

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146751.5 + phase: 0 /pseudo
(455 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
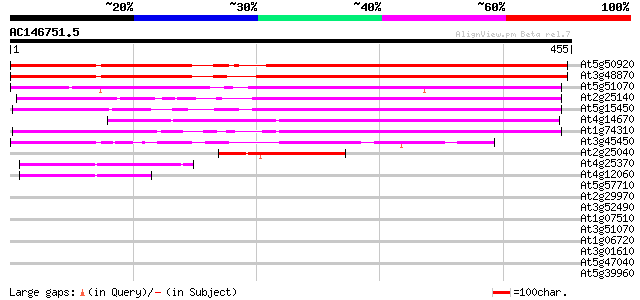
Score E
Sequences producing significant alignments: (bits) Value
At5g50920 ATP-dependent Clp protease, ATP-binding subunit 416 e-116
At3g48870 AtClpC 412 e-115
At5g51070 Erd1 protein precursor (sp|P42762) 263 2e-70
At2g25140 putative ATP-dependent CLPB protein 250 1e-66
At5g15450 clpB heat shock protein-like 248 4e-66
At4g14670 heat shock protein like 228 4e-60
At1g74310 heat shock protein 101 224 8e-59
At3g45450 clpC-like protein 189 2e-48
At2g25040 hypothetical protein 96 5e-20
At4g25370 unknown protein 45 8e-05
At4g12060 unknown protein 44 2e-04
At5g57710 101 kDa heat shock protein; HSP101-like protein 39 0.008
At2g29970 unknown protein 38 0.013
At3g52490 putative protein 35 0.065
At1g07510 unknown protein 34 0.19
At3g51070 putative protein 33 0.25
At1g06720 unknown protein 33 0.32
At3g01610 putative cell division control protein 32 0.55
At5g47040 Lon protease homolog 1 precursor 32 0.72
At5g39960 putative protein 32 0.72
>At5g50920 ATP-dependent Clp protease, ATP-binding subunit
Length = 929
Score = 416 bits (1070), Expect = e-116
Identities = 234/453 (51%), Positives = 302/453 (66%), Gaps = 44/453 (9%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AAK+LKS GI+ +R
Sbjct: 94 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLKSMGINLKDAR 153
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + + HLLLGLL+
Sbjct: 154 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGE 210
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N GAD + IR QVIR + EN +V NV SS +
Sbjct: 211 GVAARVLENLGADPSNIRTQVIRMVGEN----------------NEVTANVGGGSSSN-- 252
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
K P L E +GTNLTKLA+EGKL P VGR+ Q+ERV+QI+
Sbjct: 253 -KMPTL---------------------EEYGTNLTKLAEEGKLDPVVGRQPQIERVVQIL 290
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNPCL+GEPGVGKT+I +GLAQRI SG VPE ++GKKV+ LD+ + +G
Sbjct: 291 GRRTKNNPCLIGEPGVGKTAIAEGLAQRIASGDVPETIEGKKVITLDMGLLVAGTKYRGE 350
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ L++EI +ILF+ EVH + A A GA A ILK AL RG +QCI ATT
Sbjct: 351 FEERLKKLMEEIRQSDEIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATT 410
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQPVKV EP+V+ETI+ILKGLR YE H+KL YTDE+LVAAA
Sbjct: 411 LDEYRKHIEKDPALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDESLVAAAQ 470
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
LS QY+S+RFLPDKAIDLIDEAGS V+L HA+V
Sbjct: 471 LSYQYISDRFLPDKAIDLIDEAGSRVRLRHAQV 503
>At3g48870 AtClpC
Length = 952
Score = 412 bits (1060), Expect = e-115
Identities = 229/453 (50%), Positives = 299/453 (65%), Gaps = 43/453 (9%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM +Q+EAR +GH ++ TE ILLG++G+ G+AAK+LKS GI+ SR
Sbjct: 114 MFERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLKSMGINLKDSR 173
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + + HLLLGLL+
Sbjct: 174 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGE 230
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N GAD + IR QVIR + EN +V +V SS
Sbjct: 231 GVAARVLENLGADPSNIRTQVIRMVGEN----------------NEVTASVGGGSS---- 270
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
N + LE +GTNLTKLA+EGKL P VGR+ Q+ER++QI+
Sbjct: 271 -------------------GNSKMPTLEEYGTNLTKLAEEGKLDPVVGRQPQIERMVQIL 311
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNPCL+GEPGVGKT+I +GLAQRI SG VPE ++GK V+ LD+ + +G
Sbjct: 312 ARRTKNNPCLIGEPGVGKTAIAEGLAQRIASGDVPETIEGKTVITLDMGLLVAGTKYRGE 371
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ L++EI +ILF+ EVH + A A GA A ILK AL RG +QCI ATT
Sbjct: 372 FEERLKKLMEEIRQSDEIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATT 431
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQPVKV EP+VEE I+IL+GLR YE H+KL YTDEALVAAA
Sbjct: 432 IDEYRKHIEKDPALERRFQPVKVPEPTVEEAIQILQGLRERYEIHHKLRYTDEALVAAAQ 491
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
LS QY+S+RFLPDKAIDLIDEAGS V+L HA++
Sbjct: 492 LSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQL 524
>At5g51070 Erd1 protein precursor (sp|P42762)
Length = 945
Score = 263 bits (671), Expect = 2e-70
Identities = 160/463 (34%), Positives = 250/463 (53%), Gaps = 40/463 (8%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
+ E TE A + I+ +QKEA+ +G + T+H+LLG++ + + GI + +R
Sbjct: 79 VFERFTERAIRAIIFSQKEAKSLGKDMVYTQHLLLGLIAEDRDPQGFL--GSGITIDKAR 136
Query: 61 EQVMKLVGRGGG---------CSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHL 111
E V + S D+ F+ K V + ++++++++ + H+
Sbjct: 137 EAVWSIWDEANSDSKQEEASSTSYSKSTDMPFSISTKRVFEAAVEYSRTMDCQYIAPEHI 196
Query: 112 LLGLLQGSNGTVTQLIQNQGADVNKIREQVIRQIEENVHQVNL-PAEGSKNQMGGQVEEN 170
+GL +G+ ++++ GA++N + + +++ + + P+ SK
Sbjct: 197 AVGLFTVDDGSAGRVLKRLGANMNLLTAAALTRLKGEIAKDGREPSSSSKGSF------- 249
Query: 171 VTAESSKSNQIKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGRE 230
ES S +I A K K+ LE F +LT A EG + P +GRE
Sbjct: 250 ---ESPPSGRI------------AGSGPGGKKAKNVLEQFCVDLTARASEGLIDPVIGRE 294
Query: 231 EQVERVIQIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVAD 290
++V+RVIQI+CRR KNNP L+GE GVGKT+I +GLA I S P L K++++LD+
Sbjct: 295 KEVQRVIQILCRRTKNNPILLGEAGVGKTAIAEGLAISIAEASAPGFLLTKRIMSLDIGL 354
Query: 291 FLYVISNQGSSEERIRCLIKEIELCGNVILFVKEVHHIFDAATSG------ARSFAYILK 344
+ +G E R+ LI E++ G VILF+ EVH + + T G A +LK
Sbjct: 355 LMAGAKERGELEARVTALISEVKKSGKVILFIDEVHTLIGSGTVGRGNKGSGLDIANLLK 414
Query: 345 HALERGVIQCIFATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESH 404
+L RG +QCI +TT++E R E D L R FQPV + EPS E+ ++IL GLR YE+H
Sbjct: 415 PSLGRGELQCIASTTLDEFRSQFEKDKALARRFQPVLINEPSEEDAVKILLGLREKYEAH 474
Query: 405 YKLHYTDEALVAAANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
+ YT EA+ AA LS +Y+++RFLPDKAIDLIDEAGS ++
Sbjct: 475 HNCKYTMEAIDAAVYLSSRYIADRFLPDKAIDLIDEAGSRARI 517
>At2g25140 putative ATP-dependent CLPB protein
Length = 964
Score = 250 bits (638), Expect = 1e-66
Identities = 157/444 (35%), Positives = 245/444 (54%), Gaps = 50/444 (11%)
Query: 6 TECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQVMK 65
TE A + ++ A AR + + +EH++ +L +G+A KI GID + +
Sbjct: 89 TEMAWEGLINAFDAARESKQQIVESEHLMKALLEQKDGMARKIFTKAGIDNSSVLQATDL 148
Query: 66 LVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGTVTQ 125
+ + S S + + + +L+ + +H K + V H LL T+
Sbjct: 149 FISKQPTVSDASGQRLGSSLSV--ILENAKRHKKDMLDSYVSVEHFLLAYYSD-----TR 201
Query: 126 LIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQIKFPV 185
Q D+ K+ QV++ ++V ++ VT
Sbjct: 202 FGQEFFRDM-KLDIQVLKDAIKDVRG----------------DQRVT------------- 231
Query: 186 LTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICRRMK 245
D N + ALE +G +LT++A+ GKL P +GR++++ R IQI+CRR K
Sbjct: 232 -----------DRNPESKYQALEKYGNDLTEMARRGKLDPVIGRDDEIRRCIQILCRRTK 280
Query: 246 NNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERI 305
NNP ++GEPGVGKT+I +GLAQRI+ G VPE L +K+++LD+ L +G EER+
Sbjct: 281 NNPVIIGEPGVGKTAIAEGLAQRIVRGDVPEPLMNRKLISLDMGSLLAGAKFRGDFEERL 340
Query: 306 RCLIKEIELC-GNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATTVNEH 363
+ ++KE+ G ILF+ E+H + A A GA + +LK L RG ++CI ATT+ E+
Sbjct: 341 KAVMKEVSASNGQTILFIDEIHTVVGAGAMDGAMDASNLLKPMLGRGELRCIGATTLTEY 400
Query: 364 RMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAANLSQQ 423
R ++E D L+R FQ V V+PSVE+TI IL+GLR YE H+ + +D ALV+AA L+ +
Sbjct: 401 RKYIEKDPALERRFQQVLCVQPSVEDTISILRGLRERYELHHGVTISDSALVSAAVLADR 460
Query: 424 YVSERFLPDKAIDLIDEAGSHVQL 447
Y++ERFLPDKAIDL+DEAG+ +++
Sbjct: 461 YITERFLPDKAIDLVDEAGAKLKM 484
>At5g15450 clpB heat shock protein-like
Length = 968
Score = 248 bits (634), Expect = 4e-66
Identities = 155/447 (34%), Positives = 244/447 (53%), Gaps = 49/447 (10%)
Query: 3 ENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQ 62
+ TE A + I+++ A+ + + TEH++ +L NGLA +I G+D E
Sbjct: 80 QEFTEMAWQSIVSSPDVAKENKQQIVETEHLMKALLEQKNGLARRIFSKIGVDNTKVLEA 139
Query: 63 VMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGT 122
K + R G + + D + + + + K L V HL+L
Sbjct: 140 TEKFIQRQPKVYGDAAGSM-LGRDLEALFQRARQFKKDLKDSYVSVEHLVLAF------- 191
Query: 123 VTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQIK 182
AD + +Q+ + Q+ E + +S + K
Sbjct: 192 ---------ADDKRFGKQLFKDF--------------------QISERSLKSAIESIRGK 222
Query: 183 FPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICR 242
V+ D + + ALE +G +LT +A+EGKL P +GR++++ R IQI+ R
Sbjct: 223 QSVI----------DQDPEGKYEALEKYGKDLTAMAREGKLDPVIGRDDEIRRCIQILSR 272
Query: 243 RMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSE 302
R KNNP L+GEPGVGKT+I +GLAQRI+ G VP+ L +K+++LD+ + +G E
Sbjct: 273 RTKNNPVLIGEPGVGKTAISEGLAQRIVQGDVPQALMNRKLISLDMGALIAGAKYRGEFE 332
Query: 303 ERIRCLIKEI-ELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATTV 360
+R++ ++KE+ + G +ILF+ E+H + A AT+GA +LK L RG ++CI ATT+
Sbjct: 333 DRLKAVLKEVTDSEGQIILFIDEIHTVVGAGATNGAMDAGNLLKPMLGRGELRCIGATTL 392
Query: 361 NEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAANL 420
+E+R ++E D L+R FQ V V +P+VE+TI IL+GLR YE H+ + +D ALV AA L
Sbjct: 393 DEYRKYIEKDPALERRFQQVYVDQPTVEDTISILRGLRERYELHHGVRISDSALVEAAIL 452
Query: 421 SQQYVSERFLPDKAIDLIDEAGSHVQL 447
S +Y+S RFLPDKAIDL+DEA + +++
Sbjct: 453 SDRYISGRFLPDKAIDLVDEAAAKLKM 479
>At4g14670 heat shock protein like
Length = 668
Score = 228 bits (582), Expect = 4e-60
Identities = 134/369 (36%), Positives = 214/369 (57%), Gaps = 5/369 (1%)
Query: 80 DIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGTVTQLIQNQGADVNKIRE 139
D++F + K +L + HA SL + V +HL + L+ + I + G D + +
Sbjct: 3 DLKFDPNVKLILASARSHAMSLSHGQVTPLHLGVTLISDLTSVFYRAITSAG-DGDISAQ 61
Query: 140 QVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQIKFPVLTQNNNLGARKDDN 199
V+ I ++++++ G + ++ +S S+ +K + +
Sbjct: 62 SVVNVINQSLYKLTKRNLGDTKVGVAVLVISLLEDSQISDVLKEAGVVPEKVKSEVEKLR 121
Query: 200 ANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICRRMKNNPCLVGEPGVGKT 259
AL+ +GT+L + Q GKL P +GR ++ RVI+++ RR KNNP L+GEPGVGKT
Sbjct: 122 GEVILRALKTYGTDLVE--QAGKLDPVIGRHREIRRVIEVLSRRTKNNPVLIGEPGVGKT 179
Query: 260 SIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIRCLIKEIELC-GNV 318
++++GLAQRIL G VP L G K+++L+ + + +G EER++ ++K +E G V
Sbjct: 180 AVVEGLAQRILKGDVPINLTGVKLISLEFGAMVAGTTLRGQFEERLKSVLKAVEEAQGKV 239
Query: 319 ILFVKEVHHIFDAA-TSGARSFAYILKHALERGVIQCIFATTVNEHRMHMENDTTLKRIF 377
+LF+ E+H A SG+ A +LK L RG ++ I ATT+ E+R H+E D +R F
Sbjct: 240 VLFIDEIHMALGACKASGSTDAAKLLKPMLARGQLRFIGATTLEEYRTHVEKDAAFERRF 299
Query: 378 QPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAANLSQQYVSERFLPDKAIDL 437
Q V V EPSV +TI IL+GL+ YE H+ + D ALV +A LS++Y++ R LPDKAIDL
Sbjct: 300 QQVFVAEPSVPDTISILRGLKEKYEGHHGVRIQDRALVLSAQLSERYITGRRLPDKAIDL 359
Query: 438 IDEAGSHVQ 446
+DE+ +HV+
Sbjct: 360 VDESCAHVK 368
>At1g74310 heat shock protein 101
Length = 911
Score = 224 bits (571), Expect = 8e-59
Identities = 148/448 (33%), Positives = 233/448 (51%), Gaps = 50/448 (11%)
Query: 3 ENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNV-SRE 61
E T + I A + A GH + H+ ++ D G+ + + S G + S E
Sbjct: 4 EKFTHKTNETIATAHELAVNAGHAQFTPLHLAGALISDPTGIFPQAISSAGGENAAQSAE 63
Query: 62 QVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNG 121
+V+ + DI + V+ + KS G ++ L++GLL+ S
Sbjct: 64 RVINQALKKLPSQSPPPDDIPASSSLIKVIRRAQAAQKSRGDTHLAVDQLIMGLLEDSQ- 122
Query: 122 TVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQI 181
+ L+ G +++ +V E + + G +VE S S
Sbjct: 123 -IRDLLNEVGVATARVKSEV---------------EKLRGKEGKKVE-------SASGDT 159
Query: 182 KFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIIC 241
F AL+ +G +L + Q GKL P +GR+E++ RV++I+
Sbjct: 160 NF---------------------QALKTYGRDLVE--QAGKLDPVIGRDEEIRRVVRILS 196
Query: 242 RRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSS 301
RR KNNP L+GEPGVGKT++++GLAQRI+ G VP L ++++LD+ + +G
Sbjct: 197 RRTKNNPVLIGEPGVGKTAVVEGLAQRIVKGDVPNSLTDVRLISLDMGALVAGAKYRGEF 256
Query: 302 EERIRCLIKEIELC-GNVILFVKEVHHIFDAA-TSGARSFAYILKHALERGVIQCIFATT 359
EER++ ++KE+E G VILF+ E+H + A T G+ A + K L RG ++CI ATT
Sbjct: 257 EERLKSVLKEVEDAEGKVILFIDEIHLVLGAGKTEGSMDAANLFKPMLARGQLRCIGATT 316
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
+ E+R ++E D +R FQ V V EPSV +TI IL+GL+ YE H+ + D AL+ AA
Sbjct: 317 LEEYRKYVEKDAAFERRFQQVYVAEPSVPDTISILRGLKEKYEGHHGVRIQDRALINAAQ 376
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS +Y++ R LPDKAIDL+DEA ++V++
Sbjct: 377 LSARYITGRHLPDKAIDLVDEACANVRV 404
>At3g45450 clpC-like protein
Length = 341
Score = 189 bits (481), Expect = 2e-48
Identities = 146/398 (36%), Positives = 200/398 (49%), Gaps = 94/398 (23%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVI AQ+EAR +G+ + TEHILL ++G+ G+AAK+LKS GI+ +R
Sbjct: 1 MFERFTEKAIKVITLAQEEARRLGYNFFGTEHILLSLIGEGTGIAAKVLKSMGINLKDAR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT K F + + + T H
Sbjct: 61 VEVEKIIGRG---SGFVVVEIPFT-PRKACAGFVTRGSST-------TRH---------K 100
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++ GAD + IR QV I N S+ + +V E T + +
Sbjct: 101 GIAVRVLEILGADPSNIRTQVNAPILVN----------SQQALVAEVMETATWQVGNT-- 148
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ GKL P VGR+ Q++RV+QI+
Sbjct: 149 --------------------------------------RRGKLDPVVGRQPQIKRVVQIL 170
Query: 241 CRR-MKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQG 299
RR +NN CL+G+PGVGK +I +G+AQRI SG VPE +KGK VA N G
Sbjct: 171 ARRTCRNNACLIGKPGVGKRAIAEGIAQRIASGDVPETIKGKMNVA----------GNCG 220
Query: 300 SSEERIRCLIKEIELCG---NVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCI 355
+E R R K E+ G ++ILF+ E+H + A A GA A ILK ALER +Q
Sbjct: 221 WNEIRWRSRGKIEEVYGQSDDIILFIDEMHLLIGAGAVEGAIDAANILKPALERCELQ-- 278
Query: 356 FATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEI 393
+R H+END L+R FQPVKV EP+VEE I+I
Sbjct: 279 -------YRKHIENDPALERRFQPVKVPEPTVEEAIQI 309
>At2g25040 hypothetical protein
Length = 135
Score = 95.5 bits (236), Expect = 5e-20
Identities = 48/106 (45%), Positives = 75/106 (70%), Gaps = 4/106 (3%)
Query: 170 NVTAESSKSNQIKFPVLTQNNNLGARKDDNANK---QKSALEIFGTNLTKLAQEGKLHPF 226
+ T +S+S P+ Q +L R D AN+ + ALE +G++LTK+A++GKL P
Sbjct: 30 SATQAASRSRGQLLPLSLQFPSL-RRFSDKANQTDAKHEALETYGSDLTKMARQGKLPPL 88
Query: 227 VGREEQVERVIQIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSG 272
+GR+++V R IQI+CR ++NP ++GEPGVGKT+I++GLA+RI+ G
Sbjct: 89 IGRDDEVNRCIQILCRMTESNPVIIGEPGVGKTAIVEGLAERIVKG 134
>At4g25370 unknown protein
Length = 238
Score = 45.1 bits (105), Expect = 8e-05
Identities = 36/142 (25%), Positives = 68/142 (47%), Gaps = 3/142 (2%)
Query: 9 AKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQVMKLVG 68
A K + + EAR + + TE IL+GIL + AK L+ G+ R++ + L+G
Sbjct: 90 AIKSLAMGELEARKLKYPSTGTEAILMGILVEGTSTVAKFLRGNGVTLFKVRDETLSLLG 149
Query: 69 RGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYD-NVDTMHLLLGLLQGSNGTVTQLI 127
+ FS + T A+ + +++ D + T +LLLG+ + Q++
Sbjct: 150 K-SDMYFFSPEHPPLTEPAQKAIAWAIDEKNKSDVDGELTTAYLLLGVWSQKDSAGRQIL 208
Query: 128 QNQGADVNKIREQVIRQIEENV 149
+ G + +K +E V + + E+V
Sbjct: 209 EKLGFNEDKAKE-VEKSMNEDV 229
Score = 30.4 bits (67), Expect = 2.1
Identities = 17/69 (24%), Positives = 34/69 (48%)
Query: 74 SGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGTVTQLIQNQGAD 133
+G S K +++ A L A+ L Y + T +L+G+L TV + ++ G
Sbjct: 77 NGSSDKIPKWSARAIKSLAMGELEARKLKYPSTGTEAILMGILVEGTSTVAKFLRGNGVT 136
Query: 134 VNKIREQVI 142
+ K+R++ +
Sbjct: 137 LFKVRDETL 145
>At4g12060 unknown protein
Length = 241
Score = 43.9 bits (102), Expect = 2e-04
Identities = 34/108 (31%), Positives = 55/108 (50%), Gaps = 2/108 (1%)
Query: 9 AKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQVMKLVG 68
A K + EAR + + TE +L+GIL + +K L++ I RE+ +KL+G
Sbjct: 98 AIKSFAMGELEARKLKYPNTGTEALLMGILIEGTSFTSKFLRANKIMLYKVREETVKLLG 157
Query: 69 RGGGCSGFSCKDIRFTFDAKNVLDFSL-KHAKSLGYDNVDTMHLLLGL 115
+ FS + T DA+ LD +L ++ K+ G V H+LLG+
Sbjct: 158 K-ADMYFFSPEHPPLTEDAQRALDSALDQNLKAGGIGEVMPAHILLGI 204
>At5g57710 101 kDa heat shock protein; HSP101-like protein
Length = 990
Score = 38.5 bits (88), Expect = 0.008
Identities = 30/103 (29%), Positives = 49/103 (47%), Gaps = 23/103 (22%)
Query: 186 LTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICRRMK 245
+T+N+ L R NA+ +S V + + VERV+ I+ R K
Sbjct: 192 MTRNSYLNPRLQQNASSVQSG--------------------VSKNDDVERVMDILGRAKK 231
Query: 246 NNPCLVGEPGVGKTSIIQGLAQRILSGSVPE-KLKGKKVVALD 287
NP LVG+ G+ +I+ + ++I G V +K KVV+L+
Sbjct: 232 KNPVLVGDSEPGR--VIREILKKIEVGEVGNLAVKNSKVVSLE 272
>At2g29970 unknown protein
Length = 1002
Score = 37.7 bits (86), Expect = 0.013
Identities = 20/68 (29%), Positives = 34/68 (49%)
Query: 225 PFVGREEQVERVIQIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVV 284
PF +E R+ +++ R+ K NP LVG GV R G +P ++ G VV
Sbjct: 217 PFGDLDENCRRIGEVLARKDKKNPLLVGVCGVEALKTFTDSINRGKFGFLPLEISGLSVV 276
Query: 285 ALDVADFL 292
++ +++ L
Sbjct: 277 SIKISEVL 284
>At3g52490 putative protein
Length = 815
Score = 35.4 bits (80), Expect = 0.065
Identities = 44/196 (22%), Positives = 84/196 (42%), Gaps = 14/196 (7%)
Query: 202 KQKSALEIFG-TNLTKLAQEGKLHPFVGREEQVERVIQIICRRMKNNPCLVGEPGVGKTS 260
+Q +LEI T + +EGKL V R E V VI + + + N +VGE
Sbjct: 165 EQAVSLEICSKTTSSSKPKEGKLLTPV-RNEDVMNVINNLVDKKRRNFVIVGECLATIDG 223
Query: 261 IIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIRCLIKEIELC--GNV 318
+++ + +++ VPE LK K + L + F ++ E ++ L ++ C V
Sbjct: 224 VVKTVMEKVDKKDVPEVLKDVKFITLSFSSFGQ--PSRADVERKLEELETLVKSCVGKGV 281
Query: 319 ILFVKEVHHIFDAATSGAR------SFAYILKHALERGVIQCIFATTVNEHRMHMENDTT 372
IL + +++ ++ T G+ S+ + +E G + C + +H
Sbjct: 282 ILNLGDLNWFVESRTRGSSLYNNNDSYCVVEHMIMEIGKLAC--GLVMGDHGRFWLMGLA 339
Query: 373 LKRIFQPVKVVEPSVE 388
+ + K +PS+E
Sbjct: 340 TSQTYVRCKSGQPSLE 355
>At1g07510 unknown protein
Length = 813
Score = 33.9 bits (76), Expect = 0.19
Identities = 27/89 (30%), Positives = 41/89 (45%), Gaps = 12/89 (13%)
Query: 250 LVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIRCLI 309
LVG PG GKT + + A +VP +++ +DF+ + G S R+R L
Sbjct: 365 LVGPPGTGKTLLAKATAGE---SAVP-------FLSISGSDFMEMFVGVGPS--RVRNLF 412
Query: 310 KEIELCGNVILFVKEVHHIFDAATSGARS 338
+E C I+F+ E+ I A G S
Sbjct: 413 QEARQCAPSIIFIDEIDAIGRARGRGGFS 441
>At3g51070 putative protein
Length = 895
Score = 33.5 bits (75), Expect = 0.25
Identities = 30/111 (27%), Positives = 47/111 (42%), Gaps = 5/111 (4%)
Query: 117 QGSNGTVTQLIQN-QGADVNKIREQVIRQIEENV--HQVNLPAEGSKNQMGGQVEENVTA 173
QG ++ +N Q + N E+ +Q EE + N E S GQ EE+ TA
Sbjct: 228 QGETSETSKNEENGQPEEQNSGNEETGQQNEEKTTASEENGKGEKSMKDENGQQEEHTTA 287
Query: 174 ESSKSNQIKFPVLTQNN--NLGARKDDNANKQKSALEIFGTNLTKLAQEGK 222
E N+ + N RKD+ ++Q S FG+ + K + E +
Sbjct: 288 EEESGNKEEESTSKDENMEQQEERKDEKKHEQGSEASGFGSGIPKESAESQ 338
>At1g06720 unknown protein
Length = 1147
Score = 33.1 bits (74), Expect = 0.32
Identities = 26/97 (26%), Positives = 42/97 (42%), Gaps = 6/97 (6%)
Query: 184 PVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICRR 243
P + + L +K + ++ L+ FG A++ K H +EQ + I R
Sbjct: 20 PTARKKSELDKKKRGISVDKQKNLKAFGVKSVVHAKKAKHH--AAEKEQKRLHLPKIDRN 77
Query: 244 MKNNPCLV----GEPGVGKTSIIQGLAQRILSGSVPE 276
P V G PGVGK+ +I+ L + +VPE
Sbjct: 78 YGEAPPFVVVVQGPPGVGKSLVIKSLVKEFTKQNVPE 114
>At3g01610 putative cell division control protein
Length = 703
Score = 32.3 bits (72), Expect = 0.55
Identities = 27/104 (25%), Positives = 47/104 (44%), Gaps = 15/104 (14%)
Query: 250 LVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIRCLI 309
L G PG GKT I + A +G+ +KG +++ V G SE IR L
Sbjct: 450 LYGPPGCGKTLIAKAAANE--AGANFMHIKGAELLNKYV----------GESELAIRTLF 497
Query: 310 KEIELCGNVILFVKEVHHIFDAATSGARSFAYILKHALERGVIQ 353
+ C ++F EV + TS + A++++ L + +++
Sbjct: 498 QRARTCAPCVIFFDEVDAL---TTSRGKEGAWVVERLLNQFLVE 538
Score = 29.3 bits (64), Expect = 4.7
Identities = 48/202 (23%), Positives = 80/202 (38%), Gaps = 43/202 (21%)
Query: 145 IEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQIKFPVLTQNNNLGARKDDNANKQK 204
+ +N ++N SK +G E+NV E+ SN+ + + T +GARK+ +
Sbjct: 45 LRDNYAKLN---SSSKKPIGSPAEKNVEVETV-SNKGRSKLAT----MGARKEAKVSLSL 96
Query: 205 SA------LEIFGTN---------LTKLAQE---GKLHPFVGREEQVERVIQIICRRMKN 246
S LE+ GT + K+ E L P + E + I + +
Sbjct: 97 SGATGNGDLEVEGTKGPTFKDFGGIKKILDELEMNVLFPILNPEP-----FKKIGVKPPS 151
Query: 247 NPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIR 306
G PG GKT + +A +G K+ +V++ G+SEE IR
Sbjct: 152 GILFHGPPGCGKTKLANAIANE--AGVPFYKISATEVIS----------GVSGASEENIR 199
Query: 307 CLIKEIELCGNVILFVKEVHHI 328
L + I+F+ E+ I
Sbjct: 200 ELFSKAYRTAPSIVFIDEIDAI 221
>At5g47040 Lon protease homolog 1 precursor
Length = 888
Score = 32.0 bits (71), Expect = 0.72
Identities = 33/112 (29%), Positives = 52/112 (45%), Gaps = 11/112 (9%)
Query: 216 KLAQEGKLHPFVGREEQVERVIQIIC-RRMKNNP-----CLVGEPGVGKTSIIQGLAQRI 269
K A+E G + +R+I+ + R++K + C VG PGVGKTS+ +A +
Sbjct: 366 KAAKERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIAAAL 425
Query: 270 LSGSVPEKLKGKKVVALDVADFL-YVISNQGSSEERIRCLIKEIELCGNVIL 320
V L G K D AD + + GS R+ +K + +C V+L
Sbjct: 426 GRKFVRLSLGGVK----DEADIRGHRRTYIGSMPGRLIDGLKRVGVCNPVML 473
>At5g39960 putative protein
Length = 614
Score = 32.0 bits (71), Expect = 0.72
Identities = 33/145 (22%), Positives = 59/145 (39%), Gaps = 13/145 (8%)
Query: 250 LVGEPGVGKTSIIQGL--AQRILSG----------SVPEKLKGKKVVALDVADFLYVISN 297
+VG+P VGK++++ L +R+L G V + +G+ V +D A +L
Sbjct: 315 IVGKPNVGKSTLLNALLEEERVLVGPEAGLTRDAVRVQFEFQGRTVYLVDTAGWLERTER 374
Query: 298 QGSSEERIRCLIKEIELCGNVILFVKEVHHIFDAATSGARSFAYILKHALERGVIQCIFA 357
++ + +VI V + I A S S I + A+E G +
Sbjct: 375 DKGPASLSIMQSRKSLMRAHVIALVLDAEEIIKAKCSMTHSEVVIARRAVEEGRGLVVIV 434
Query: 358 TTVNEHRMHMENDTTLKRIFQPVKV 382
++ R EN K+I + V +
Sbjct: 435 NKMDRLR-GRENSEMYKKIKEAVPI 458
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,552,331
Number of Sequences: 26719
Number of extensions: 403866
Number of successful extensions: 1641
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 1567
Number of HSP's gapped (non-prelim): 76
length of query: 455
length of database: 11,318,596
effective HSP length: 103
effective length of query: 352
effective length of database: 8,566,539
effective search space: 3015421728
effective search space used: 3015421728
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146751.5


