

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.15 + phase: 0 /pseudo
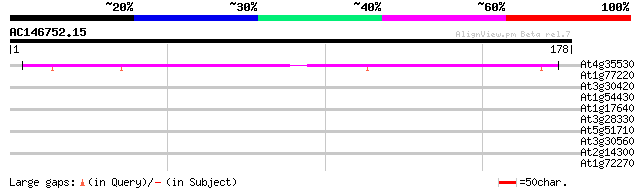
(178 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g35530 unknown protein 113 6e-26
At1g77220 unknown protei 32 0.18
At3g30420 hypothetical protein 29 1.5
At1g54430 hypothetical protein 29 1.5
At1g17640 RNA-binding protein, putative 28 3.4
At3g28330 hypothetical protein 27 5.8
At5g51710 potassium/proton antiporter-like protein 27 7.6
At3g30560 hypothetical protein 27 7.6
At2g14300 pseudogene; similar to MURA transposase of maize Muta... 27 7.6
At1g72270 hypothetical protein 27 7.6
>At4g35530 unknown protein
Length = 195
Score = 113 bits (282), Expect = 6e-26
Identities = 69/181 (38%), Positives = 103/181 (56%), Gaps = 16/181 (8%)
Query: 5 RYNYVHDQ--KYPSEDVDIHHIVLRSNGGKYF---FVYASALIVLACGIYLYLLEEKSIS 59
RY Y+H+ K E +DIHH+++ + G + + L+ LA +Y L ++
Sbjct: 10 RYTYIHESGSKSTREAIDIHHVIINGSSGTGYARRWGLGFFLVFLASSMYFLLGKDNPAR 69
Query: 60 LVYYSLLFDIFLVKLLLRLPVIKVLKLILVILTKLSFSCRVCCDYASFWSTA*----DSL 115
+ + L FLV L R V K +IL +F ++ Y S + + D +
Sbjct: 70 TLSWGCLLSGFLVMLHSRKFVKKESVIILP-----TFGIQLETQYLSGKTVSRFIPIDKI 124
Query: 116 YEPVLLECVTPVTCYWTLSLIVREESEMVLVYKNLRPPVKMLVHVWKALCAA--TDNKGE 173
+PVL+ECVTP+TCYW+LSL +R E ++ LV+K LRPP+KMLV +WKALCAA TD++ E
Sbjct: 125 LKPVLVECVTPITCYWSLSLFLRGEEQLTLVFKELRPPLKMLVPIWKALCAAIGTDHQSE 184
Query: 174 T 174
T
Sbjct: 185 T 185
>At1g77220 unknown protei
Length = 484
Score = 32.0 bits (71), Expect = 0.18
Identities = 26/89 (29%), Positives = 45/89 (50%), Gaps = 6/89 (6%)
Query: 23 HIVLRSNGGKYF---FVYASALIVLACGIYLYLLEEKSISLVYYSLLFDIFLVKLLLRLP 79
H L + G+Y + AS +V+A + +YL+ E S Y FL+ L+L +P
Sbjct: 28 HPNLGVDSGQYLTWPILSASVFVVIAILLPMYLIFEHLAS--YNQPEEQKFLIGLILMVP 85
Query: 80 VIKVLKLILVILTKLSFSCRVCCD-YASF 107
V V + ++ ++ +F+C V D Y +F
Sbjct: 86 VYAVESFLSLVNSEAAFNCEVIRDCYEAF 114
>At3g30420 hypothetical protein
Length = 837
Score = 28.9 bits (63), Expect = 1.5
Identities = 9/34 (26%), Positives = 21/34 (61%)
Query: 131 WTLSLIVREESEMVLVYKNLRPPVKMLVHVWKAL 164
W+ S ++R ++L+Y + P+K+ H W+++
Sbjct: 229 WSTSYLLRSLFVLILIYCEVSEPLKLWSHCWESM 262
>At1g54430 hypothetical protein
Length = 1639
Score = 28.9 bits (63), Expect = 1.5
Identities = 9/34 (26%), Positives = 21/34 (61%)
Query: 131 WTLSLIVREESEMVLVYKNLRPPVKMLVHVWKAL 164
W+ S ++R ++L+Y + P+K+ H W+++
Sbjct: 1062 WSTSYLLRSLFVLILIYCEVSEPLKLWSHCWESM 1095
>At1g17640 RNA-binding protein, putative
Length = 369
Score = 27.7 bits (60), Expect = 3.4
Identities = 10/29 (34%), Positives = 18/29 (61%)
Query: 3 DTRYNYVHDQKYPSEDVDIHHIVLRSNGG 31
D+ +++VHD++ P D ++ R NGG
Sbjct: 11 DSYHHHVHDEQLPQSDFEVETFDDRRNGG 39
>At3g28330 hypothetical protein
Length = 349
Score = 26.9 bits (58), Expect = 5.8
Identities = 16/49 (32%), Positives = 25/49 (50%), Gaps = 7/49 (14%)
Query: 118 PVLLECVT-----PVTCYWTLSLIVREESEMVLVYK--NLRPPVKMLVH 159
PV +CV + C+W L + R E+ +VL YK L+P L++
Sbjct: 87 PVSRQCVEILPSQKLDCFWILGIATRVENGVVLGYKVVLLKPNFTFLIY 135
>At5g51710 potassium/proton antiporter-like protein
Length = 568
Score = 26.6 bits (57), Expect = 7.6
Identities = 20/54 (37%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query: 19 VDIHHIVLRSNGGKYFFVY---ASALIVLACGIYLYLLEEKSISLVYYSLLFDI 69
+ H VL + G++ FV AS L V+ +YL LL ++SLV LLF +
Sbjct: 476 ISFHVGVLLAQIGEFAFVLLSRASNLHVIEGKMYLLLLGTTALSLVTTPLLFKL 529
>At3g30560 hypothetical protein
Length = 1473
Score = 26.6 bits (57), Expect = 7.6
Identities = 10/35 (28%), Positives = 19/35 (53%)
Query: 130 YWTLSLIVREESEMVLVYKNLRPPVKMLVHVWKAL 164
+W VR+ ++L++++L P + H WK L
Sbjct: 920 FWCFHKYVRKLFVIMLIFESLSSPAVVWEHTWKIL 954
>At2g14300 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 1230
Score = 26.6 bits (57), Expect = 7.6
Identities = 11/35 (31%), Positives = 19/35 (53%)
Query: 130 YWTLSLIVREESEMVLVYKNLRPPVKMLVHVWKAL 164
+W VR+ ++L+ ++L PV + H WK L
Sbjct: 727 FWCSPKYVRKSFVIMLISESLSSPVVVWEHTWKIL 761
>At1g72270 hypothetical protein
Length = 2777
Score = 26.6 bits (57), Expect = 7.6
Identities = 21/74 (28%), Positives = 30/74 (40%), Gaps = 14/74 (18%)
Query: 48 IYLYLLEEKSISLVYYSLLFDIFLVKL------------LLRLPVIKVLKLILVILTKLS 95
I YL+E + L + +F L KL ++ LP VL + + KL
Sbjct: 1668 ILFYLVESSPLHLSVFGHIFFSMLSKLQGDSALITDDQFVMLLP--PVLLFLASVFAKLE 1725
Query: 96 FSCRVCCDYASFWS 109
SC C D S +S
Sbjct: 1726 KSCSKCLDITSLYS 1739
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.331 0.144 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,015,007
Number of Sequences: 26719
Number of extensions: 149159
Number of successful extensions: 372
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 363
Number of HSP's gapped (non-prelim): 11
length of query: 178
length of database: 11,318,596
effective HSP length: 93
effective length of query: 85
effective length of database: 8,833,729
effective search space: 750866965
effective search space used: 750866965
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146752.15


