sbjct: 570 DCWVLSVVPVSG-PNTLPVIYDRGLLGVMHDWCEPFDTYPRTYDFLHASGLFSIERKRCE 628
Query: 554 ITDIVIEMHRILRPEGTVIIRDSRDVILKVKEITDKMRW 592
++ I++EM RILRP G IRDS DV+ +++EIT M W
Sbjct: 629 MSTILLEMDRILRPGGRAYIRDSIDVMDEIQEITKAMGW 667
>At1g04430 unknown protein
Length = 623
Score = 421 bits (1081), Expect = e-118
Identities = 237/541 (43%), Positives = 305/541 (55%), Gaps = 34/541 (6%)
Query: 92 FQFCSTNYTNYCPCEDP----KRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPF 147
F C ++ PC D + + K ERHCP R CLIP P GYK P
Sbjct: 84 FPVCDDRHSEIIPCLDRNFIYQMRLKLDLSLMEHYERHCPPPERRFNCLIPPPSGYKVPI 143
Query: 148 PWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLL 207
WPKS+D W +N+P T L + K QNW+ G++ FPGGGT F G Y+ + +L
Sbjct: 144 KWPKSRDEVWKANIPHTHLAKEKSDQNWMVEKGEKISFPGGGTHFHYGADKYIASIANML 203
Query: 208 ----PVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPA 263
V D GR+RTVLDVGCGVASFGA L+ DI+TMS+AP+D H Q+ FALERG+PA
Sbjct: 204 NFSNDVLNDEGRLRTVLDVGCGVASFGAYLLASDIMTMSLAPNDVHQNQIQFALERGIPA 263
Query: 264 MLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPP--IN 321
LGV T RL +PS+SF+ AHCSRC + W+ DGL L E+DR+LRPGG++ S P
Sbjct: 264 YLGVLGTKRLPYPSRSFEFAHCSRCRIDWLQRDGLLLLELDRVLRPGGYFAYSSPEAYAQ 323
Query: 322 WRVNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLS 381
N K W KE + L E +MCW + Q +WQKP+++ C + +
Sbjct: 324 DEENLKIW--------KEMSALVE---RMCWRIAVKRNQTVVWQKPLSN-DCYLEREPGT 371
Query: 382 SPKFCNS-SDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHD 440
P C S +D DA M ACI P K + G L WP RL SPPRL
Sbjct: 372 QPPLCRSDADPDAVAGVSMEACITPYS--KHDHKTKGSGLAPWPARLTSSPPRLADFG-- 427
Query: 441 VFSLKTYSEDNMIWKKRV-SYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMN 499
+S + +D +WK++V SY+ +M + S RN+MDM A G FAAAL VWVMN
Sbjct: 428 -YSTDMFEKDTELWKQQVDSYWNLMSSKVKSNTVRNIMDMKAHMGSFAAALKDKDVWVMN 486
Query: 500 VVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDK-CDITDIV 558
VV D N L +IY+RGLIGT +WCE FSTYPRTYDL+HA+++FS K C D++
Sbjct: 487 VVSPDG-PNTLKLIYDRGLIGTNHNWCEAFSTYPRTYDLLHAWSIFSDIKSKGCSAEDLL 545
Query: 559 IEMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMVLNNN 618
IEM RILRP G VIIRD + V+ +K+ + WE V ++ N SS + N
Sbjct: 546 IEMDRILRPTGFVIIRDKQSVVESIKKYLQALHWE---TVASEKVNTSSELDQDSEDGEN 602
Query: 619 N 619
N
Sbjct: 603 N 603
>At1g77260 unknown protein
Length = 655
Score = 415 bits (1067), Expect = e-116
Identities = 225/538 (41%), Positives = 313/538 (57%), Gaps = 36/538 (6%)
Query: 88 KISHFQFCSTNYTNYCPCEDPKRQKKFPK-----KNYFRKERHCPQNNERLTCLIPKPIG 142
+I + C +Y PC D + + K +NY ERHCP+ + L CLIP P G
Sbjct: 139 QIEKLKLCDKTKIDYIPCLDNEEEIKRLNNTDRGENY---ERHCPKQS--LDCLIPPPDG 193
Query: 143 YKNPFPWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDD 202
YK P WP+S+D WF+NVP T+LVE K QNWI D+FVFPGGGT F G Y+D
Sbjct: 194 YKKPIQWPQSRDKIWFNNVPHTRLVEDKGGQNWIRREKDKFVFPGGGTQFIHGADQYLDQ 253
Query: 203 LKKLLPVNLDSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLP 262
+ +++P R R LD+GCGVASFGA LM + T+S+AP D H+ Q+ FALERG+P
Sbjct: 254 ISQMIPDITFGSRTRVALDIGCGVASFGAFLMQRNTTTLSVAPKDVHENQIQFALERGVP 313
Query: 263 AMLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINW 322
AM+ VF+T RL +PS+SF++ HCSRC + W +DG+ L E++R+LR GG++V + P+
Sbjct: 314 AMVAVFATRRLLYPSQSFEMIHCSRCRINWTRDDGILLLEVNRMLRAGGYFVWAAQPV-- 371
Query: 323 RVNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSS 382
YK L+++ + +L ++CWE + + G IA+W+KP+N+ C +
Sbjct: 372 ---YK----HEDNLQEQWKEMLDLTNRICWELIKKEGYIAVWRKPLNN-SCYVSREAGTK 423
Query: 383 PKFCN-SSDSDAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDV 441
P C D D WY M CI LP D G + WP RL+D P RL+ D
Sbjct: 424 PPLCRPDDDPDDVWYVDMKPCITRLP-----DNGYGANVSTWPARLHDPPERLQSIQMDA 478
Query: 442 F--SLKTYSEDNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAAL--VKYPVWV 497
+ + ++ W + V Y V + K RNV+DM AGFGGFAAAL + WV
Sbjct: 479 YISRKEIMKAESRFWLEVVESY-VRVFRWKEFKLRNVLDMRAGFGGFAAALNDLGLDCWV 537
Query: 498 MNVVPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDI 557
MN+VP N L +IY+RGL G DWCEPF TYPRTYDLIHA LFS+ +C+IT+I
Sbjct: 538 MNIVPVSG-FNTLPVIYDRGLQGAMHDWCEPFDTYPRTYDLIHAAFLFSVEKKRCNITNI 596
Query: 558 VIEMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMVL 615
++EM R+LRP G V IRDS ++ +++++ + W G D E H + +++
Sbjct: 597 MLEMDRMLRPGGHVYIRDSLSLMDQLQQVAKAIGWTAG----VHDTGEGPHASVRILI 650
>At5g64030 ankyrin-like protein
Length = 829
Score = 412 bits (1060), Expect = e-115
Identities = 220/504 (43%), Positives = 302/504 (59%), Gaps = 26/504 (5%)
Query: 101 NYCPCEDPKRQ-KKFPK-KNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPKSKDNAWF 158
+Y PC D + + P K+Y +ERHCP + TCL+P P GYK P WPKS++ W+
Sbjct: 308 DYIPCLDNVQAIRSLPSTKHYEHRERHCPDSPP--TCLVPLPDGYKRPIEWPKSREKIWY 365
Query: 159 SNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNLDSGRIRT 218
+NVP TKL EYK QNW+ + G+ FPGGGT F G Y+D +++ +P R R
Sbjct: 366 TNVPHTKLAEYKGHQNWVKVTGEYLTFPGGGTQFKHGALHYIDFIQESVPAIAWGKRSRV 425
Query: 219 VLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTHRLTFPSK 278
VLDVGCGVASFG L D D++TMS+AP DEH+AQV FALERG+PA+ V T RL FP +
Sbjct: 426 VLDVGCGVASFGGFLFDRDVITMSLAPKDEHEAQVQFALERGIPAISAVMGTTRLPFPGR 485
Query: 279 SFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQTEPTVLEK 338
FD+ HC+RC VPW G L E++R+LRPGGF+V S P+ + +TE + K
Sbjct: 486 VFDIVHCARCRVPWHIEGGKLLLELNRVLRPGGFFVWSATPVYQK------KTEDVEIWK 539
Query: 339 EQNNLEELAMQMCWEKVA------EGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSSDSD 392
+ EL +MCWE V+ G +A ++KP ++ +C + + P +S D +
Sbjct: 540 A---MSELIKKMCWELVSINKDTINGVGVATYRKPTSN-ECYKNRSEPVPPICADSDDPN 595
Query: 393 AGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVF---SLKTYSE 449
A W + AC+ PE D + E+WP RL +P L V+ + + +S
Sbjct: 596 ASWKVPLQACMHTAPE--DKTQRGSQWPEQWPARLEKAPFWLSSSQTGVYGKAAPEDFSA 653
Query: 450 DNMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSNN 509
D WK+ V+ + ++ RNVMDM A +GGFAAAL VWVMNVVP D+ +
Sbjct: 654 DYEHWKRVVTKSYLNGLGINWASVRNVMDMRAVYGGFAAALRDLKVWVMNVVPIDS-PDT 712
Query: 510 LGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPEG 569
L IIYERGL G Y DWCE FSTYPR+YDL+HA LFS +C++T ++ E+ R+LRPEG
Sbjct: 713 LAIIYERGLFGIYHDWCESFSTYPRSYDLLHADHLFSKLKQRCNLTAVIAEVDRVLRPEG 772
Query: 570 TVIIRDSRDVILKVKEITDKMRWE 593
+I+RD + I +V+ + M+WE
Sbjct: 773 KLIVRDDAETIQQVEGMVKAMKWE 796
>At4g14360
Length = 608
Score = 412 bits (1058), Expect = e-115
Identities = 229/535 (42%), Positives = 305/535 (56%), Gaps = 30/535 (5%)
Query: 92 FQFCSTNYTNYCPCEDP----KRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPF 147
F C ++ PC D + + K ERHCP R CLIP P GYK P
Sbjct: 76 FPVCDDRHSELIPCLDRNLIYQMRLKLDLSLMEHYERHCPPPERRFNCLIPPPNGYKVPI 135
Query: 148 PWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLL 207
WPKS+D W N+P T L K QNW+ + GD+ FPGGGT F G Y+ + +L
Sbjct: 136 KWPKSRDEVWKVNIPHTHLAHEKSDQNWMVVKGDKINFPGGGTHFHYGADKYIASMANML 195
Query: 208 --PVNL--DSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPA 263
P N+ + GR+RTV DVGCGVASFG L+ DILTMS+AP+D H Q+ FALERG+PA
Sbjct: 196 NYPNNVLNNGGRLRTVFDVGCGVASFGGYLLSSDILTMSLAPNDVHQNQIQFALERGIPA 255
Query: 264 MLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWR 323
LGV T RL +PS+SF+++HCSRC + W+ DG+ L E+DR+LRPGG++ S P
Sbjct: 256 SLGVLGTKRLPYPSRSFELSHCSRCRIDWLQRDGILLLELDRVLRPGGYFAYSSP----- 310
Query: 324 VNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSP 383
A E + +E + L E +MCW+ A+ Q IWQKP+ + C + + P
Sbjct: 311 -EAYAQDEEDLRIWREMSALVE---RMCWKIAAKRNQTVIWQKPLTN-DCYLEREPGTQP 365
Query: 384 KFCNS-SDSDAGWYTKMTACIFPLPEVKDID-EIAGGVLEKWPIRLNDSPPRLRKENHDV 441
C S +D DA W M ACI D D + G L WP RL PPRL
Sbjct: 366 PLCRSDNDPDAVWGVNMEACI---TSYSDHDHKTKGSGLAPWPARLTSPPPRLADFG--- 419
Query: 442 FSLKTYSEDNMIWKKRV-SYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNV 500
+S + +D +W++RV +Y++++ + S RN+MDM A G FAAAL + VWVMNV
Sbjct: 420 YSTGMFEKDTELWRQRVDTYWDLLSPRIESDTVRNIMDMKASMGSFAAALKEKDVWVMNV 479
Query: 501 VPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDK-CDITDIVI 559
VP D N L +IY+RGL+G WCE FSTYPRTYDL+HA+ + S K C D+++
Sbjct: 480 VPEDG-PNTLKLIYDRGLMGAVHSWCEAFSTYPRTYDLLHAWDIISDIKKKGCSEVDLLL 538
Query: 560 EMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEG-GTVVVADDQNESSHPEMIM 613
EM RILRP G +IIRD + V+ VK+ + WE GT +D +S + I+
Sbjct: 539 EMDRILRPSGFIIIRDKQRVVDFVKKYLKALHWEEVGTKTDSDSDQDSDNVVFIV 593
>At2g40280 unknown protein
Length = 589
Score = 409 bits (1052), Expect = e-114
Identities = 221/503 (43%), Positives = 300/503 (58%), Gaps = 39/503 (7%)
Query: 101 NYCPCEDPKRQKKFPK--KNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPKSKDNAWF 158
+Y PC D K K ++ +ERHCP+ + + CL+P P YK P PWPKS+D W+
Sbjct: 90 DYIPCLDNYAAIKQLKSRRHMEHRERHCPEPSPK--CLLPLPDNYKPPVPWPKSRDMIWY 147
Query: 159 SNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNLDSGRIRT 218
NVP KLVEYKK QNW+ G+ VFPGGGT F GV YV+ ++K LP IR
Sbjct: 148 DNVPHPKLVEYKKEQNWVKKEGEFLVFPGGGTQFKFGVTHYVEFIEKALPSIKWGKNIRV 207
Query: 219 VLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTHRLTFPSK 278
VLDVGCGVASFG SL+D D++TMS AP DEH+AQ+ FALERG+PA L V T +LTFPS
Sbjct: 208 VLDVGCGVASFGGSLLDKDVITMSFAPKDEHEAQIQFALERGIPATLSVIGTQQLTFPSN 267
Query: 279 SFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQTEPTVLEK 338
+FD+ HC+RC V W A+ G L E++R+LRPGGF++ S P+ +R N + +
Sbjct: 268 AFDLIHCARCRVHWDADGGKPLLELNRVLRPGGFFIWSATPV-YRDNDRD--------SR 318
Query: 339 EQNNLEELAMQMCWEKV-----AEGGQIAIWQKPINHIKCMQKLNTLSSPKFCNSSDSDA 393
N + L +CW+ V + G + I+QKP + C K +T P C+ +++
Sbjct: 319 IWNEMVSLTKSICWKVVTKTVDSSGIGLVIYQKPTSE-SCYNKRST-QDPPLCDKKEANG 376
Query: 394 GWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLK--TYSEDN 451
WY + C+ LP G ++ WP P RL S+K T +D
Sbjct: 377 SWYVPLAKCLSKLP---------SGNVQSWP---ELWPKRLVSVKPQSISVKAETLKKDT 424
Query: 452 MIWKKRVSYYEVMLKSLSS--GKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSNN 509
W VS +V LK L+ RNVMDMNAGFGGFAAAL+ P+WVMNVVP D K +
Sbjct: 425 EKWSASVS--DVYLKHLAVNWSTVRNVMDMNAGFGGFAAALINLPLWVMNVVPVD-KPDT 481
Query: 510 LGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPEG 569
L ++Y+RGLIG Y DWCE +TYPRTYDL+H+ L +C+I +V E+ RI+RP G
Sbjct: 482 LSVVYDRGLIGVYHDWCESVNTYPRTYDLLHSSFLLGDLTQRCEIVQVVAEIDRIVRPGG 541
Query: 570 TVIIRDSRDVILKVKEITDKMRW 592
++++D+ + I+K++ I + W
Sbjct: 542 YLVVQDNMETIMKLESILGSLHW 564
>At5g14430 unknown protein
Length = 612
Score = 408 bits (1048), Expect = e-114
Identities = 221/510 (43%), Positives = 298/510 (58%), Gaps = 27/510 (5%)
Query: 95 CSTNYTNYCPCEDPKRQKKFPKKNYF----RKERHCPQNNERLTCLIPKPIGYKNPFPWP 150
C + ++ PC D + K E HCP + R CL+P P+GYK P WP
Sbjct: 83 CDSRHSELIPCLDRNLHYQLKLKLNLSLMEHYEHHCPPSERRFNCLVPPPVGYKIPLRWP 142
Query: 151 KSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVN 210
S+D W +N+P T L + K QNW+ + GD+ FPGGGT F +G Y+ L ++L
Sbjct: 143 VSRDEVWKANIPHTHLAQEKSDQNWMVVNGDKINFPGGGTHFHNGADKYIVSLAQMLKFP 202
Query: 211 LDS----GRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLG 266
D G IR VLDVGCGVASFGA L+ +DI+ MS+AP+D H Q+ FALERG+P+ LG
Sbjct: 203 GDKLNNGGSIRNVLDVGCGVASFGAYLLSHDIIAMSLAPNDVHQNQIQFALERGIPSTLG 262
Query: 267 VFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNY 326
V T RL +PS+SF++AHCSRC + W+ DG+ L E+DR+LRPGG++V S P
Sbjct: 263 VLGTKRLPYPSRSFELAHCSRCRIDWLQRDGILLLELDRLLRPGGYFVYSSP-------- 314
Query: 327 KAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSPKFC 386
+A+ +P K N + +L +MCW+ VA+ Q IW KPI++ C K + P C
Sbjct: 315 EAYAHDPE-NRKIGNAMHDLFKRMCWKVVAKRDQSVIWGKPISN-SCYLKRDPGVLPPLC 372
Query: 387 NSSDS-DAGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKENHDVFSLK 445
S D DA W M ACI P + +G L WP RL PPRL + + +
Sbjct: 373 PSGDDPDATWNVSMKACISPYSVRMHKERWSG--LVPWPRRLTAPPPRLEEIG---VTPE 427
Query: 446 TYSEDNMIWKKRV-SYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFD 504
+ ED W+ RV Y++++ + RNVMDM++ GGFAAAL VWVMNV+P
Sbjct: 428 QFREDTETWRLRVIEYWKLLKPMVQKNSIRNVMDMSSNLGGFAAALNDKDVWVMNVMPVQ 487
Query: 505 AKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFS-MYIDKCDITDIVIEMHR 563
+ S + IIY+RGLIG DWCE F TYPRT+DLIHA+ F+ C D++IEM R
Sbjct: 488 S-SPRMKIIYDRGLIGATHDWCEAFDTYPRTFDLIHAWNTFTETQARGCSFEDLLIEMDR 546
Query: 564 ILRPEGTVIIRDSRDVILKVKEITDKMRWE 593
ILRPEG VIIRD+ D I +K+ ++W+
Sbjct: 547 ILRPEGFVIIRDTTDNISYIKKYLTLLKWD 576
>At3g23300 ankyrin repeat protein
Length = 611
Score = 405 bits (1042), Expect = e-113
Identities = 222/535 (41%), Positives = 304/535 (56%), Gaps = 29/535 (5%)
Query: 92 FQFCSTNYTNYCPCEDP----KRQKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPF 147
F C ++ PC D + + K ERHCP R CLIP P GYK P
Sbjct: 79 FPVCDDRHSELIPCLDRNLIYQMRLKLDLSLMEHYERHCPPPERRFNCLIPPPPGYKIPI 138
Query: 148 PWPKSKDNAWFSNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLL 207
WPKS+D W N+P T L K QNW+ + G++ FPGGGT F G Y+ + +L
Sbjct: 139 KWPKSRDEVWKVNIPHTHLAHEKSDQNWMVVKGEKINFPGGGTHFHYGADKYIASMANML 198
Query: 208 --PVNL--DSGRIRTVLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPA 263
P N+ + GR+RT LDVGCGVASFG L+ +I+TMS+AP+D H Q+ FALERG+PA
Sbjct: 199 NFPNNVLNNGGRLRTFLDVGCGVASFGGYLLASEIMTMSLAPNDVHQNQIQFALERGIPA 258
Query: 264 MLGVFSTHRLTFPSKSFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWR 323
LGV T RL +PS+SF++AHCSRC + W+ DG+ L E+DR+LRPGG++ S P
Sbjct: 259 YLGVLGTKRLPYPSRSFELAHCSRCRIDWLQRDGILLLELDRVLRPGGYFAYSSP----- 313
Query: 324 VNYKAWQTEPTVLEKEQNNLEELAMQMCWEKVAEGGQIAIWQKPINHIKCMQKLNTLSSP 383
+A+ + L + + L +MCW A+ Q IWQKP+ + C + P
Sbjct: 314 ---EAYAQDEEDL-RIWREMSALVGRMCWTIAAKRNQTVIWQKPLTN-DCYLGREPGTQP 368
Query: 384 KFCNS-SDSDAGWYTKMTACIFPLPEVKDID-EIAGGVLEKWPIRLNDSPPRLRKENHDV 441
CNS SD DA + M ACI + D D + G L WP RL PPRL
Sbjct: 369 PLCNSDSDPDAVYGVNMEACI---TQYSDHDHKTKGSGLAPWPARLTSPPPRLADFG--- 422
Query: 442 FSLKTYSEDNMIWKKRV-SYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNV 500
+S + +D W++RV +Y++++ + S RN+MDM A G FAAAL + VWVMNV
Sbjct: 423 YSTDIFEKDTETWRQRVDTYWDLLSPKIQSDTVRNIMDMKASMGSFAAALKEKDVWVMNV 482
Query: 501 VPFDAKSNNLGIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDK-CDITDIVI 559
VP D N L +IY+RGL+G WCE FSTYPRTYDL+HA+ + S + C D+++
Sbjct: 483 VPEDG-PNTLKLIYDRGLMGAVHSWCEAFSTYPRTYDLLHAWDIISDIKKRGCSAEDLLL 541
Query: 560 EMHRILRPEGTVIIRDSRDVILKVKEITDKMRWEGGTVVVADDQNESSHPEMIMV 614
EM RILRP G ++IRD + V+ VK+ + WE A + ++ S +++V
Sbjct: 542 EMDRILRPSGFILIRDKQSVVDLVKKYLKALHWEAVETKTASESDQDSDNVILIV 596
>At1g29470 unknown protein
Length = 768
Score = 390 bits (1001), Expect = e-108
Identities = 212/502 (42%), Positives = 293/502 (58%), Gaps = 25/502 (4%)
Query: 101 NYCPCEDPKR--QKKFPKKNYFRKERHCPQNNERLTCLIPKPIGYKNPFPWPKSKDNAWF 158
+Y PC D + +K K+Y +ERHCP+ + R CL+ P GYK WPKS++ W+
Sbjct: 248 DYIPCLDNWQAIRKLHSTKHYEHRERHCPEESPR--CLVSLPEGYKRSIKWPKSREKIWY 305
Query: 159 SNVPFTKLVEYKKSQNWITLVGDRFVFPGGGTSFPDGVKGYVDDLKKLLPVNLDSGRIRT 218
+N+P TKL E K QNW+ + G+ FPGGGT F +G Y+D L++ P R R
Sbjct: 306 TNIPHTKLAEVKGHQNWVKMSGEYLTFPGGGTQFKNGALHYIDFLQESYPDIAWGNRTRV 365
Query: 219 VLDVGCGVASFGASLMDYDILTMSIAPSDEHDAQVMFALERGLPAMLGVFSTHRLTFPSK 278
+LDVGCGVASFG L D D+L +S AP DEH+AQV FALERG+PAM V T RL FP
Sbjct: 366 ILDVGCGVASFGGYLFDRDVLALSFAPKDEHEAQVQFALERGIPAMSNVMGTKRLPFPGS 425
Query: 279 SFDVAHCSRCLVPWIANDGLYLREIDRILRPGGFWVLSGPPINWRVNYKAWQTEPTVLEK 338
FD+ HC+RC VPW G L E++R LRPGGF+V S P+ Y+ + + + +
Sbjct: 426 VFDLIHCARCRVPWHIEGGKLLLELNRALRPGGFFVWSATPV-----YRKTEEDVGIWKA 480
Query: 339 EQNNLEELAMQMCWE----KVAEGGQI--AIWQKPINHIKCMQKLNTLSSPKFCNSSDSD 392
+ +L MCWE K E ++ AI+QKP+++ KC + + P +S D +
Sbjct: 481 ----MSKLTKAMCWELMTIKKDELNEVGAAIYQKPMSN-KCYNERSQNEPPLCKDSDDQN 535
Query: 393 AGWYTKMTACIFPLPEVKDIDEIAGGVLEKWPIRLNDSPPRLRKEN--HDVFSLKTYSED 450
A W + ACI + E D + E WP R+ P L + + + + ++ D
Sbjct: 536 AAWNVPLEACIHKVTE--DSSKRGAVWPESWPERVETVPQWLDSQEGVYGKPAQEDFTAD 593
Query: 451 NMIWKKRVSYYEVMLKSLSSGKYRNVMDMNAGFGGFAAALVKYPVWVMNVVPFDAKSNNL 510
+ WK VS + + RNVMDM A +GGFAAAL +WVMNVVP D+ + L
Sbjct: 594 HERWKTIVSKSYLNGMGIDWSYVRNVMDMRAVYGGFAAALKDLKLWVMNVVPIDS-PDTL 652
Query: 511 GIIYERGLIGTYMDWCEPFSTYPRTYDLIHAYALFSMYIDKCDITDIVIEMHRILRPEGT 570
IIYERGL G Y DWCE FSTYPRTYDL+HA LFS +C++ ++ E+ RILRP+GT
Sbjct: 653 PIIYERGLFGIYHDWCESFSTYPRTYDLLHADHLFSSLKKRCNLVGVMAEVDRILRPQGT 712
Query: 571 VIIRDSRDVILKVKEITDKMRW 592
I+RD + I +++++ M+W
Sbjct: 713 FIVRDDMETIGEIEKMVKSMKW 734
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.140 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,309,549
Number of Sequences: 26719
Number of extensions: 717204
Number of successful extensions: 2165
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1914
Number of HSP's gapped (non-prelim): 54
length of query: 620
length of database: 11,318,596
effective HSP length: 105
effective length of query: 515
effective length of database: 8,513,101
effective search space: 4384247015
effective search space used: 4384247015
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146752.3


