

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146751.12 - phase: 0
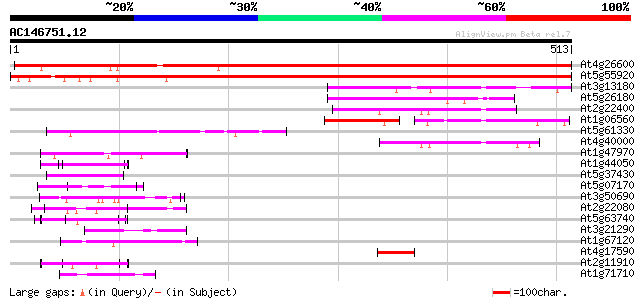
(513 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g26600 unknown protein 697 0.0
At5g55920 nucleolar protein-like 660 0.0
At3g13180 putative sun (fmu) protein 115 8e-26
At5g26180 unknown protein 85 1e-16
At2g22400 unknown protein 79 8e-15
At1g06560 unknown protein 63 4e-10
At5g61330 unknown protein 57 2e-08
At4g40000 proliferating-cell nucleolar antigen - like protein 57 2e-08
At1g47970 Unknown protein (T2J15.12) 52 8e-07
At1g44050 unknown protein 52 1e-06
At5g37430 contains similarity to unknown protein (pir||C71432) 51 2e-06
At5g07170 putative protein 50 2e-06
At3g50690 putative protein 50 3e-06
At2g22080 En/Spm-like transposon protein 50 4e-06
At5g63740 unknown protein 49 5e-06
At3g21290 unknown protein 47 2e-05
At1g67120 hypothetical protein 47 3e-05
At4g17590 putative protein 46 4e-05
At2g11910 unknown protein 46 4e-05
At1g71710 RIBOSOMAL PROTEIN, putative (At1g71710) 46 4e-05
>At4g26600 unknown protein
Length = 671
Score = 697 bits (1798), Expect = 0.0
Identities = 363/526 (69%), Positives = 423/526 (80%), Gaps = 21/526 (3%)
Query: 5 TKKKP-SKPVKPQKAPKKVQIVPI-------KDDSTSESEEEEAPPLQHLELDGSDSDLS 56
TK P K K QK P K Q I K + +++ EEEE Q E S SDL
Sbjct: 23 TKASPLKKAAKTQKPPLKKQRKCISEKKPLKKPEVSTDEEEEEEENEQSDEGSESGSDLF 82
Query: 57 SDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESG--SGSDEDE-----DSDSESGEEDIA 109
SDGD++ +D DD+D+D+D+D++ L GSD +E D DS+SG +
Sbjct: 83 SDGDEEGNNDSDDDDDDDDDDDDDDEDAEPLAEDFLDGSDNEEVTMGSDLDSDSGGSKLE 142
Query: 110 RKSKLIDRKRERDSQAAADELQTNIQTNIQDESDEFTLPTKQELEEEALRPPDLSNLQRR 169
RKS+ IDRKR+++ Q A DE + NI+ ++ DEF LPT++ELEEEA RPPDL +LQ R
Sbjct: 143 RKSRAIDRKRKKEVQDADDEFKMNIK----EKPDEFQLPTQKELEEEARRPPDLPSLQMR 198
Query: 170 IKEIVRVLSNFKALRQDGAT--RKDYVDQLKTDIRSYYGYNEFLIGALVEMFPVVELMEL 227
I+EIVR+LSNFK L+ G R DYV QLK D+ SYYGYNEFLIG L+EMFPVVELMEL
Sbjct: 199 IREIVRILSNFKDLKPKGDKHERNDYVGQLKADLSSYYGYNEFLIGTLIEMFPVVELMEL 258
Query: 228 IEAFEKPRPICLRTNTLKTRRRDLADVLINRGVNLDPLSKWSKVGLVVYDSQVPIGATPE 287
IEAFEK RP +RTNTLKTRRRDLAD+L+NRGVNLDPLSKWSKVGL+VYDSQVPIGATPE
Sbjct: 259 IEAFEKKRPTSIRTNTLKTRRRDLADILLNRGVNLDPLSKWSKVGLIVYDSQVPIGATPE 318
Query: 288 YMAGFYMLQSASSFLPVMALAPQEKERIVDMAAAPGGKTTYIAALMKNSGIIFANEMKVP 347
Y+AGFYMLQSASSFLPVMALAP+EKER+VDMAAAPGGKTTY+AALMKN+GII+ANEMKVP
Sbjct: 319 YLAGFYMLQSASSFLPVMALAPREKERVVDMAAAPGGKTTYVAALMKNTGIIYANEMKVP 378
Query: 348 RLKSLTANLHRMGVSNTVVSNYDGKELPKVLGFNSVDRVLLDAPCSGTGVISKDESVKTS 407
RLKSL+ANLHRMGV+NT+V NYDG+EL KVLG +SVDRVLLDAPCSGTGVISKDESVKTS
Sbjct: 379 RLKSLSANLHRMGVTNTIVCNYDGRELTKVLGQSSVDRVLLDAPCSGTGVISKDESVKTS 438
Query: 408 KNLEDIKKCAHLQKELLLAAIDMVDSYSKSGGYVVYSTCSIMVAENEAVIDYVLKRRDVK 467
K+ +DIKK AHLQK+L+L AID+VD+ SK+GGY+VYSTCS+M+ ENEAVIDY LK RDVK
Sbjct: 439 KSADDIKKFAHLQKQLILGAIDLVDANSKTGGYIVYSTCSVMIPENEAVIDYALKNRDVK 498
Query: 468 LVPCGLDFGRPGFTKFREQRFHPSLDKTRRFYPHVHNMDGFFVAKV 513
LVPCGLDFGRPGF+ FRE RFHPSL+KTRRFYPHVHNMDGFFVAK+
Sbjct: 499 LVPCGLDFGRPGFSSFREHRFHPSLEKTRRFYPHVHNMDGFFVAKL 544
>At5g55920 nucleolar protein-like
Length = 682
Score = 660 bits (1704), Expect = 0.0
Identities = 356/549 (64%), Positives = 413/549 (74%), Gaps = 39/549 (7%)
Query: 1 MAPATK---KKPSKPVKPQ-----KAPKKVQIVPIKDDSTSESEEEEAPPLQHLEL--DG 50
+ P TK K + P+KPQ K K P+K EEE PL+ E+ D
Sbjct: 16 ITPPTKQLTKSKTPPMKPQTSMLKKGAKSQNKPPLKKQKKEVVEEE---PLEDYEVTDDS 72
Query: 51 SDSDLSSDGDDQ-------------LADDFLQGSD----DEDNDNDNDDEGSDLESGSGS 93
+ D SDG D+ +DD GS+ D++ +ND + G D GSG
Sbjct: 73 DEDDEVSDGSDEDDISPAVESEEIDESDDGENGSNQLFSDDEEENDEETLGDDFLEGSGD 132
Query: 94 DEDE-----DSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNI-QTNIQDES---DE 144
+++E DSD++S ++DI KS IDR + AA EL+ I Q ++ DE D
Sbjct: 133 EDEEGSLDADSDADSDDDDIVAKSDAIDRDLAMQKKDAAAELEDFIKQDDVHDEEPEHDA 192
Query: 145 FTLPTKQELEEEALRPPDLSNLQRRIKEIVRVLSNFKALRQDGATRKDYVDQLKTDIRSY 204
F LPT++ELEEEA PPDL L+ RI+EIVR L NFKA R TRK V+QLK D+ SY
Sbjct: 193 FRLPTEEELEEEARGPPDLPLLKTRIEEIVRALKNFKAFRPKDTTRKACVEQLKADLGSY 252
Query: 205 YGYNEFLIGALVEMFPVVELMELIEAFEKPRPICLRTNTLKTRRRDLADVLINRGVNLDP 264
YGYN FLIG LVEMFP ELMELIEAFEK RP +RTNTLKTRRRDLADVL+NRGVNLDP
Sbjct: 253 YGYNSFLIGTLVEMFPPGELMELIEAFEKQRPTSIRTNTLKTRRRDLADVLLNRGVNLDP 312
Query: 265 LSKWSKVGLVVYDSQVPIGATPEYMAGFYMLQSASSFLPVMALAPQEKERIVDMAAAPGG 324
LSKWSKVGLV+YDSQVPIGATPEY+AG+YMLQ ASSFLPVMALAP+E ERIVD+AAAPGG
Sbjct: 313 LSKWSKVGLVIYDSQVPIGATPEYLAGYYMLQGASSFLPVMALAPRENERIVDVAAAPGG 372
Query: 325 KTTYIAALMKNSGIIFANEMKVPRLKSLTANLHRMGVSNTVVSNYDGKELPKVLGFNSVD 384
KTTYIAALMKN+G+I+ANEMKVPRLKSLTANLHRMGV+NT+V NYDG+ELPKVLG N+VD
Sbjct: 373 KTTYIAALMKNTGLIYANEMKVPRLKSLTANLHRMGVTNTIVCNYDGRELPKVLGQNTVD 432
Query: 385 RVLLDAPCSGTGVISKDESVKTSKNLEDIKKCAHLQKELLLAAIDMVDSYSKSGGYVVYS 444
RVLLDAPCSGTG+ISKDESVK +K +++IKK AHLQK+LLLAAIDMVD+ SK+GGY+VYS
Sbjct: 433 RVLLDAPCSGTGIISKDESVKITKTMDEIKKFAHLQKQLLLAAIDMVDANSKTGGYIVYS 492
Query: 445 TCSIMVAENEAVIDYVLKRRDVKLVPCGLDFGRPGFTKFREQRFHPSLDKTRRFYPHVHN 504
TCSIMV ENEAVIDY LK+RDVKLV CGLDFGR GFT+FRE RF PSLDKTRRFYPHVHN
Sbjct: 493 TCSIMVTENEAVIDYALKKRDVKLVTCGLDFGRKGFTRFREHRFQPSLDKTRRFYPHVHN 552
Query: 505 MDGFFVAKV 513
MDGFFVAK+
Sbjct: 553 MDGFFVAKL 561
>At3g13180 putative sun (fmu) protein
Length = 523
Score = 115 bits (287), Expect = 8e-26
Identities = 87/232 (37%), Positives = 124/232 (52%), Gaps = 31/232 (13%)
Query: 291 GFYMLQSASSFLPVMALAPQEKERIVDMAAAPGGKTTYIAALMKNSGIIFANEMKVPRLK 350
G +Q S+ L V + PQ ERI+D AAPGGKT ++A+ +K G+I+A ++ RL+
Sbjct: 310 GICSVQDESAGLIVSVVKPQPGERIMDACAAPGGKTLFMASCLKGQGMIYAMDVNEGRLR 369
Query: 351 SL--TANLHRM-GVSNTVVSNYDGKELPKVLGFNSV--DRVLLDAPCSGTGVISKDESVK 405
L TA H++ G+ T+ S +L N V D+VLLDAPCSG GV+SK ++
Sbjct: 370 ILGETAKSHQVDGLITTIHS-----DLRVFAETNEVQYDKVLLDAPCSGLGVLSKRADLR 424
Query: 406 TSKNLEDIKKCAHLQKELLLAAIDMVDSYSKSGGYVVYSTCSIMVAENEAVIDYVLKRRD 465
++ LED+ + LQ ELL +A +V K GG +VYSTCSI ENE ++ L R
Sbjct: 425 WNRKLEDMLELTKLQDELLDSASKLV----KHGGVLVYSTCSIDPEENEGRVEAFLLR-- 478
Query: 466 VKLVPCGLDFGRPGFTKFREQRFHPSLDKTRRFY----PHVHNMDGFFVAKV 513
P FT F PS T + + P H++DG F A++
Sbjct: 479 -----------HPEFTIDPVTSFVPSSFVTSQGFFLSNPVKHSLDGAFAARL 519
>At5g26180 unknown protein
Length = 567
Score = 84.7 bits (208), Expect = 1e-16
Identities = 66/181 (36%), Positives = 90/181 (49%), Gaps = 15/181 (8%)
Query: 291 GFYMLQSASSFLPVMALAPQEKERIVDMAAAPGGKTTYIAALMKNSGIIFANEMKVPRLK 350
G LQ +S + AL PQ ++D +APG KT ++AALM+ G I A E+ R+K
Sbjct: 284 GRIFLQGKASSMVAAALQPQAGWEVLDACSAPGNKTIHLAALMEGQGKIIACELNEERVK 343
Query: 351 SLTANLHRMGVSNTVVSNYDGKEL-PKVLGFNSVDRVLLDAPCSGTGVIS-------KDE 402
L + G SN V + D L PK F + +LLD CSG+G I+
Sbjct: 344 RLEHTIKLSGASNIEVCHGDFLGLNPKDPSFAKIRAILLDPSCSGSGTITDRLDHLLPSH 403
Query: 403 SVKTSKNLEDIK--KCAHLQKELLLAAIDMVDSYSKSGGYVVYSTCSIMVAENEAVIDYV 460
S + N + ++ K A QK+ L A+ S+ K VVYSTCSI ENE V+ V
Sbjct: 404 SEDNNMNYDSMRLHKLAVFQKKALAHAL----SFPKV-ERVVYSTCSIYQIENEDVVSSV 458
Query: 461 L 461
L
Sbjct: 459 L 459
>At2g22400 unknown protein
Length = 808
Score = 78.6 bits (192), Expect = 8e-15
Identities = 57/193 (29%), Positives = 90/193 (46%), Gaps = 29/193 (15%)
Query: 296 QSASSFLPVMALAPQEKERIVDMAAAPGGKTTYIAALMKNS--------GIIFANEMKVP 347
Q A S +P + L ++DM AAPG KT + ++ + G++ AN++
Sbjct: 170 QEAVSMVPPLFLDVHPDHFVLDMCAAPGSKTFQLLEIIHEASEPGSLPNGLVVANDVDFK 229
Query: 348 RLKSLTANLHRMGVSNTVVSNYDGKELP-----------KVLGFN------SVDRVLLDA 390
R L RM SN +V+N++G++ P K + N + DRVL D
Sbjct: 230 RSNLLIHQTKRMCTSNLIVTNHEGQQFPGCRLNKSRASEKGISENMPINQLAFDRVLCDV 289
Query: 391 PCSGTGVISKDESVKTSKNLEDIKKCAHLQKELLLAAIDMVDSYSKSGGYVVYSTCSIMV 450
PCSG G + K + N LQ L + + ++ K GG ++YSTCS+
Sbjct: 290 PCSGDGTLRKAPDIWRKWNSGMGNGLHSLQIILAMRGLSLL----KVGGKMIYSTCSMNP 345
Query: 451 AENEAVIDYVLKR 463
E+EAV+ +L+R
Sbjct: 346 VEDEAVVAEILRR 358
>At1g06560 unknown protein
Length = 599
Score = 62.8 bits (151), Expect = 4e-10
Identities = 51/154 (33%), Positives = 76/154 (49%), Gaps = 18/154 (11%)
Query: 371 GKELPKVLGF--NSVDRVLLDAPCSGTGVISKDESVKTSKNLEDIKKCAHLQKELLLAAI 428
G K GF NS DRVLLDAPCS G+ + + + ++ Q+++L A+
Sbjct: 445 GGRAGKSQGFPPNSFDRVLLDAPCSALGL--RPRLFAGLETVVSLRNHGWYQRKMLDQAV 502
Query: 429 DMVDSYSKSGGYVVYSTCSIMVAENEAVIDYVL-KRRDVKLVPCGLDFGRPGFT------ 481
+V + GG +VYSTC+I +ENEAV+ Y L K R + L P G PG
Sbjct: 503 QLV----RVGGILVYSTCTINPSENEAVVRYALDKYRFLSLAPQHPRIGGPGLVGRCEFP 558
Query: 482 -KFREQRFHPSLDKTRRFYPHVHNMD--GFFVAK 512
+ E+ P ++ + + +D GFF+AK
Sbjct: 559 DGYIEEWLKPGEEELVQKFDPSSELDTIGFFIAK 592
Score = 52.8 bits (125), Expect = 5e-07
Identities = 32/71 (45%), Positives = 44/71 (61%), Gaps = 3/71 (4%)
Query: 289 MAGFYMLQSASSFLPVMALAPQEKERIVDMAAAPGGKTTYIAALMKNSGIIFA---NEMK 345
+ G LQ+ S + AL PQ+ ERI+DM AAPGGKTT IA LM + G I A + K
Sbjct: 274 LEGEIFLQNLPSIIVAHALDPQKGERILDMCAAPGGKTTAIAILMNDEGEIVAADRSHNK 333
Query: 346 VPRLKSLTANL 356
V +++L+A +
Sbjct: 334 VLVVQNLSAEM 344
>At5g61330 unknown protein
Length = 436
Score = 57.4 bits (137), Expect = 2e-08
Identities = 49/236 (20%), Positives = 106/236 (44%), Gaps = 25/236 (10%)
Query: 34 ESEEEEAPPLQHLELDGSDSD------LSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDL 87
+SE E+ ++L+ + + D + D D + DD + +D++ D + DDEG
Sbjct: 14 DSESEDISDQENLKAESDNEDDQLPDGIEDDEVDSMEDDEGESEEDDEGDTEEDDEGDSE 73
Query: 88 ESGSGSD-EDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDESDEFT 146
E G + EDED +SE E+ ++S+ D + + A +EL+ ++ ++ + +
Sbjct: 74 EDDEGENKEDEDGESEDFEDGNDKESESGDEGNDDNKDAQMEELEKEVK-ELRSQEQDIL 132
Query: 147 LPTKQELEEEALRPPDLSNLQRRIKEIVRVLSNFKALRQDGATRKDYVDQLKTDIRSYY- 205
K++ E+A++ + N + +I+ F+ L Q R + + Q ++S +
Sbjct: 133 KNLKRDKGEDAVKGQAVKNQKALWDKIL----EFRFLLQKAFDRSNRLPQ--EPVKSLFC 186
Query: 206 --------GYNEFLIGALVEMFPVVELMELIEAFEKPRPICLRTNTLKTRRRDLAD 253
Y + + + + ++EL E + FEK + + N + + +D
Sbjct: 187 SEDEDVSTAYTDLVTSSKKTLDSLLELQEAL--FEKNPSVDQQVNATASEESNKSD 240
>At4g40000 proliferating-cell nucleolar antigen - like protein
Length = 682
Score = 57.0 bits (136), Expect = 2e-08
Identities = 50/168 (29%), Positives = 74/168 (43%), Gaps = 26/168 (15%)
Query: 339 IFANEMKVPRLKSLTANLHRMGVSNTVVSNYDGKELP-----KVLGFNS----------- 382
+ AN++ R L R +N +V+N +G+ P + L S
Sbjct: 113 VVANDVDYKRSNLLIHQTKRTCTTNLMVTNNEGQHFPSCNTKRTLSVASETNPHPIDQLL 172
Query: 383 VDRVLLDAPCSGTGVISKDESVKTSKNLEDIKKCAHLQKELLLAAIDMVDSYSKSGGYVV 442
DRVL D PCSG G + K + N LQ L + + ++ K GG +V
Sbjct: 173 FDRVLCDVPCSGDGTLRKAPDIWRRWNSGSGNGLHSLQVVLAMRGLSLL----KVGGRMV 228
Query: 443 YSTCSIMVAENEAVIDYVLKR--RDVKLVPCGLD----FGRPGFTKFR 484
YSTCS+ E+EAV+ +L+R V+LV RPG TK++
Sbjct: 229 YSTCSMNPIEDEAVVAEILRRCGCSVELVDVSDKLPELIRRPGLTKWK 276
>At1g47970 Unknown protein (T2J15.12)
Length = 198
Score = 52.0 bits (123), Expect = 8e-07
Identities = 33/135 (24%), Positives = 63/135 (46%), Gaps = 7/135 (5%)
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLE 88
DD E ++++ +Q L+ G S++ +D+ D+ G DD+D+ +D+DD+ D
Sbjct: 53 DDEEEEEDDDDDDDVQVLQSLGGPPVQSAEDEDEEGDEDGNGDDDDDDGDDDDDDDDD-- 110
Query: 89 SGSGSDEDEDSDSESGEEDIARK-SKLIDRKRERDSQAAADELQTNIQTNIQDESDEFTL 147
DED + + + G E + R + D + D + + ++ +I DE+D
Sbjct: 111 ----EDEDVEDEGDLGTEYLVRPVGRAEDEEDASDFEPEENGVEEDIDEGEDDENDNSGG 166
Query: 148 PTKQELEEEALRPPD 162
K E + R P+
Sbjct: 167 AGKSEAPPKRKRAPE 181
Score = 43.5 bits (101), Expect = 3e-04
Identities = 42/160 (26%), Positives = 63/160 (39%), Gaps = 40/160 (25%)
Query: 29 DDSTSESEEEE--------------APPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDE 74
DD E EEE+ PP+Q E + + D +GDD D DD+
Sbjct: 50 DDDDDEEEEEDDDDDDDVQVLQSLGGPPVQSAEDEDEEGDEDGNGDDDDDD-----GDDD 104
Query: 75 DNDNDNDDEGSDLES-----------GSGSDEDEDSDSESGEEDIARKSKLIDRKR--ER 121
D+D+D++DE + E G DE++ SD E E + ID E
Sbjct: 105 DDDDDDEDEDVEDEGDLGTEYLVRPVGRAEDEEDASDFEPEENGVEED---IDEGEDDEN 161
Query: 122 DSQAAADELQTNIQTNIQDESDEFTLPTKQELEEEALRPP 161
D+ A + + + E DE + +E+ RPP
Sbjct: 162 DNSGGAGKSEAPPKRKRAPEEDE-----EDSGDEDDDRPP 196
Score = 38.5 bits (88), Expect = 0.009
Identities = 28/118 (23%), Positives = 47/118 (39%), Gaps = 10/118 (8%)
Query: 27 IKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSD 86
+ DD +EE P +Q L D SD + D ++++D+ DD+D +DD+ +
Sbjct: 4 VNDDDDKNFKEE--PEIQVLSDDDSDEEQVKDVGEEVSDE-----DDDDGSEGDDDDDEE 56
Query: 87 LESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDESDE 144
E D+D G + D E D D+ + + D+ DE
Sbjct: 57 EEEDDDDDDDVQVLQSLGGPPVQSAE---DEDEEGDEDGNGDDDDDDGDDDDDDDDDE 111
>At1g44050 unknown protein
Length = 734
Score = 51.6 bits (122), Expect = 1e-06
Identities = 23/61 (37%), Positives = 36/61 (58%), Gaps = 1/61 (1%)
Query: 49 DGSDSDLSSDGDDQLADDFLQG-SDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEED 107
DG D + DGDD DD +DD+D+D+D+DD+ D + G D++ED D + + D
Sbjct: 86 DGDDDNKGDDGDDDNEDDNEDDDNDDDDDDDDDDDDDDDDDDDDGDDDNEDGDCDDDDGD 145
Query: 108 I 108
+
Sbjct: 146 L 146
Score = 46.6 bits (109), Expect = 3e-05
Identities = 22/77 (28%), Positives = 37/77 (47%), Gaps = 1/77 (1%)
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLE 88
DD +++ + + D D D +D DD DD+D+D+D+DD+ D +
Sbjct: 70 DDDDDAEDDDNGDDKEDGDDDNKGDDGDDDNEDDNEDDDNDDDDDDDDDDDDDDDDDD-D 128
Query: 89 SGSGSDEDEDSDSESGE 105
G +ED D D + G+
Sbjct: 129 DGDDDNEDGDCDDDDGD 145
Score = 45.8 bits (107), Expect = 6e-05
Identities = 20/63 (31%), Positives = 34/63 (53%)
Query: 45 HLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESG 104
H D D++ +GDD+ D DD D+DN++D+E D + D+D+D D +
Sbjct: 67 HNHDDDDDAEDDDNGDDKEDGDDDNKGDDGDDDNEDDNEDDDNDDDDDDDDDDDDDDDDD 126
Query: 105 EED 107
++D
Sbjct: 127 DDD 129
Score = 43.1 bits (100), Expect = 4e-04
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 1/60 (1%)
Query: 49 DGSDSDLSSDGDDQLADDFLQGSDDEDNDN-DNDDEGSDLESGSGSDEDEDSDSESGEED 107
D + D DGDD D +++DN++ DNDD+ D + D+D+D D + ED
Sbjct: 77 DDDNGDDKEDGDDDNKGDDGDDDNEDDNEDDDNDDDDDDDDDDDDDDDDDDDDGDDDNED 136
Score = 39.3 bits (90), Expect = 0.005
Identities = 17/62 (27%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query: 46 LELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGE 105
L++ D D ++ DD D + DD++ +D DD+ D +D+D+D D + +
Sbjct: 64 LKMHNHDDDDDAEDDDN--GDDKEDGDDDNKGDDGDDDNEDDNEDDDNDDDDDDDDDDDD 121
Query: 106 ED 107
+D
Sbjct: 122 DD 123
Score = 35.0 bits (79), Expect = 0.099
Identities = 19/59 (32%), Positives = 27/59 (45%), Gaps = 10/59 (16%)
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDL 87
DD+ ++E++ D D D D DD DD DD++ D D DD+ DL
Sbjct: 98 DDNEDDNEDD----------DNDDDDDDDDDDDDDDDDDDDDGDDDNEDGDCDDDDGDL 146
>At5g37430 contains similarity to unknown protein (pir||C71432)
Length = 607
Score = 50.8 bits (120), Expect = 2e-06
Identities = 25/71 (35%), Positives = 37/71 (51%)
Query: 34 ESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGS 93
+ EE++ P + L+ D D DD DD G DD D+D+D+ D+G D + G
Sbjct: 536 KEEEKQLPETEFDWLNLHDDDGDDGDDDDGDDDDDDGDDDGDDDDDDGDDGDDDDDDDGD 595
Query: 94 DEDEDSDSESG 104
D D+D D + G
Sbjct: 596 DGDDDDDDDDG 606
Score = 35.8 bits (81), Expect = 0.058
Identities = 21/65 (32%), Positives = 32/65 (48%), Gaps = 10/65 (15%)
Query: 27 IKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDND-NDNDDEGS 85
+ DD + ++++ D D D DGDD DD G DD+D+D +D DD+
Sbjct: 552 LHDDDGDDGDDDDG--------DDDDDDGDDDGDDD-DDDGDDGDDDDDDDGDDGDDDDD 602
Query: 86 DLESG 90
D + G
Sbjct: 603 DDDGG 607
>At5g07170 putative protein
Length = 542
Score = 50.4 bits (119), Expect = 2e-06
Identities = 32/92 (34%), Positives = 49/92 (52%), Gaps = 12/92 (13%)
Query: 26 PIKDDSTSESEEE-EAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEG 84
P+ DDS+S+ E++ E+ + D D+D D DD DD DD+D+D+D+DD+
Sbjct: 85 PLIDDSSSDEEDDSESTHCYAADDDADDTDDDEDDDDDDDDD-----DDDDDDDDDDDDD 139
Query: 85 SDLESGSGSDEDEDSDSESGEEDIARKSKLID 116
D DE +DS+ E E D + + ID
Sbjct: 140 DD------DDESKDSEVEEEEGDDDLRMRKID 165
Score = 42.7 bits (99), Expect = 5e-04
Identities = 18/69 (26%), Positives = 34/69 (49%)
Query: 54 DLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSK 113
D SSD +D +DD+ +D D+D++ D + D+D+D D + ++D K
Sbjct: 89 DSSSDEEDDSESTHCYAADDDADDTDDDEDDDDDDDDDDDDDDDDDDDDDDDDDDESKDS 148
Query: 114 LIDRKRERD 122
++ + D
Sbjct: 149 EVEEEEGDD 157
Score = 41.6 bits (96), Expect = 0.001
Identities = 17/71 (23%), Positives = 37/71 (51%)
Query: 52 DSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARK 111
D S + DD + DD D+ +D++D+ D + D+D+D D + ++D ++
Sbjct: 88 DDSSSDEEDDSESTHCYAADDDADDTDDDEDDDDDDDDDDDDDDDDDDDDDDDDDDESKD 147
Query: 112 SKLIDRKRERD 122
S++ + + + D
Sbjct: 148 SEVEEEEGDDD 158
Score = 38.5 bits (88), Expect = 0.009
Identities = 20/73 (27%), Positives = 34/73 (46%), Gaps = 19/73 (26%)
Query: 55 LSSDGDDQL-------------------ADDFLQGSDDEDNDNDNDDEGSDLESGSGSDE 95
+ S DD L ADD +DD+++D+D+DD+ D + D+
Sbjct: 78 IPSSQDDPLIDDSSSDEEDDSESTHCYAADDDADDTDDDEDDDDDDDDDDDDDDDDDDDD 137
Query: 96 DEDSDSESGEEDI 108
D+D D ES + ++
Sbjct: 138 DDDDDDESKDSEV 150
Score = 28.5 bits (62), Expect = 9.3
Identities = 18/59 (30%), Positives = 26/59 (43%), Gaps = 2/59 (3%)
Query: 51 SDSDLSSDGDD--QLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEED 107
SD D D+ ++A DD D+ + DE D ES D+D+D +ED
Sbjct: 60 SDDHYCGDDDENMKIAAAIPSSQDDPLIDDSSSDEEDDSESTHCYAADDDADDTDDDED 118
>At3g50690 putative protein
Length = 447
Score = 50.1 bits (118), Expect = 3e-06
Identities = 43/140 (30%), Positives = 65/140 (45%), Gaps = 16/140 (11%)
Query: 28 KDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDND--NDDEGS 85
+DD E EEE + L GS S L + G+ D QG DDED D + DD+G
Sbjct: 297 EDDEEDEEEEEVDNADRGLGGSGSTSRLMNAGE---IDGHEQGDDDEDGDGETGEDDQGV 353
Query: 86 DLESGSGSDEDED---SDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDES 142
+ + G +DED+D D ESGE + + ++ D A ++++ + N DE
Sbjct: 354 E-DDGEFADEDDDVEEEDEESGEGYLVQPVSQVE-----DHDAVGNDIEPINEDNDPDEE 407
Query: 143 DEF--TLPTKQELEEEALRP 160
+E LP + + RP
Sbjct: 408 EEVEDDLPIPDQSLASSSRP 427
Score = 46.6 bits (109), Expect = 3e-05
Identities = 38/148 (25%), Positives = 69/148 (45%), Gaps = 32/148 (21%)
Query: 28 KDDSTSESEEEEAPPLQHLELDG---SDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDE- 83
++D E E +E P E+DG +++ S+G + D + +DE++D ++D+
Sbjct: 173 EEDEEEEEEGDEEDPGSG-EIDGDERAEAPRMSNGHSERVDGVVDVDEDEESDAEDDESE 231
Query: 84 -------------GSDLESGSGSD--EDEDSDSESGEEDIARKSKLIDRKRERDSQAAAD 128
G LE+ +G + ED+ DSESGEE++ + +++ DS+
Sbjct: 232 QATGVNGTSYRANGFRLEAVNGEEVREDDGDDSESGEEEVGEDNDVVEVHEIEDSE---- 287
Query: 129 ELQTNIQTNIQDESDEFTLPTKQELEEE 156
N + + DE D+ + E EEE
Sbjct: 288 ----NEEDGVDDEEDD----EEDEEEEE 307
Score = 41.2 bits (95), Expect = 0.001
Identities = 30/94 (31%), Positives = 45/94 (46%), Gaps = 20/94 (21%)
Query: 29 DDSTSESEE--EEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEG-- 84
DDS S EE E+ ++ E++ DS+ DG D DD D+E+ + DN D G
Sbjct: 262 DDSESGEEEVGEDNDVVEVHEIE--DSENEEDGVDDEEDD---EEDEEEEEVDNADRGLG 316
Query: 85 -----------SDLESGSGSDEDEDSDSESGEED 107
+++ D+DED D E+GE+D
Sbjct: 317 GSGSTSRLMNAGEIDGHEQGDDDEDGDGETGEDD 350
Score = 35.0 bits (79), Expect = 0.099
Identities = 28/145 (19%), Positives = 60/145 (41%), Gaps = 14/145 (9%)
Query: 24 IVPIKDDSTSESEEEEAPPLQ------------HLELDGSDSDLSSDGDDQLADDFLQGS 71
+V + +D S++E++E+ LE + DGDD + + G
Sbjct: 214 VVDVDEDEESDAEDDESEQATGVNGTSYRANGFRLEAVNGEEVREDDGDDSESGEEEVGE 273
Query: 72 DDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQ 131
D++ + ++ + E G +ED++ D E EE++ + + A E+
Sbjct: 274 DNDVVEVHEIEDSENEEDGVDDEEDDEEDEE--EEEVDNADRGLGGSGSTSRLMNAGEID 331
Query: 132 TNIQTNIQDESDEFTLPTKQELEEE 156
+ Q + ++ D T Q +E++
Sbjct: 332 GHEQGDDDEDGDGETGEDDQGVEDD 356
Score = 29.6 bits (65), Expect = 4.2
Identities = 24/91 (26%), Positives = 44/91 (47%), Gaps = 16/91 (17%)
Query: 43 LQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGS-DEDEDSDS 101
L + +G++ S D DD+ D+ED + ++EG + + GSG D DE +++
Sbjct: 151 LDKTDAEGNERPESDDEDDE--------EDEEDE--EEEEEGDEEDPGSGEIDGDERAEA 200
Query: 102 ---ESGEEDIARKSKLIDRKRERDSQAAADE 129
+G + R ++D + +S A DE
Sbjct: 201 PRMSNGHSE--RVDGVVDVDEDEESDAEDDE 229
Score = 29.6 bits (65), Expect = 4.2
Identities = 21/79 (26%), Positives = 35/79 (43%), Gaps = 6/79 (7%)
Query: 67 FLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRE--RDSQ 124
+L +D E N+ D+ D E +EDE+ + E EED D + E R S
Sbjct: 150 YLDKTDAEGNERPESDDEDDEE----DEEDEEEEEEGDEEDPGSGEIDGDERAEAPRMSN 205
Query: 125 AAADELQTNIQTNIQDESD 143
++ + + + +ESD
Sbjct: 206 GHSERVDGVVDVDEDEESD 224
>At2g22080 En/Spm-like transposon protein
Length = 140
Score = 49.7 bits (117), Expect = 4e-06
Identities = 29/81 (35%), Positives = 46/81 (55%), Gaps = 14/81 (17%)
Query: 33 SESEEEEAPPLQHLELDGS--DSDLSSDGDDQLADDFLQGSDDEDNDN----DNDDEGSD 86
+E +E + P+ +L+G D+D +GDD G+DDED+DN D D+E +
Sbjct: 49 AEGKETKKGPVSDPDLNGEAGDNDDEPEGDD--------GNDDEDDDNHENDDEDEEEDE 100
Query: 87 LESGSGSDEDEDSDSESGEED 107
E+ G +ED+D D+E EE+
Sbjct: 101 DENDDGGEEDDDEDAEVEEEE 121
Score = 44.7 bits (104), Expect = 1e-04
Identities = 34/143 (23%), Positives = 63/143 (43%), Gaps = 13/143 (9%)
Query: 21 KVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDG--DDQLADDFLQGSDDEDNDN 78
K+Q+ +K D E + +E +G++ D DG D++ + D +G + +
Sbjct: 4 KLQVTVLKGDVIENGYEV----VDAVEGNGNNDDSGDDGGEDEEGSADDAEGKETKKGPV 59
Query: 79 DNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNI 138
+ D E+G DE E D E+D ++ D + + D E + +
Sbjct: 60 SDPDLNG--EAGDNDDEPEGDDGNDDEDDDNHENDDEDEEEDEDENDDGGEEDDDEDAEV 117
Query: 139 QDESDEFTLPTKQELEEEALRPP 161
++E +E + E +EEAL+PP
Sbjct: 118 EEEEEE-----EDEDDEEALQPP 135
>At5g63740 unknown protein
Length = 226
Score = 49.3 bits (116), Expect = 5e-06
Identities = 25/77 (32%), Positives = 38/77 (48%), Gaps = 4/77 (5%)
Query: 30 DSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLES 89
D + +E+E E + D D D DD DD +DD D+D D+DDE D +
Sbjct: 66 DGDGDEDEDEDADADEDEDEDEDEDDDDDDDDDDDDD----ADDADDDEDDDDEDDDEDE 121
Query: 90 GSGSDEDEDSDSESGEE 106
D+D+++D E +E
Sbjct: 122 DDDDDDDDENDEECDDE 138
Score = 48.5 bits (114), Expect = 9e-06
Identities = 24/59 (40%), Positives = 33/59 (55%), Gaps = 3/59 (5%)
Query: 52 DSDLSSDGD---DQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEED 107
D D DGD D+ AD +DED D+D+DD+ D + +D+DED D E +ED
Sbjct: 62 DGDGDGDGDEDEDEDADADEDEDEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDED 120
Score = 46.6 bits (109), Expect = 3e-05
Identities = 23/85 (27%), Positives = 42/85 (49%), Gaps = 2/85 (2%)
Query: 23 QIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDD 82
+ + + DD ++ E + E + D+D D D+ +D DD+D+D+D+ D
Sbjct: 47 EYIKVYDDHNNDGEGDGDGDGDGDEDEDEDADADEDEDEDEDED--DDDDDDDDDDDDAD 104
Query: 83 EGSDLESGSGSDEDEDSDSESGEED 107
+ D E D+DED D + ++D
Sbjct: 105 DADDDEDDDDEDDDEDEDDDDDDDD 129
Score = 44.7 bits (104), Expect = 1e-04
Identities = 26/78 (33%), Positives = 38/78 (48%), Gaps = 11/78 (14%)
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLE 88
D E E+ +A + + D D D D DD ADD D+D D+D++D+ D
Sbjct: 68 DGDEDEDEDADADEDEDEDEDEDDDDDDDDDDDDDADDA-----DDDEDDDDEDDDED-- 120
Query: 89 SGSGSDEDEDSDSESGEE 106
D+D+D D E+ EE
Sbjct: 121 ----EDDDDDDDDENDEE 134
Score = 42.4 bits (98), Expect = 6e-04
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 11/79 (13%)
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLE 88
D E E+E+A D+D D D+ DD DD+D+ +D DD+ D +
Sbjct: 66 DGDGDEDEDEDA-----------DADEDEDEDEDEDDDDDDDDDDDDDADDADDDEDDDD 114
Query: 89 SGSGSDEDEDSDSESGEED 107
DED+D D + ++
Sbjct: 115 EDDDEDEDDDDDDDDENDE 133
Score = 42.0 bits (97), Expect = 8e-04
Identities = 21/71 (29%), Positives = 39/71 (54%), Gaps = 2/71 (2%)
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLE 88
+D ++++E+E + D D D D DD ADD D++D+++++DD+ D E
Sbjct: 73 EDEDADADEDEDEDEDEDDDDDDDDDDDDDADD--ADDDEDDDDEDDDEDEDDDDDDDDE 130
Query: 89 SGSGSDEDEDS 99
+ D++ DS
Sbjct: 131 NDEECDDEYDS 141
Score = 41.2 bits (95), Expect = 0.001
Identities = 23/87 (26%), Positives = 43/87 (48%), Gaps = 15/87 (17%)
Query: 65 DDFLQGSDDEDNDNDND-------DEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDR 117
D++++ DD +ND + D DE D ++ + DEDED D + ++D D
Sbjct: 46 DEYIKVYDDHNNDGEGDGDGDGDGDEDEDEDADADEDEDEDEDEDDDDDD--------DD 97
Query: 118 KRERDSQAAADELQTNIQTNIQDESDE 144
+ D+ A D+ + + + +DE D+
Sbjct: 98 DDDDDADDADDDEDDDDEDDDEDEDDD 124
Score = 41.2 bits (95), Expect = 0.001
Identities = 20/59 (33%), Positives = 30/59 (49%)
Query: 28 KDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSD 86
+D E E+E+ + D D D + D DD DD +DED+D+D+DDE +
Sbjct: 75 EDADADEDEDEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDEDDDDDDDDENDE 133
Score = 38.1 bits (87), Expect = 0.012
Identities = 27/96 (28%), Positives = 40/96 (41%), Gaps = 15/96 (15%)
Query: 49 DGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDI 108
D ++D DGD G DED D D D +D + DED+D D + ++D
Sbjct: 53 DDHNNDGEGDGDGD-------GDGDEDEDEDAD---ADEDEDEDEDEDDDDDDDDDDDDD 102
Query: 109 ARKSKLIDRKRERDSQAAADELQTNIQTNIQDESDE 144
A D + D D+ + + DE+DE
Sbjct: 103 AD-----DADDDEDDDDEDDDEDEDDDDDDDDENDE 133
Score = 33.9 bits (76), Expect = 0.22
Identities = 15/56 (26%), Positives = 28/56 (49%)
Query: 28 KDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDE 83
+D+ E ++++ + D +D D D +D D+ DD++ND + DDE
Sbjct: 83 EDEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDEDDDDDDDDENDEECDDE 138
>At3g21290 unknown protein
Length = 1848
Score = 47.4 bits (111), Expect = 2e-05
Identities = 30/93 (32%), Positives = 44/93 (47%), Gaps = 21/93 (22%)
Query: 69 QGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAAD 128
+ S D D+D+DN D GSD +S +GSD SDSE+ +S+ +D
Sbjct: 516 RSSSDSDSDSDNSDSGSDSKSAAGSDSGSSSDSEASS----------------NSKDGSD 559
Query: 129 ELQTNIQTNIQDESDEFTLPTKQELEEEALRPP 161
E +I + D L T Q LE++A+ P
Sbjct: 560 E-----DVDIMSDGDREPLLTTQSLEQDAIDLP 587
Score = 37.0 bits (84), Expect = 0.026
Identities = 21/79 (26%), Positives = 34/79 (42%)
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLE 88
++ SE+ E +A + D +SD SD D SD E + N D D++
Sbjct: 504 EEKRSENREAQARSSSDSDSDSDNSDSGSDSKSAAGSDSGSSSDSEASSNSKDGSDEDVD 563
Query: 89 SGSGSDEDEDSDSESGEED 107
S D + ++S E+D
Sbjct: 564 IMSDGDREPLLTTQSLEQD 582
Score = 31.6 bits (70), Expect = 1.1
Identities = 26/77 (33%), Positives = 42/77 (53%), Gaps = 11/77 (14%)
Query: 32 TSESEEEEAPPL-----QHLELDGSDSD-LSSDGDDQLADDFL-QGSDDEDNDNDNDDEG 84
T++S E++A L +E++G +SD + DG D A D GSD D + ++ DEG
Sbjct: 575 TTQSLEQDAIDLPGHGSSAVEIEGHNSDAVDIDGHDSDAVDIDGHGSDTVDVEGNSSDEG 634
Query: 85 SDLESGSGSDEDEDSDS 101
GS +D ++SD+
Sbjct: 635 ----HGSDADRKKNSDN 647
>At1g67120 hypothetical protein
Length = 5138
Score = 46.6 bits (109), Expect = 3e-05
Identities = 34/128 (26%), Positives = 62/128 (47%), Gaps = 8/128 (6%)
Query: 47 ELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGS--DEDEDSDSESG 104
E DG + +S D ++ D+ GS+DE DN D GSD E ++DE+ + E+
Sbjct: 4343 EFDGKEYSVSEDEEEDKEDE---GSEDEPLDNGIGDVGSDAEKADEKPWNKDEEDEEENM 4399
Query: 105 EEDIARKSKLIDR-KRERDSQAAADELQTNIQTNIQDESDEFTLPTKQELEEEALRPPDL 163
E ++D+ R R+ +A D ++T + + SD+ + +E++ D
Sbjct: 4400 NEKNESGPSIVDKDTRSRELRAKDDGVETADEPEESNTSDKPEEGNDENVEQDDF--DDT 4457
Query: 164 SNLQRRIK 171
NL+ +I+
Sbjct: 4458 DNLEEKIQ 4465
Score = 35.4 bits (80), Expect = 0.076
Identities = 39/144 (27%), Positives = 67/144 (46%), Gaps = 12/144 (8%)
Query: 28 KDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDL 87
K+ S SE EEE+ + E + D+ + G D D + DE+++ +N +E +
Sbjct: 4347 KEYSVSEDEEEDKED-EGSEDEPLDNGIGDVGSDAEKADEKPWNKDEEDEEENMNEKN-- 4403
Query: 88 ESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQ--DESDEF 145
ESG S D+D+ S ++ K ++ E + +D+ + N++ D D
Sbjct: 4404 ESGP-SIVDKDTRS----RELRAKDDGVETADEPEESNTSDKPEEGNDENVEQDDFDDTD 4458
Query: 146 TLPTKQELEEEAL--RPPDLSNLQ 167
L K + +EEAL PD+ N Q
Sbjct: 4459 NLEEKIQTKEEALGGLTPDVDNEQ 4482
Score = 31.2 bits (69), Expect = 1.4
Identities = 31/153 (20%), Positives = 61/153 (39%), Gaps = 20/153 (13%)
Query: 28 KDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDD---------QLADDFLQG-SDDEDND 77
KDD ++E E +G+D ++ D D Q ++ L G + D DN+
Sbjct: 4422 KDDGVETADEPEESNTSDKPEEGNDENVEQDDFDDTDNLEEKIQTKEEALGGLTPDVDNE 4481
Query: 78 NDNDD------EGSDLESGSGSDE---DEDSDSESGEEDIARKSKLIDRKRERDSQAAAD 128
+DD E + E + +E ++ E GE D + + E +++
Sbjct: 4482 QIDDDMEMDKTEEVEKEDANQQEEPCSEDQKHPEEGENDQEETQEPSEENMEAEAEDRCG 4541
Query: 129 ELQTNIQTN-IQDESDEFTLPTKQELEEEALRP 160
Q N ++ E + + K+ + E+ ++P
Sbjct: 4542 SPQKEEPGNDLEQEPETEPIEGKEVMSEDMMKP 4574
Score = 31.2 bits (69), Expect = 1.4
Identities = 24/90 (26%), Positives = 42/90 (46%), Gaps = 14/90 (15%)
Query: 63 LADDFLQGSDDEDNDNDNDD--------EGSDLESGSGSDEDEDSDSESGEEDIARKSKL 114
LAD F +G N+ D+D EG+ + G G+ ++E+ E ++D+ K+K
Sbjct: 4281 LADLFTKGFGISKNEEDDDSKVDKSEAAEGTGMGDGVGAKDEEE---EKEQDDVLGKNKG 4337
Query: 115 IDRKRERDSQ---AAADELQTNIQTNIQDE 141
I+ E D + + DE + +DE
Sbjct: 4338 IEMSDEFDGKEYSVSEDEEEDKEDEGSEDE 4367
>At4g17590 putative protein
Length = 109
Score = 46.2 bits (108), Expect = 4e-05
Identities = 23/34 (67%), Positives = 26/34 (75%)
Query: 337 GIIFANEMKVPRLKSLTANLHRMGVSNTVVSNYD 370
GIIFAN L SL ANLHRMG++NTVVSNY+
Sbjct: 55 GIIFANASTEHLLGSLYANLHRMGITNTVVSNYN 88
>At2g11910 unknown protein
Length = 168
Score = 46.2 bits (108), Expect = 4e-05
Identities = 18/59 (30%), Positives = 30/59 (50%)
Query: 49 DGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEED 107
D D+D D +D D+ G + ++ + + D E + +G G +DED D E G+ D
Sbjct: 80 DDEDADEDDDDEDDANDEDFSGGEGDEGEEEADPEDDPVTNGGGGSDDEDDDDEEGDND 138
Score = 45.1 bits (105), Expect = 1e-04
Identities = 29/84 (34%), Positives = 41/84 (48%), Gaps = 13/84 (15%)
Query: 30 DSTSESEEEEAPPLQHLELDGSDSDLS----------SDGDDQLADDFLQGSDDEDNDN- 78
DS + ++E+A E D +D D S +D +D + GSDDED+D+
Sbjct: 74 DSDDDDDDEDADEDDDDEDDANDEDFSGGEGDEGEEEADPEDDPVTNGGGGSDDEDDDDE 133
Query: 79 --DNDDEGSDLESGSGSDEDEDSD 100
DNDDE D E D++ED D
Sbjct: 134 EGDNDDEDEDNEDEEEDDDEEDDD 157
Score = 43.9 bits (102), Expect = 2e-04
Identities = 16/80 (20%), Positives = 40/80 (50%)
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLE 88
DD ++ ++++ + G + D + D D G D+++D+D+EG + +
Sbjct: 80 DDEDADEDDDDEDDANDEDFSGGEGDEGEEEADPEDDPVTNGGGGSDDEDDDDEEGDNDD 139
Query: 89 SGSGSDEDEDSDSESGEEDI 108
++++E+ D E ++D+
Sbjct: 140 EDEDNEDEEEDDDEEDDDDV 159
Score = 35.8 bits (81), Expect = 0.058
Identities = 18/95 (18%), Positives = 45/95 (46%), Gaps = 3/95 (3%)
Query: 50 GSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIA 109
GS + + D D DD +D D D+D++D+ +D + G ++ + +++ ++ +
Sbjct: 63 GSRIEENKDASDSDDDD---DDEDADEDDDDEDDANDEDFSGGEGDEGEEEADPEDDPVT 119
Query: 110 RKSKLIDRKRERDSQAAADELQTNIQTNIQDESDE 144
D + + D + D+ + + +D+ +E
Sbjct: 120 NGGGGSDDEDDDDEEGDNDDEDEDNEDEEEDDDEE 154
Score = 30.0 bits (66), Expect = 3.2
Identities = 21/93 (22%), Positives = 40/93 (42%), Gaps = 11/93 (11%)
Query: 69 QGSDDEDNDNDNDDEGSDLESGSGSDEDE-----DSDSESGEEDIARKSKLIDRKRERDS 123
+ D D+D+D+DDE +D D+D+ D D GE D + + +
Sbjct: 68 ENKDASDSDDDDDDEDAD------EDDDDEDDANDEDFSGGEGDEGEEEADPEDDPVTNG 121
Query: 124 QAAADELQTNIQTNIQDESDEFTLPTKQELEEE 156
+D+ + + D+ DE +++ +EE
Sbjct: 122 GGGSDDEDDDDEEGDNDDEDEDNEDEEEDDDEE 154
>At1g71710 RIBOSOMAL PROTEIN, putative (At1g71710)
Length = 664
Score = 46.2 bits (108), Expect = 4e-05
Identities = 31/88 (35%), Positives = 44/88 (49%), Gaps = 14/88 (15%)
Query: 46 LELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGE 105
L + G D + +D D++ S++ED DNDD SD E GSGS E E+ E
Sbjct: 31 LNISGRDPEYGADTDNE--------SENEDAREDNDDSSSDEEGGSGSRGRESKVYENAE 82
Query: 106 EDIARKSKLIDRKRERDSQAAADELQTN 133
+ IA S ++ D+ AAA E +N
Sbjct: 83 DAIAAASAVV------DAAAAAAEFISN 104
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,151,773
Number of Sequences: 26719
Number of extensions: 594773
Number of successful extensions: 9376
Number of sequences better than 10.0: 670
Number of HSP's better than 10.0 without gapping: 274
Number of HSP's successfully gapped in prelim test: 417
Number of HSP's that attempted gapping in prelim test: 4027
Number of HSP's gapped (non-prelim): 2305
length of query: 513
length of database: 11,318,596
effective HSP length: 104
effective length of query: 409
effective length of database: 8,539,820
effective search space: 3492786380
effective search space used: 3492786380
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146751.12


