

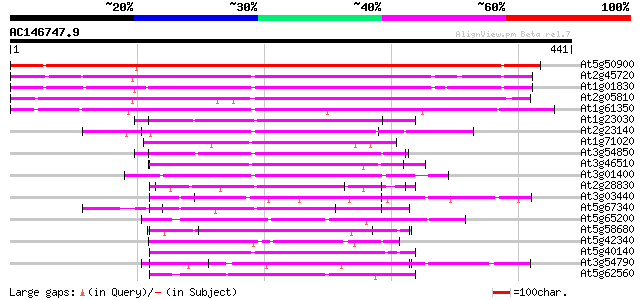
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146747.9 - phase: 2 /pseudo
(441 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g50900 unknown protein 475 e-134
At2g45720 unknown protein 193 2e-49
At1g01830 unknown protein 185 5e-47
At2g05810 hypothetical protein 151 8e-37
At1g61350 unknown 145 4e-35
At1g23030 unknown protein 80 2e-15
At2g23140 hypothetical protein 77 1e-14
At1g71020 unknown protein 74 2e-13
At3g54850 putative protein 73 4e-13
At3g46510 arm repeat containing protein homolog 70 3e-12
At3g01400 unknown protein 68 1e-11
At2g28830 unknown protein 68 1e-11
At3g03440 unknown protein 62 5e-10
At5g67340 unknown proteins 62 6e-10
At5g65200 unknown protein 60 2e-09
At5g58680 unknown protein 59 4e-09
At5g42340 arm repeat containing protein 57 2e-08
At5g40140 putative protein 56 5e-08
At3g54790 unknown protein 55 8e-08
At5g62560 unknown protein 54 2e-07
>At5g50900 unknown protein
Length = 555
Score = 475 bits (1222), Expect = e-134
Identities = 256/425 (60%), Positives = 323/425 (75%), Gaps = 12/425 (2%)
Query: 1 WSIFGVKLTQLQTHLTDFSTEYSKSSTTNPLSLHLLHSVLQTLNDAVSLSHHCQSQILPH 60
WS KL L+T L+DFS SS N L++ LL SV +TLNDAV+++ C+ L
Sbjct: 34 WSSIRAKLADLKTQLSDFSDFAGSSS--NKLAVDLLVSVRETLNDAVAVAARCEGPDLAE 91
Query: 61 GKLQTQSQIDSLIATLHRHVTDCDVLFRSGLLLETPAF-------SKREAVRSLTRNLIA 113
GKL+TQS++DS++A L RHV D +VL +SGLL++ SK+EAVR RNL+
Sbjct: 92 GKLKTQSEVDSVMARLDRHVKDAEVLIKSGLLIDNGIVVSGFSISSKKEAVRLEARNLVI 151
Query: 114 RLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSS-DMKEKTVAAIS 172
RLQIG ES+ +AIDSL+ LL +DDKNV I VAQGVVPVLVRLLDS S MKEKTVA IS
Sbjct: 152 RLQIGGVESKNSAIDSLIELLQEDDKNVMICVAQGVVPVLVRLLDSCSLVMKEKTVAVIS 211
Query: 173 RVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGG 232
R+S VES K+ L+AEGL LLNHL+RVL+SGSG A EKAC+ALQALSLS++NARAIG RGG
Sbjct: 212 RISMVESSKHVLIAEGLSLLNHLLRVLESGSGFAKEKACVALQALSLSKENARAIGCRGG 271
Query: 233 ISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLARENA 292
ISSLL ICQGG+PGSQ +AA VLRNLA F E KENFVEENA+ VL+ + SSGT LA+ENA
Sbjct: 272 ISSLLEICQGGSPGSQAFAAGVLRNLALFGETKENFVEENAIFVLISMVSSGTSLAQENA 331
Query: 293 IGCVANLISEDESMRVLAVKEGGVECLKNYWDSVTMIQSLEVGVEMLRYLAMTGPIDEVL 352
+GC+ANL S DE + + V+EGG++CLK++WDSV+ ++SLEVGV +L+ LA+ + EV+
Sbjct: 332 VGCLANLTSGDEDLMISVVREGGIQCLKSFWDSVSSVKSLEVGVVLLKNLALCPIVREVV 391
Query: 353 VGEGFVGRVIGVLDCDVLTVRIAAARAVYAMGLNGGNKTRKEMGECGCVPSLIKMLDGKG 412
+ EGF+ R++ VL C VL VRIAAA AV ++G + +K+RKEMGE GC+ LI MLDGK
Sbjct: 392 ISEGFIPRLVPVLSCGVLGVRIAAAEAVSSLGFS--SKSRKEMGESGCIVPLIDMLDGKA 449
Query: 413 VEEKD 417
+EEK+
Sbjct: 450 IEEKE 454
>At2g45720 unknown protein
Length = 553
Score = 193 bits (490), Expect = 2e-49
Identities = 134/416 (32%), Positives = 233/416 (55%), Gaps = 18/416 (4%)
Query: 1 WSIFGVKLTQLQTHLTDFSTEYSKSSTTNPLSLHLLHSVLQTLNDAVSLSHHCQSQILPH 60
W + +L ++ T L+D S+ S T L L +VL+TL + + L++ C S+
Sbjct: 44 WRVIISRLEKIPTCLSDLSSHPCFSKHT--LCKEQLQAVLETLKETIELANVCVSE-KQE 100
Query: 61 GKLQTQSQIDSLIATLHRHVTDCDVLFRSGLLLET--PAFSKREAVRSLT-RNLIARLQI 117
GKL+ QS +DSL A + + DC +L ++G+L E P S + + + + R L+ARLQI
Sbjct: 101 GKLKMQSDLDSLSAKIDLSLKDCGLLMKTGVLGEVTKPLSSSTQDLETFSVRELLARLQI 160
Query: 118 GSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSS-DMKEKTVAAISRVST 176
G ES+ A++ L+ ++ +D+K V A+ + V LV+LL ++S ++E V I ++
Sbjct: 161 GHLESKRKALEQLVEVMKEDEKAVITALGRTNVASLVQLLTATSPSVRENAVTVICSLAE 220
Query: 177 VESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGGISSL 236
+N L++E L L+R+L+SGS +A EKA I+LQ +S+S + +R+I GG+ L
Sbjct: 221 SGGCENWLISENAL--PSLIRLLESGSIVAKEKAVISLQRMSISSETSRSIVGHGGVGPL 278
Query: 237 LGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGL-ARENAIGC 295
+ IC+ G SQ +A L+N++ E+++N EE V V++ + + G L ++E A C
Sbjct: 279 IEICKTGDSVSQSASACTLKNISAVPEVRQNLAEEGIVKVMINILNCGILLGSKEYAAEC 338
Query: 296 VANLISEDESMRVLAVKEGGVECLKNYWDSVTMIQSLEVGVEMLRYLAMTGPIDEVLVGE 355
+ NL S +E++R + E G++ L Y D +S GV +R L + ++
Sbjct: 339 LQNLTSSNETLRRSVISENGIQTLLAYLDGPLPQES---GVAAIRNLVGSVSVETYF--- 392
Query: 356 GFVGRVIGVLDCDVLTVRIAAARAVYAMGLNGGNKTRKEMGECGCVPSLIKMLDGK 411
+ ++ VL + + AAA + + + N+T++ +GE GC+P LI+ML+ K
Sbjct: 393 KIIPSLVHVLKSGSIGAQQAAASTICRIATS--NETKRMIGESGCIPLLIRMLEAK 446
>At1g01830 unknown protein
Length = 574
Score = 185 bits (469), Expect = 5e-47
Identities = 141/420 (33%), Positives = 227/420 (53%), Gaps = 21/420 (5%)
Query: 1 WSIFGVKLTQLQTHLTDFSTEYSKSSTTNPLSLHLLHSVLQTLNDAVSLSHHCQSQILPH 60
W K+ Q+ L+D S+ S N L L SV +TL++ + L+ C +
Sbjct: 60 WKTIISKIEQIPACLSDLSSHPCFSK--NKLCNEQLQSVAKTLSEVIELAEQCSTDKY-E 116
Query: 61 GKLQTQSQIDSLIATLHRHVTDCDVLFRSGLLLETP------AFSKREAVRSLTRNLIAR 114
GKL+ QS +DSL L ++ DC VL ++G+L E + S+ + SL + L+AR
Sbjct: 117 GKLRMQSDLDSLSGKLDLNLRDCGVLIKTGVLGEATLPLYISSSSETPKISSL-KELLAR 175
Query: 115 LQIGSPESRATAIDSLLSLLNQDDKNVTIA-VAQGVVPVLVRLLDSSSD-MKEKTVAAIS 172
LQIG ES+ A++SLL + +D+K V + + + V LV+LL ++S ++EK V IS
Sbjct: 176 LQIGHLESKHNALESLLGAMQEDEKMVLMPLIGRANVAALVQLLTATSTRIREKAVNLIS 235
Query: 173 RVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGG 232
++ L++EG+L LVR+++SGS EKA IA+Q LS++ +NAR I GG
Sbjct: 236 VLAESGHCDEWLISEGVL--PPLVRLIESGSLETKEKAAIAIQRLSMTEENAREIAGHGG 293
Query: 233 ISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGL-AREN 291
I+ L+ +C+ G SQ +AA L+N++ +E+++ EE + V + L + G L +RE+
Sbjct: 294 ITPLIDLCKTGDSVSQAASAAALKNMSAVSELRQLLAEEGIIRVSIDLLNHGILLGSREH 353
Query: 292 AIGCVANLISEDESMRVLAVKEGGVECLKNYWDSVTMIQSLEVGVEMLRYLAMTGPIDEV 351
C+ NL + +++R V EGGV L Y D Q V LR L + E+
Sbjct: 354 MAECLQNLTAASDALREAIVSEGGVPSLLAYLDGPLPQQP---AVTALRNL-IPSVNPEI 409
Query: 352 LVGEGFVGRVIGVLDCDVLTVRIAAARAVYAMGLNGGNKTRKEMGECGCVPSLIKMLDGK 411
V + R+ VL L + AAA A+ + +T++ +GE GC+P ++K+L+ K
Sbjct: 410 WVALNLLPRLRHVLKSGSLGAQQAAASAICRFACS--PETKRLVGESGCIPEIVKLLESK 467
>At2g05810 hypothetical protein
Length = 580
Score = 151 bits (381), Expect = 8e-37
Identities = 123/423 (29%), Positives = 208/423 (49%), Gaps = 23/423 (5%)
Query: 1 WSIFGVKLTQLQTHLTDFSTEYSKSSTTNPLSLHLLHSVLQTLNDAVSLSHHCQSQILPH 60
W I KL L + L+ S S + NPL LL S+L L SLS C S
Sbjct: 48 WQILRSKLFTLNSSLSSLSE--SPHWSQNPLLHTLLPSLLSNLQRLSSLSDQCSSASFSG 105
Query: 61 GKLQTQSQIDSLIATLHRHVTDCDVLFRSGLLLET--------PAFSKREAVRSLTRNLI 112
GKL QS +D ++L H++D D+L RSG+L + P S ++ + R+L
Sbjct: 106 GKLLMQSDLDIASSSLSTHISDLDLLLRSGVLHQQNAIVLSLPPPTSDKDDIAFFIRDLF 165
Query: 113 ARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSD--MKEKTVAA 170
RLQIG E + +++SLL LL ++K+ I +G V LV LLD ++E +AA
Sbjct: 166 TRLQIGGAEFKKKSLESLLQLLTDNEKSARIIAKEGNVGYLVTLLDLHHHPLIREHALAA 225
Query: 171 ISRV--STVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIG 228
+S + S+ +S K GL L+R+L++GS +A IA++A++ A AI
Sbjct: 226 VSLLTSSSADSRKTVFEQGGL---GPLLRLLETGSSPFKTRAAIAIEAITADPATAWAIS 282
Query: 229 SRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLA 288
+ GG++ L+ C+ G+ Q + A + N+A EI+ EE A+ VL+ L SG+
Sbjct: 283 AYGGVTVLIEACRSGSKQVQEHIAGAISNIAAVEEIRTTLAEEGAIPVLIQLLISGSSSV 342
Query: 289 RENAIGCVANLISEDESMRVLAVKE-GGVECLKNYWDSVTMIQSLEVGVEMLRYLAMTGP 347
+E ++ + S E R L V+E GG++ L + + ++E + L ++
Sbjct: 343 QEKTANFISLISSSGEYYRDLIVRERGGLQILIHLVQESSNPDTIEHCLLALSQISAMET 402
Query: 348 IDEVLVGE-GFVGRVIGVLDCDVLTVRIAAARAVYAMGLNGGNKTRKEMGECGCVPSLIK 406
+ VL F+ R+ ++ + ++ + + + ++ GNK C+ SLI+
Sbjct: 403 VSRVLSSSTRFIIRLGELIKHGNVILQQISTSLLSNLTISDGNK----RAVADCLSSLIR 458
Query: 407 MLD 409
+++
Sbjct: 459 LME 461
>At1g61350 unknown
Length = 573
Score = 145 bits (366), Expect = 4e-35
Identities = 115/444 (25%), Positives = 222/444 (49%), Gaps = 23/444 (5%)
Query: 1 WSIFGVKLTQLQTHLTDFSTEYSKSSTTNPLSLHLLHSVLQTLNDAVSLSHHCQSQILPH 60
W + KL +L + L + +S +P L+ ++L +L D L+ C + +
Sbjct: 38 WQLIRTKLQELYSGLDSLR---NLNSGFDPSLSSLISAILISLKDTYDLATRCVN-VSFS 93
Query: 61 GKLQTQSQIDSLIATLHRHVTDCDVLFRSGLL--------LETPAFSKREAVRSLTRNLI 112
GKL QS +D + H + ++ +G+L L+ + ++ +R R+L+
Sbjct: 94 GKLLMQSDLDVMAGKFDGHTRNLSRIYSAGILSHGFAIVVLKPNGNACKDDMRFYIRDLL 153
Query: 113 ARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVA-QGVVPVLVRLLDSSSDMKEKTVAAI 171
R++IG E + A+ L + +DD+ V I + +V VLV LDS ++E++ A+
Sbjct: 154 TRMKIGDLEMKKQALVKLNEAMEEDDRYVKILIEISDMVNVLVGFLDSEIGIQEESAKAV 213
Query: 172 SRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRG 231
+S S ++ L+ G++ LVRVL++G+G+ E + L L+ + +NA ++ + G
Sbjct: 214 FFISGFGSYRDVLIRSGVI--GPLVRVLENGNGVGREASARCLMKLTENSENAWSVSAHG 271
Query: 232 GISSLLGICQGGTPGSQ--GYAAAVLRNLAKFNEIKENFVEE-NAVIVLLGLASSGTGLA 288
G+S+LL IC G + G + VLRNL EIK +EE + V + L S +
Sbjct: 272 GVSALLKICSCSDFGGELIGTSCGVLRNLVGVEEIKRFMIEEDHTVATFIKLIGSKEEIV 331
Query: 289 RENAIGCVANLISEDESMRVLAVKEGGVECLKNYW---DSVTMIQSLEVGVEMLRYLAM- 344
+ N+I + ++ +DE R + V+EGG++ L + +S++ +S E+ + + L
Sbjct: 332 QVNSIDLLLSMCCKDEQTRDILVREGGIQELVSVLSDPNSLSSSKSKEIALRAIDNLCFG 391
Query: 345 TGPIDEVLVGEGFVGRVIGVLDCDVLTVRIAAARAVYAMGLNGGNKTRKEMGECGCVPSL 404
+ L+G F+ ++ +L ++V+ +A + + + + ++ MGE G +P L
Sbjct: 392 SAGCLNALMGCKFLDHLLNLLRNGEISVQESALKVTSRL-CSLQEEVKRIMGEAGFMPEL 450
Query: 405 IKMLDGKGVEEKDLLQWRCRCCCS 428
+K LD K ++ +++ C S
Sbjct: 451 VKFLDAKSIDVREMASVALYCLIS 474
>At1g23030 unknown protein
Length = 612
Score = 80.5 bits (197), Expect = 2e-15
Identities = 60/222 (27%), Positives = 102/222 (45%), Gaps = 3/222 (1%)
Query: 99 SKREAVRSLTRNLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLD 158
+K S+ R L+ RL S E R A+ + SL + N + G +PVLV LL
Sbjct: 324 TKNSGDMSVIRALVQRLSSRSTEDRRNAVSEIRSLSKRSTDNRILIAEAGAIPVLVNLLT 383
Query: 159 SSS-DMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQAL 217
S +E + + +S E+ K ++ G + +V+VL +G+ A E A L +L
Sbjct: 384 SEDVATQENAITCVLNLSIYENNKELIMFAGAV--TSIVQVLRAGTMEARENAAATLFSL 441
Query: 218 SLSRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVL 277
SL+ +N IG G I +L+ + + GTP + AA L NL ++ K V V L
Sbjct: 442 SLADENKIIIGGSGAIPALVDLLENGTPRGKKDAATALFNLCIYHGNKGRAVRAGIVTAL 501
Query: 278 LGLASSGTGLARENAIGCVANLISEDESMRVLAVKEGGVECL 319
+ + S T + + ++++ ++ + VK + L
Sbjct: 502 VKMLSDSTRHRMVDEALTILSVLANNQDAKSAIVKANTLPAL 543
Score = 57.8 bits (138), Expect = 1e-08
Identities = 51/187 (27%), Positives = 92/187 (48%), Gaps = 6/187 (3%)
Query: 110 NLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVA 169
+++ L+ G+ E+R A +L SL D+ + I G +P LV LL++ + +K A
Sbjct: 418 SIVQVLRAGTMEARENAAATLFSLSLADENKIIIG-GSGAIPALVDLLENGTPRGKKDAA 476
Query: 170 -AISRVSTVESGKNNLLAEGLLLLNHLVRVL-DSGSGLAIEKACIALQALSLSRDNARAI 227
A+ + K + G++ LV++L DS +++A L L+ ++D AI
Sbjct: 477 TALFNLCIYHGNKGRAVRAGIVTA--LVKMLSDSTRHRMVDEALTILSVLANNQDAKSAI 534
Query: 228 GSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEE-NAVIVLLGLASSGTG 286
+ +L+GI Q ++ AAA+L +L K + K + AV+ L+ L+ +GT
Sbjct: 535 VKANTLPALIGILQTDQTRNRENAAAILLSLCKRDTEKLITIGRLGAVVPLMDLSKNGTE 594
Query: 287 LARENAI 293
+ AI
Sbjct: 595 RGKRKAI 601
Score = 38.9 bits (89), Expect = 0.006
Identities = 32/144 (22%), Positives = 66/144 (45%), Gaps = 3/144 (2%)
Query: 266 ENFVEENAVIVLLGLASSGTGLARENAIGCVANLISEDESMRVLAVKEGGVECLKNYWDS 325
+N + + + L+ SS + R NA+ + +L R+L + G + L N S
Sbjct: 325 KNSGDMSVIRALVQRLSSRSTEDRRNAVSEIRSLSKRSTDNRILIAEAGAIPVLVNLLTS 384
Query: 326 VTMIQSLEVGVEMLRYLAMTGPIDEVLVGEGFVGRVIGVLDCDVLTVRIAAARAVYAMGL 385
+ + E + + L++ E+++ G V ++ VL + R AA ++++ L
Sbjct: 385 ED-VATQENAITCVLNLSIYENNKELIMFAGAVTSIVQVLRAGTMEARENAAATLFSLSL 443
Query: 386 NGGNKTRKEMGECGCVPSLIKMLD 409
NK +G G +P+L+ +L+
Sbjct: 444 ADENKI--IIGGSGAIPALVDLLE 465
>At2g23140 hypothetical protein
Length = 924
Score = 77.4 bits (189), Expect = 1e-14
Identities = 67/262 (25%), Positives = 118/262 (44%), Gaps = 5/262 (1%)
Query: 104 VRSLTRNLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSS-SD 162
V + + L+ L+ S +++ A L L + N + G + +LV LL S+ S
Sbjct: 618 VETQVKKLVEELKSSSLDTQRQATAELRLLAKHNMDNRIVIGNSGAIVLLVELLYSTDSA 677
Query: 163 MKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRD 222
+E V A+ +S ++ K + G + L+ VL++GS A E + L +LS+ +
Sbjct: 678 TQENAVTALLNLSINDNNKKAIADAGAI--EPLIHVLENGSSEAKENSAATLFSLSVIEE 735
Query: 223 NARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLAS 282
N IG G I L+ + GTP + AA L NL+ E K V+ AV L+ L
Sbjct: 736 NKIKIGQSGAIGPLVDLLGNGTPRGKKDAATALFNLSIHQENKAMIVQSGAVRYLIDLMD 795
Query: 283 SGTGLARENAIGCVANLISEDESMRVLAVKEGGVECLKNYWDSVTMIQSLEVGVEMLRYL 342
G+ + A+ +ANL + E + +EGG+ L + + +L+
Sbjct: 796 PAAGMV-DKAVAVLANLATIPEGRNAIG-QEGGIPLLVEVVELGSARGKENAAAALLQLS 853
Query: 343 AMTGPIDEVLVGEGFVGRVIGV 364
+G +++ EG V ++ +
Sbjct: 854 TNSGRFCNMVLQEGAVPPLVAL 875
Score = 62.4 bits (150), Expect = 5e-10
Identities = 62/260 (23%), Positives = 113/260 (42%), Gaps = 30/260 (11%)
Query: 58 LPHGKLQTQSQIDSLIATLHRHVTDCDVLFRSG--------LLLETPAFSKREAVRSLTR 109
L L TQ Q + + L +H D ++ + LL T + ++ AV +L
Sbjct: 629 LKSSSLDTQRQATAELRLLAKHNMDNRIVIGNSGAIVLLVELLYSTDSATQENAVTALLN 688
Query: 110 ------------------NLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVP 151
LI L+ GS E++ + +L SL ++ + I + + P
Sbjct: 689 LSINDNNKKAIADAGAIEPLIHVLENGSSEAKENSAATLFSLSVIEENKIKIGQSGAIGP 748
Query: 152 VLVRLLDSSSDMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKAC 211
++ L + + K+ A+ +S + K ++ G + +L+ ++D +G+ ++KA
Sbjct: 749 LVDLLGNGTPRGKKDAATALFNLSIHQENKAMIVQSGAV--RYLIDLMDPAAGM-VDKAV 805
Query: 212 IALQALSLSRDNARAIGSRGGISSLLGICQ-GGTPGSQGYAAAVLRNLAKFNEIKENFVE 270
L L+ + AIG GGI L+ + + G G + AAA+L+ ++
Sbjct: 806 AVLANLATIPEGRNAIGQEGGIPLLVEVVELGSARGKENAAAALLQLSTNSGRFCNMVLQ 865
Query: 271 ENAVIVLLGLASSGTGLARE 290
E AV L+ L+ SGT ARE
Sbjct: 866 EGAVPPLVALSQSGTPRARE 885
>At1g71020 unknown protein
Length = 628
Score = 73.6 bits (179), Expect = 2e-13
Identities = 65/224 (29%), Positives = 102/224 (45%), Gaps = 27/224 (12%)
Query: 106 SLTRNLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLL--DSSSDM 163
S R L+ +L S E R TA+ + SL + N + G +PVLV+LL D ++
Sbjct: 341 SAIRALVCKLSSQSIEDRRTAVSEIRSLSKRSTDNRILIAEAGAIPVLVKLLTSDGDTET 400
Query: 164 KEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDN 223
+E V I +S E K ++ G + +V VL +GS A E A L +LSL+ +N
Sbjct: 401 QENAVTCILNLSIYEHNKELIMLAG--AVTSIVLVLRAGSMEARENAAATLFSLSLADEN 458
Query: 224 ARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVE------------- 270
IG+ G I +L+ + Q G+ + AA L NL + K V
Sbjct: 459 KIIIGASGAIMALVDLLQYGSVRGKKDAATALFNLCIYQGNKGRAVRAGIVKPLVKMLTD 518
Query: 271 -------ENAVIVLLGLAS---SGTGLARENAIGCVANLISEDE 304
+ A+ +L LAS + T + R NAI + + + +D+
Sbjct: 519 SSSERMADEALTILSVLASNQVAKTAILRANAIPPLIDCLQKDQ 562
Score = 32.7 bits (73), Expect = 0.41
Identities = 46/196 (23%), Positives = 75/196 (37%), Gaps = 46/196 (23%)
Query: 220 SRDNARAIGSRGGISSLLGICQG-GTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLL 278
S DN I G I L+ + G +Q A + NL+ + KE + AV ++
Sbjct: 372 STDNRILIAEAGAIPVLVKLLTSDGDTETQENAVTCILNLSIYEHNKELIMLAGAVTSIV 431
Query: 279 GLASSGTGLARENAIGCVANLISEDESMRVLAVKEGGVECLKNYWDSVTMIQSLEVGVEM 338
+ +G+ ARENA + +L DE+ +++ G + L V++
Sbjct: 432 LVLRAGSMEARENAAATLFSLSLADEN-KIIIGASGAIMAL----------------VDL 474
Query: 339 LRYLAMTGPIDEVLVGEGFVGRVIGVLDCDVLTVRIAAARAVYAMGLNGGNKTRKEMGEC 398
L+Y ++ G D AA A++ + + GNK R
Sbjct: 475 LQYGSVRGKKD--------------------------AATALFNLCIYQGNKGRAV--RA 506
Query: 399 GCVPSLIKMLDGKGVE 414
G V L+KML E
Sbjct: 507 GIVKPLVKMLTDSSSE 522
>At3g54850 putative protein
Length = 632
Score = 72.8 bits (177), Expect = 4e-13
Identities = 60/216 (27%), Positives = 97/216 (44%), Gaps = 3/216 (1%)
Query: 99 SKREAVRSLTRNLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLD 158
S + R+ +L+ +L G+ E + A L L ++ N G +P+LV LL
Sbjct: 338 SSSDCDRTFVLSLLEKLANGTTEQQRAAAGELRLLAKRNVDNRVCIAEAGAIPLLVELLS 397
Query: 159 SSSDM-KEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQAL 217
S +E +V A+ +S E K ++ G + +V VL +GS A E A L +L
Sbjct: 398 SPDPRTQEHSVTALLNLSINEGNKGAIVDAGAI--TDIVEVLKNGSMEARENAAATLFSL 455
Query: 218 SLSRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVL 277
S+ +N AIG+ G I +L+ + + GT + AA + NL + K V+ V L
Sbjct: 456 SVIDENKVAIGAAGAIQALISLLEEGTRRGKKDAATAIFNLCIYQGNKSRAVKGGIVDPL 515
Query: 278 LGLASSGTGLARENAIGCVANLISEDESMRVLAVKE 313
L G + A+ +A L + E +A E
Sbjct: 516 TRLLKDAGGGMVDEALAILAILSTNQEGKTAIAEAE 551
Score = 63.9 bits (154), Expect = 2e-10
Identities = 58/204 (28%), Positives = 97/204 (47%), Gaps = 6/204 (2%)
Query: 110 NLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVA 169
+++ L+ GS E+R A +L SL D+ V I A G + L+ LL+ + +K A
Sbjct: 432 DIVEVLKNGSMEARENAAATLFSLSVIDENKVAIGAA-GAIQALISLLEEGTRRGKKDAA 490
Query: 170 -AISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIG 228
AI + + K+ + G++ + L R+L G +++A L LS +++ AI
Sbjct: 491 TAIFNLCIYQGNKSRAVKGGIV--DPLTRLLKDAGGGMVDEALAILAILSTNQEGKTAIA 548
Query: 229 SRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEE-NAVIVLLGLASSGTGL 287
I L+ I + G+P ++ AAA+L L N + N E A + L L +GT
Sbjct: 549 EAESIPVLVEIIRTGSPRNRENAAAILWYLCIGNIERLNVAREVGADVALKELTENGTDR 608
Query: 288 ARENAIGCVANLISEDESMRVLAV 311
A+ A + LI + E + V V
Sbjct: 609 AKRKA-ASLLELIQQTEGVAVTTV 631
>At3g46510 arm repeat containing protein homolog
Length = 660
Score = 69.7 bits (169), Expect = 3e-12
Identities = 58/202 (28%), Positives = 98/202 (47%), Gaps = 5/202 (2%)
Query: 110 NLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDS-SSDMKEKTV 168
+L+ RL G+PE + +A + L ++ N G +P+LV LL + S ++E +V
Sbjct: 356 DLMWRLAYGNPEDQRSAAGEIRLLAKRNADNRVAIAEAGAIPLLVGLLSTPDSRIQEHSV 415
Query: 169 AAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIG 228
A+ +S E+ K +++ G + +V+VL GS A E A L +LS+ +N IG
Sbjct: 416 TALLNLSICENNKGAIVSAG--AIPGIVQVLKKGSMEARENAAATLFSLSVIDENKVTIG 473
Query: 229 SRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASS-GTGL 287
+ G I L+ + GT + AA L NL + K + + L L + G+G+
Sbjct: 474 ALGAIPPLVVLLNEGTQRGKKDAATALFNLCIYQGNKGKAIRAGVIPTLTRLLTEPGSGM 533
Query: 288 ARENAIGCVANLISEDESMRVL 309
E A+ +A L S E ++
Sbjct: 534 VDE-ALAILAILSSHPEGKAII 554
Score = 60.8 bits (146), Expect = 1e-09
Identities = 57/221 (25%), Positives = 103/221 (45%), Gaps = 9/221 (4%)
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVAA 170
++ L+ GS E+R A +L SL D+ VTI + P++V L + + K+ A
Sbjct: 440 IVQVLKKGSMEARENAAATLFSLSVIDENKVTIGALGAIPPLVVLLNEGTQRGKKDAATA 499
Query: 171 ISRVSTVESGKNNLLAEGLLLLNHLVRVL-DSGSGLAIEKACIALQALSLSRDNARAIGS 229
+ + + K + G ++ L R+L + GSG+ +++A L LS + IGS
Sbjct: 500 LFNLCIYQGNKGKAIRAG--VIPTLTRLLTEPGSGM-VDEALAILAILSSHPEGKAIIGS 556
Query: 230 RGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIV---LLGLASSGTG 286
+ SL+ + G+P ++ AAAVL +L + ++ VE + + L+ LA +GT
Sbjct: 557 SDAVPSLVEFIRTGSPRNRENAAAVLVHLCSGD--PQHLVEAQKLGLMGPLIDLAGNGTD 614
Query: 287 LARENAIGCVANLISEDESMRVLAVKEGGVECLKNYWDSVT 327
+ A + + E + AV + E + +S T
Sbjct: 615 RGKRKAAQLLERISRLAEQQKETAVSQPEEEAEPTHPESTT 655
>At3g01400 unknown protein
Length = 355
Score = 67.8 bits (164), Expect = 1e-11
Identities = 65/256 (25%), Positives = 117/256 (45%), Gaps = 16/256 (6%)
Query: 91 LLLETPAFSKREAVRSLTRNLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVV 150
LLL + + + + L +L + I E + A++ L N+ + + IA A +
Sbjct: 51 LLLSCASENSDDLINHLVSHLDSSYSID--EQKQAAMEIRLLSKNKPENRIKIAKAGAIK 108
Query: 151 PVLVRLLDSSSDMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKA 210
P++ + S ++E V AI +S + K ++ + G + LVR L G+ A E A
Sbjct: 109 PLISLISSSDLQLQEYGVTAILNLSLCDENKESIASSGAI--KPLVRALKMGTPTAKENA 166
Query: 211 CIALQALSLSRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVE 270
AL LS +N AIG G I L+ + + G ++ A+ L +L E K V+
Sbjct: 167 ACALLRLSQIEENKVAIGRSGAIPLLVNLLETGGFRAKKDASTALYSLCSAKENKIRAVQ 226
Query: 271 ENAVIVLLGL-ASSGTGLARENAIGCVANLISEDESMRVLAVKEGGVECLKNYWDSVTMI 329
+ L+ L A G+ + ++A V +L+ + V+EGGV L +
Sbjct: 227 SGIMKPLVELMADFGSNMVDKSAF--VMSLLMSVPESKPAIVEEGGVPVL---------V 275
Query: 330 QSLEVGVEMLRYLAMT 345
+ +EVG + + +A++
Sbjct: 276 EIVEVGTQRQKEMAVS 291
>At2g28830 unknown protein
Length = 654
Score = 67.8 bits (164), Expect = 1e-11
Identities = 61/203 (30%), Positives = 91/203 (44%), Gaps = 16/203 (7%)
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMK--EKTV 168
L+ +L PE R +A + L Q++ N A G +P+LV LL S+D + E V
Sbjct: 360 LLLKLTSQQPEDRRSAAGEIRLLAKQNNHNRVAIAASGAIPLLVNLLTISNDSRTQEHAV 419
Query: 169 AAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIG 228
+I +S + K ++ + +V VL GS A E A L +LS+ +N IG
Sbjct: 420 TSILNLSICQENKGKIVYSSGAVPG-IVHVLQKGSMEARENAAATLFSLSVIDENKVTIG 478
Query: 229 SRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLA 288
+ G I L+ + G+ + AA L NL F K V V VL+ L +
Sbjct: 479 AAGAIPPLVTLLSEGSQRGKKDAATALFNLCIFQGNKGKAVRAGLVPVLMRLLT------ 532
Query: 289 RENAIGCVANLISEDESMRVLAV 311
E G V DES+ +LA+
Sbjct: 533 -EPESGMV------DESLSILAI 548
Score = 61.6 bits (148), Expect = 8e-10
Identities = 51/182 (28%), Positives = 89/182 (48%), Gaps = 9/182 (4%)
Query: 115 LQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVA-AISR 173
LQ GS E+R A +L SL D+ VTI A G +P LV LL S +K A A+
Sbjct: 449 LQKGSMEARENAAATLFSLSVIDENKVTIGAA-GAIPPLVTLLSEGSQRGKKDAATALFN 507
Query: 174 VSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGGI 233
+ + K + GL+ + L+R+L ++++ L LS D +G+ +
Sbjct: 508 LCIFQGNKGKAVRAGLVPV--LMRLLTEPESGMVDESLSILAILSSHPDGKSEVGAADAV 565
Query: 234 SSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIV---LLGLASSGTGLARE 290
L+ + G+P ++ +AAVL +L +N+ ++ +E + + L+ +A +GT +
Sbjct: 566 PVLVDFIRSGSPRNKENSAAVLVHLCSWNQ--QHLIEAQKLGIMDLLIEMAENGTDRGKR 623
Query: 291 NA 292
A
Sbjct: 624 KA 625
Score = 49.7 bits (117), Expect = 3e-06
Identities = 52/212 (24%), Positives = 93/212 (43%), Gaps = 5/212 (2%)
Query: 111 LIARLQIGSPESRAT--AIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSS-DMKEKT 167
L+ L S +SR A+ S+L+L + I + G VP +V +L S + +E
Sbjct: 401 LLVNLLTISNDSRTQEHAVTSILNLSICQENKGKIVYSSGAVPGIVHVLQKGSMEARENA 460
Query: 168 VAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAI 227
A + +S ++ K + A G + LV +L GS + A AL L + + N
Sbjct: 461 AATLFSLSVIDENKVTIGAAGAI--PPLVTLLSEGSQRGKKDAATALFNLCIFQGNKGKA 518
Query: 228 GSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGL 287
G + L+ + G + ++L L+ + K +AV VL+ SG+
Sbjct: 519 VRAGLVPVLMRLLTEPESGMVDESLSILAILSSHPDGKSEVGAADAVPVLVDFIRSGSPR 578
Query: 288 ARENAIGCVANLISEDESMRVLAVKEGGVECL 319
+EN+ + +L S ++ + A K G ++ L
Sbjct: 579 NKENSAAVLVHLCSWNQQHLIEAQKLGIMDLL 610
Score = 42.4 bits (98), Expect = 5e-04
Identities = 42/156 (26%), Positives = 79/156 (49%), Gaps = 7/156 (4%)
Query: 111 LIARLQIGSPESRATAIDSLLSL-LNQDDKNVTIAVAQGVVPVLVRLL-DSSSDMKEKTV 168
L+ L GS + A +L +L + Q +K AV G+VPVL+RLL + S M ++++
Sbjct: 486 LVTLLSEGSQRGKKDAATALFNLCIFQGNKGK--AVRAGLVPVLMRLLTEPESGMVDESL 543
Query: 169 AAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEK-ACIALQALSLSRDNARAI 227
+ ++ +S+ GK+ + A + + LV + SGS E A + + S ++ +
Sbjct: 544 SILAILSSHPDGKSEVGAADAVPV--LVDFIRSGSPRNKENSAAVLVHLCSWNQQHLIEA 601
Query: 228 GSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNE 263
G + L+ + + GT + AA +L ++FN+
Sbjct: 602 QKLGIMDLLIEMAENGTDRGKRKAAQLLNRFSRFND 637
>At3g03440 unknown protein
Length = 408
Score = 62.4 bits (150), Expect = 5e-10
Identities = 57/189 (30%), Positives = 94/189 (49%), Gaps = 10/189 (5%)
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLL-DSSSDMKEKTVA 169
+I LQ SP + A SLL+L + N I A GVVP+LV+++ S K V
Sbjct: 152 IINFLQSNSPTLQEYASASLLTL-SASANNKPIIGANGVVPLLVKVIKHGSPQAKADAVM 210
Query: 170 AISRVSTVESGKNNLLAEGLLLLNHLVRVLDSG--SGLAIEKACIALQALSLSRDNARA- 226
A+S +ST+ + +LA L+ ++ +L S S EK C ++AL +S + AR
Sbjct: 211 ALSNLSTLPDNLSMILA--TKPLSPILNLLKSSKKSSKTSEKCCSLIEALMVSGEEARTG 268
Query: 227 -IGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIK--ENFVEENAVIVLLGLASS 283
+ GG+ +++ + + G+ ++ +A VL L + + K E + E + LL L
Sbjct: 269 LVSDEGGVLAVVEVLENGSLQAREHAVGVLLTLCQSDRSKYREPILREGVIPGLLELTVQ 328
Query: 284 GTGLARENA 292
GT +R A
Sbjct: 329 GTSKSRIKA 337
Score = 46.6 bits (109), Expect = 3e-05
Identities = 65/274 (23%), Positives = 115/274 (41%), Gaps = 13/274 (4%)
Query: 146 AQGVVPVLVRL-LDSSSDMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSG 204
+Q V P++ L DS E + A+ ++ V+ KN + L ++ L S S
Sbjct: 103 SQAVEPLVSMLRFDSPESHHEAALLALLNLA-VKDEKNKVSIIEAGALEPIINFLQSNSP 161
Query: 205 LAIEKACIALQALSLSRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEI 264
E A +L LS S +N IG+ G + L+ + + G+P ++ A L NL+ +
Sbjct: 162 TLQEYASASLLTLSASANNKPIIGANGVVPLLVKVIKHGSPQAKADAVMALSNLSTLPDN 221
Query: 265 KENFVEENAVIVLLGLASSGTGLARENAIGC---VANLISEDESMRVLAVKEGGVECLKN 321
+ + +L L S ++ + C A ++S +E+ L EGGV +
Sbjct: 222 LSMILATKPLSPILNLLKSSKKSSKTSEKCCSLIEALMVSGEEARTGLVSDEGGVLAVVE 281
Query: 322 YWDSVTMIQSLEVGVEMLRYLAMT--GPIDEVLVGEGFVGRVIGVLDCDVLTVRIAAARA 379
++ + +Q+ E V +L L + E ++ EG + ++ + RI A R
Sbjct: 282 VLENGS-LQAREHAVGVLLTLCQSDRSKYREPILREGVIPGLLELTVQGTSKSRIKAQRL 340
Query: 380 VYAMGLNGGNKTRKEMGEC---GCVPSLIKMLDG 410
+ L R E+ V SLI +DG
Sbjct: 341 LCL--LRNSESPRSEVQPDTIENIVSSLISHIDG 372
Score = 34.3 bits (77), Expect = 0.14
Identities = 48/151 (31%), Positives = 70/151 (45%), Gaps = 8/151 (5%)
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDS--SSDMKEKTV 168
L+ ++ GSP+++A A+ +L +L D I + + P+L L S SS EK
Sbjct: 193 LVKVIKHGSPQAKADAVMALSNLSTLPDNLSMILATKPLSPILNLLKSSKKSSKTSEKCC 252
Query: 169 AAIS--RVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLS-RDNAR 225
+ I VS E+ + EG +L +V VL++GS A E A L L S R R
Sbjct: 253 SLIEALMVSGEEARTGLVSDEGGVLA--VVEVLENGSLQAREHAVGVLLTLCQSDRSKYR 310
Query: 226 -AIGSRGGISSLLGICQGGTPGSQGYAAAVL 255
I G I LL + GT S+ A +L
Sbjct: 311 EPILREGVIPGLLELTVQGTSKSRIKAQRLL 341
>At5g67340 unknown proteins
Length = 707
Score = 62.0 bits (149), Expect = 6e-10
Identities = 46/173 (26%), Positives = 80/173 (45%), Gaps = 4/173 (2%)
Query: 121 ESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVAAISRVSTVESG 180
E++A + +L SL ++ I A + P++ L S K+ A+ +S
Sbjct: 520 EAKANSAATLFSLSVIEEYKTEIGEAGAIEPLVDLLGSGSLSGKKDAATALFNLSIHHEN 579
Query: 181 KNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGGISSLLGIC 240
K ++ G + +LV ++D G+ +EKA + L L+ R+ AIG GGI L+ +
Sbjct: 580 KTKVIEAGAV--RYLVELMDPAFGM-VEKAVVVLANLATVREGKIAIGEEGGIPVLVEVV 636
Query: 241 Q-GGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLARENA 292
+ G G + AA+L+ + N + E + L+ L SGT +E A
Sbjct: 637 ELGSARGKENATAALLQLCTHSPKFCNNVIREGVIPPLVALTKSGTARGKEKA 689
Score = 60.1 bits (144), Expect = 2e-09
Identities = 66/259 (25%), Positives = 109/259 (41%), Gaps = 21/259 (8%)
Query: 58 LPHGKLQTQSQIDSLIATLHRHVTDCDVLFRSGLLLETPAFSKREAVRSLTRNLIARLQI 117
L L TQ + + I L R+ TD ++ ++ EA+ SL ++ L
Sbjct: 430 LKSSSLDTQREATARIRILARNSTDNRIVI-----------ARCEAIPSL----VSLLYS 474
Query: 118 GSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSS--SDMKEKTVAAISRVS 175
+A A+ LL+L D+ IA + +VP L+ +L + + K + A + +S
Sbjct: 475 TDERIQADAVTCLLNLSINDNNKSLIAESGAIVP-LIHVLKTGYLEEAKANSAATLFSLS 533
Query: 176 TVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGGISS 235
+E K + G + LV +L SGS + A AL LS+ +N + G +
Sbjct: 534 VIEEYKTEIGEAGAI--EPLVDLLGSGSLSGKKDAATALFNLSIHHENKTKVIEAGAVRY 591
Query: 236 LLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLARENAIGC 295
L+ + G A VL NLA E K EE + VL+ + G+ +ENA
Sbjct: 592 LVELMDPAF-GMVEKAVVVLANLATVREGKIAIGEEGGIPVLVEVVELGSARGKENATAA 650
Query: 296 VANLISEDESMRVLAVKEG 314
+ L + ++EG
Sbjct: 651 LLQLCTHSPKFCNNVIREG 669
Score = 45.8 bits (107), Expect = 5e-05
Identities = 43/147 (29%), Positives = 69/147 (46%), Gaps = 4/147 (2%)
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVAA 170
L+ L GS + A +L +L + +N T + G V LV L+D + M EK V
Sbjct: 551 LVDLLGSGSLSGKKDAATALFNL-SIHHENKTKVIEAGAVRYLVELMDPAFGMVEKAVVV 609
Query: 171 ISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIA-LQALSLSRDNARAIGS 229
++ ++TV GK + EG + + LV V++ GS E A A LQ + S +
Sbjct: 610 LANLATVREGKIAIGEEGGIPV--LVEVVELGSARGKENATAALLQLCTHSPKFCNNVIR 667
Query: 230 RGGISSLLGICQGGTPGSQGYAAAVLR 256
G I L+ + + GT + A +L+
Sbjct: 668 EGVIPPLVALTKSGTARGKEKAQNLLK 694
>At5g65200 unknown protein
Length = 556
Score = 60.1 bits (144), Expect = 2e-09
Identities = 68/268 (25%), Positives = 121/268 (44%), Gaps = 23/268 (8%)
Query: 104 VRSLTRNLI-ARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSS- 161
+ SL +N+I +R + + A+ ++ L+ D KN V G VP+L+ +L S S
Sbjct: 267 ILSLLKNMIVSRYSLVQTNALASLVN-----LSLDKKNKLTIVRLGFVPILIDVLKSGSR 321
Query: 162 DMKEKTVAAISRVSTVESGKNNLLAEGLLL-LNHLVRVLDSGSGLAIEKACIALQALSLS 220
+ +E I +S + K + G L L H +R +S + +AL L+L+
Sbjct: 322 EAQEHAAGTIFSLSLEDDNKMPIGVLGALQPLLHALRAAESDR--TRHDSALALYHLTLN 379
Query: 221 RDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLG- 279
+ N + G + +L + + G S+ A V+ NLA +E + ++ NAV +L+G
Sbjct: 380 QTNRSKLVRLGAVPALFSMVRSGESASR--ALLVICNLACCSEGRSAMLDANAVAILVGK 437
Query: 280 ---------LASSGTGLARENAIGCVANLISEDESMRVLAVKEGGVECLKNYWDSVTMIQ 330
+ + AREN + + L E + LA + VE LK + T +
Sbjct: 438 LREEWTEEPTEARSSSSARENCVAALFALSHESLRFKGLAKEARAVEVLKEVEERGTE-R 496
Query: 331 SLEVGVEMLRYLAMTGPIDEVLVGEGFV 358
+ E ++L+ + P D+ GEG +
Sbjct: 497 AREKAKKILQLMRERVPEDDEEDGEGSI 524
Score = 30.8 bits (68), Expect = 1.6
Identities = 29/105 (27%), Positives = 49/105 (46%), Gaps = 1/105 (0%)
Query: 208 EKACIALQALSLSRDNARAIGSRGGISSLL-GICQGGTPGSQGYAAAVLRNLAKFNEIKE 266
E+ I ++ ++ + D AR I SLL + Q A A L NL+ + K
Sbjct: 242 EQGLIMMRKMTRTNDEARVSLCSPRILSLLKNMIVSRYSLVQTNALASLVNLSLDKKNKL 301
Query: 267 NFVEENAVIVLLGLASSGTGLARENAIGCVANLISEDESMRVLAV 311
V V +L+ + SG+ A+E+A G + +L ED++ + V
Sbjct: 302 TIVRLGFVPILIDVLKSGSREAQEHAAGTIFSLSLEDDNKMPIGV 346
>At5g58680 unknown protein
Length = 357
Score = 59.3 bits (142), Expect = 4e-09
Identities = 58/251 (23%), Positives = 97/251 (38%), Gaps = 45/251 (17%)
Query: 109 RNLIARLQIGSP--ESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEK 166
RNLI L+ S E + A++ L N+ + + +A A + P++ + S ++E
Sbjct: 63 RNLITHLESSSSIEEQKQAAMEIRLLSKNKPENRIKLAKAGAIKPLVSLISSSDLQLQEY 122
Query: 167 TVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARA 226
V A+ +S + K +++ G + LV L G+ E A AL LS +N
Sbjct: 123 GVTAVLNLSLCDENKEMIVSSGAV--KPLVNALRLGTPTTKENAACALLRLSQVEENKIT 180
Query: 227 IGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKEN------------------- 267
IG G I L+ + + G ++ A+ L +L NE K
Sbjct: 181 IGRSGAIPLLVNLLENGGFRAKKDASTALYSLCSTNENKTRAVESGIMKPLVELMIDFES 240
Query: 268 ----------------------FVEENAVIVLLGLASSGTGLARENAIGCVANLISEDES 305
VEE V VL+ + +GT +E ++ + L E
Sbjct: 241 DMVDKSAFVMNLLMSAPESKPAVVEEGGVPVLVEIVEAGTQRQKEISVSILLQLCEESVV 300
Query: 306 MRVLAVKEGGV 316
R + +EG V
Sbjct: 301 YRTMVAREGAV 311
Score = 54.7 bits (130), Expect = 1e-07
Identities = 38/175 (21%), Positives = 81/175 (45%), Gaps = 2/175 (1%)
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVAA 170
L+ L++G+P ++ A +LL L ++ +TI G +P+LV LL++ + K A+
Sbjct: 149 LVNALRLGTPTTKENAACALLRLSQVEENKITIG-RSGAIPLLVNLLENGG-FRAKKDAS 206
Query: 171 ISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSR 230
+ S + +N A ++ LV ++ ++K+ + L + ++ A+
Sbjct: 207 TALYSLCSTNENKTRAVESGIMKPLVELMIDFESDMVDKSAFVMNLLMSAPESKPAVVEE 266
Query: 231 GGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGT 285
GG+ L+ I + GT + + ++L L + + + V + L S G+
Sbjct: 267 GGVPVLVEIVEAGTQRQKEISVSILLQLCEESVVYRTMVAREGAVPPLVALSQGS 321
Score = 52.0 bits (123), Expect = 7e-07
Identities = 45/167 (26%), Positives = 81/167 (47%), Gaps = 2/167 (1%)
Query: 149 VVPVLVRLLDSSSDMKEKTVAAIS-RVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAI 207
V+ L+ L+SSS ++E+ AA+ R+ + +N + + LV ++ S
Sbjct: 61 VIRNLITHLESSSSIEEQKQAAMEIRLLSKNKPENRIKLAKAGAIKPLVSLISSSDLQLQ 120
Query: 208 EKACIALQALSLSRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKEN 267
E A+ LSL +N I S G + L+ + GTP ++ AA L L++ E K
Sbjct: 121 EYGVTAVLNLSLCDENKEMIVSSGAVKPLVNALRLGTPTTKENAACALLRLSQVEENKIT 180
Query: 268 FVEENAVIVLLGLASSGTGLARENAIGCVANLISEDESMRVLAVKEG 314
A+ +L+ L +G A+++A + +L S +E+ + AV+ G
Sbjct: 181 IGRSGAIPLLVNLLENGGFRAKKDASTALYSLCSTNEN-KTRAVESG 226
>At5g42340 arm repeat containing protein
Length = 656
Score = 57.4 bits (137), Expect = 2e-08
Identities = 55/203 (27%), Positives = 98/203 (48%), Gaps = 14/203 (6%)
Query: 110 NLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVA 169
N+I L+ G+ E+R + +L SL D+ VTI ++ G+ P++ L + K+ +
Sbjct: 462 NIIEILENGNREARENSAAALFSLSMLDENKVTIGLSNGIPPLVDLLQHGTLRGKKDALT 521
Query: 170 AISRVSTVESGKNNLLAEGLL--LLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAI 227
A+ +S + K + G++ LLN L D G+ I++A L L+ + +AI
Sbjct: 522 ALFNLSLNSANKGRAIDAGIVQPLLNLL---KDKNLGM-IDEALSILLLLASHPEGRQAI 577
Query: 228 GSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFV----EENAVIVLLGLASS 283
G I +L+ + GTP ++ A +VL L N +F+ + L+ + +S
Sbjct: 578 GQLSFIETLVEFIRQGTPKNKECATSVLLELGSNN---SSFILAALQFGVYEYLVEITTS 634
Query: 284 GTGLARENAIGCVANLISEDESM 306
GT A+ A + LIS+ E +
Sbjct: 635 GTNRAQRKA-NALIQLISKSEQI 656
Score = 31.2 bits (69), Expect = 1.2
Identities = 23/93 (24%), Positives = 40/93 (42%)
Query: 222 DNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLA 281
+N I + G I L+ + G Q A L NL+ K+ E A+ ++ +
Sbjct: 408 ENRVLIANAGAIPLLVQLLSYPDSGIQENAVTTLLNLSIDEVNKKLISNEGAIPNIIEIL 467
Query: 282 SSGTGLARENAIGCVANLISEDESMRVLAVKEG 314
+G AREN+ + +L DE+ + + G
Sbjct: 468 ENGNREARENSAAALFSLSMLDENKVTIGLSNG 500
>At5g40140 putative protein
Length = 550
Score = 55.8 bits (133), Expect = 5e-08
Identities = 56/210 (26%), Positives = 98/210 (46%), Gaps = 6/210 (2%)
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSS-SDMKEKTVA 169
L+ +L+ A+ S+ + D+ + V+ L L+ S + ++ A
Sbjct: 233 LLTKLKSNRISEIEEALISIRRITRIDESSRISLCTTRVISALKSLIVSRYATVQVNVTA 292
Query: 170 AISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGS 229
+ +S +S K ++ G++ L+ VL GS A E + + +L+L +N AIG
Sbjct: 293 VLVNLSLEKSNKVKIVRSGIV--PPLIDVLKCGSVEAQEHSAGVIFSLALEDENKTAIGV 350
Query: 230 RGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLAR 289
GG+ LL + + GT ++ +A L +L+ + V+ AV +LLG+ S G + R
Sbjct: 351 LGGLEPLLHLIRVGTELTRHDSALALYHLSLVQSNRGKLVKLGAVQMLLGMVSLGQMIGR 410
Query: 290 ENAIGCVANLISEDESMRVLAVKEGGVECL 319
I C N+ S S R + GGVEC+
Sbjct: 411 VLLILC--NMASCPVS-RPALLDSGGVECM 437
Score = 28.5 bits (62), Expect = 7.7
Identities = 28/120 (23%), Positives = 55/120 (45%), Gaps = 5/120 (4%)
Query: 290 ENAIGCVANLISEDESMRVLAVKEGGVECLKNYWDSVTMIQSLEVGVE-MLRYLAMTGPI 348
E A+ + + DES R+ + LK+ V+ +++V V +L L++
Sbjct: 246 EEALISIRRITRIDESSRISLCTTRVISALKSL--IVSRYATVQVNVTAVLVNLSLEKSN 303
Query: 349 DEVLVGEGFVGRVIGVLDCDVLTVRIAAARAVYAMGLNGGNKTRKEMGECGCVPSLIKML 408
+V G V +I VL C + + +A ++++ L NKT +G G + L+ ++
Sbjct: 304 KVKIVRSGIVPPLIDVLKCGSVEAQEHSAGVIFSLALEDENKT--AIGVLGGLEPLLHLI 361
>At3g54790 unknown protein
Length = 760
Score = 55.1 bits (131), Expect = 8e-08
Identities = 58/233 (24%), Positives = 99/233 (41%), Gaps = 26/233 (11%)
Query: 104 VRSLTRNLIA-RLQIGSPESRATAIDSLLSLLNQDDK------------------NVTIA 144
+R LT N I R+ IG R AI LLSLL ++K N +
Sbjct: 496 IRHLTINSIENRVHIG----RCGAITPLLSLLYSEEKLTQEHAVTALLNLSISELNKAMI 551
Query: 145 VAQGVVPVLVRLLDSSSDM-KEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGS 203
V G + LV +L++ +D KE + A++ +S ++ + + + + LV +L G+
Sbjct: 552 VEVGAIEPLVHVLNTGNDRAKENSAASLFSLSVLQVNRERI-GQSNAAIQALVNLLGKGT 610
Query: 204 GLAIEKACIALQALSLSRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNE 263
+ A AL LS++ DN I + L+ + A A+L NL+ E
Sbjct: 611 FRGKKDAASALFNLSITHDNKARIVQAKAVKYLVELLDPDLE-MVDKAVALLANLSAVGE 669
Query: 264 IKENFVEENAVIVLLGLASSGTGLARENAIGCVANLISEDESMRVLAVKEGGV 316
++ V E + +L+ G+ +ENA + L L ++EG +
Sbjct: 670 GRQAIVREGGIPLLVETVDLGSQRGKENAASVLLQLCLNSPKFCTLVLQEGAI 722
Score = 46.6 bits (109), Expect = 3e-05
Identities = 55/209 (26%), Positives = 91/209 (43%), Gaps = 15/209 (7%)
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVA- 169
L+ L G+ ++ + SL SL I + + LV LL + +K A
Sbjct: 560 LVHVLNTGNDRAKENSAASLFSLSVLQVNRERIGQSNAAIQALVNLLGKGTFRGKKDAAS 619
Query: 170 AISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGS 229
A+ +S K ++ + +LV +LD + ++KA L LS + +AI
Sbjct: 620 ALFNLSITHDNKARIVQAKAV--KYLVELLDPDLEM-VDKAVALLANLSAVGEGRQAIVR 676
Query: 230 RGGISSLLGICQGGTPGSQGYAAAVLRNLA----KFNEIKENFVEENAVIVLLGLASSGT 285
GGI L+ G+ + AA+VL L KF + ++E A+ L+ L+ SGT
Sbjct: 677 EGGIPLLVETVDLGSQRGKENAASVLLQLCLNSPKFCTL---VLQEGAIPPLVALSQSGT 733
Query: 286 GLARENAIGCVANLISEDESMRVLAVKEG 314
A+E A L+S + R +K+G
Sbjct: 734 QRAKEKA----QQLLSHFRNQRDARMKKG 758
Score = 45.1 bits (105), Expect = 8e-05
Identities = 58/257 (22%), Positives = 112/257 (43%), Gaps = 12/257 (4%)
Query: 157 LDSSSDMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLD---SGSGLAIEKACIA 213
L+SS+++ + AA T E ++L G + +H +++++ SGS A
Sbjct: 440 LESSNNVNHEHSAA----KTYECSVHDLDDSGTMTTSHTIKLVEDLKSGSNKVKTAAAAE 495
Query: 214 LQALSL-SRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEEN 272
++ L++ S +N IG G I+ LL + +Q +A L NL+ K VE
Sbjct: 496 IRHLTINSIENRVHIGRCGAITPLLSLLYSEEKLTQEHAVTALLNLSISELNKAMIVEVG 555
Query: 273 AVIVLLGLASSGTGLARENAIGCVANLISEDESMRVLAVKEGGVECLKNYWDSVTMIQSL 332
A+ L+ + ++G A+EN+ + +L + + ++ L N T +
Sbjct: 556 AIEPLVHVLNTGNDRAKENSAASLFSLSVLQVNRERIGQSNAAIQALVNLLGKGT-FRGK 614
Query: 333 EVGVEMLRYLAMTGPIDEVLVGEGFVGRVIGVLDCDVLTVRIAAARAVYAMGLNGGNKTR 392
+ L L++T +V V ++ +LD D+ V A A L+ + R
Sbjct: 615 KDAASALFNLSITHDNKARIVQAKAVKYLVELLDPDLEMVDKAVA---LLANLSAVGEGR 671
Query: 393 KEMGECGCVPSLIKMLD 409
+ + G +P L++ +D
Sbjct: 672 QAIVREGGIPLLVETVD 688
>At5g62560 unknown protein
Length = 559
Score = 53.9 bits (128), Expect = 2e-07
Identities = 59/214 (27%), Positives = 98/214 (45%), Gaps = 13/214 (6%)
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVAA 170
L++R + + A+ ++ L+ + +N V G VP+L+ +L S + ++ VA
Sbjct: 285 LVSRYNLVQTNAAASVVN-----LSLEKQNKVKIVRSGFVPLLIDVLKSGTTEAQEHVAG 339
Query: 171 ISRVSTVESGKNNLLAEGLLLLNHLVRVLDSG-SGLAIEKACIALQALSLSRDNARAIGS 229
+ S +N ++ L + L+ L S S A + A +AL LSL N +
Sbjct: 340 -ALFSLALEDENKMVIGVLGAVEPLLHALRSSESERARQDAALALYHLSLIPSNRTRLVR 398
Query: 230 RGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLG-LASSGTG-- 286
G + +LL + + G S+ VL NLA + K ++ NAV +L+G L G G
Sbjct: 399 AGAVPTLLSMVRSGDSTSR--ILLVLCNLAACPDGKGAMLDGNAVAILVGKLREVGGGDS 456
Query: 287 -LARENAIGCVANLISEDESMRVLAVKEGGVECL 319
AREN + + L + R LA + G E L
Sbjct: 457 EAARENCVAVLLTLCQGNLRFRGLASEAGAEEVL 490
Score = 35.8 bits (81), Expect = 0.048
Identities = 36/127 (28%), Positives = 59/127 (46%), Gaps = 1/127 (0%)
Query: 248 QGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLARENAIGCVANLISEDESMR 307
Q AAA + NL+ + K V V +L+ + SGT A+E+ G + +L EDE+
Sbjct: 293 QTNAAASVVNLSLEKQNKVKIVRSGFVPLLIDVLKSGTTEAQEHVAGALFSLALEDENKM 352
Query: 308 VLAVKEGGVECLKNYWDSVTMIQSLEVGVEMLRYLAMTGPIDEVLVGEGFVGRVIGVLDC 367
V+ V G VE L + S ++ + L +L++ LV G V ++ ++
Sbjct: 353 VIGVL-GAVEPLLHALRSSESERARQDAALALYHLSLIPSNRTRLVRAGAVPTLLSMVRS 411
Query: 368 DVLTVRI 374
T RI
Sbjct: 412 GDSTSRI 418
Score = 34.7 bits (78), Expect = 0.11
Identities = 40/130 (30%), Positives = 60/130 (45%), Gaps = 2/130 (1%)
Query: 191 LLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGGISSLLGICQGGTPGSQGY 250
+L+ L +L S L A ++ LSL + N I G + L+ + + GT +Q +
Sbjct: 277 ILSFLRSLLVSRYNLVQTNAAASVVNLSLEKQNKVKIVRSGFVPLLIDVLKSGTTEAQEH 336
Query: 251 AAAVLRNLAKFNEIKENFVEENAVIVLL-GLASSGTGLARENAIGCVANLISEDESMRVL 309
A L +LA +E K AV LL L SS + AR++A + +L S S R
Sbjct: 337 VAGALFSLALEDENKMVIGVLGAVEPLLHALRSSESERARQDAALALYHL-SLIPSNRTR 395
Query: 310 AVKEGGVECL 319
V+ G V L
Sbjct: 396 LVRAGAVPTL 405
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,960,638
Number of Sequences: 26719
Number of extensions: 359592
Number of successful extensions: 1252
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 41
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 1044
Number of HSP's gapped (non-prelim): 146
length of query: 441
length of database: 11,318,596
effective HSP length: 102
effective length of query: 339
effective length of database: 8,593,258
effective search space: 2913114462
effective search space used: 2913114462
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146747.9


