

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.8 - phase: 0
(298 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
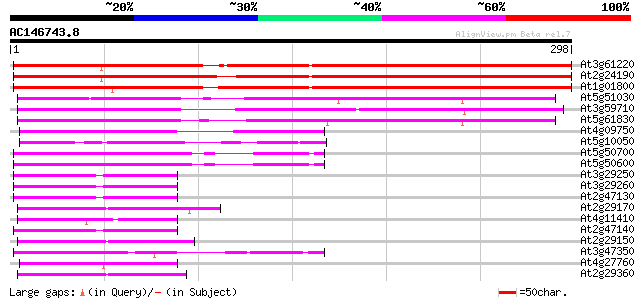
Score E
Sequences producing significant alignments: (bits) Value
At3g61220 unknown protein (At3g61220) 258 3e-69
At2g24190 putative carbonyl reductase 254 4e-68
At1g01800 putative short-chain dehydrogenase 239 1e-63
At5g51030 carbonyl reductase-like protein 179 1e-45
At3g59710 putative protein 178 4e-45
At5g61830 carbonyl reductase - like protein 173 1e-43
At4g09750 putative protein 65 6e-11
At5g10050 unknown protein 61 6e-10
At5g50700 11-beta-hydroxysteroid dehydrogenase-like 54 1e-07
At5g50600 11-beta-hydroxysteroid dehydrogenase-like 54 1e-07
At3g29250 short-chain alcohol dehydrogenase, putative 54 1e-07
At3g29260 short-chain alcohol dehydrogenase, putative 53 2e-07
At2g47130 putative alcohol dehydrogenase 52 3e-07
At2g29170 putative tropinone reductase 52 5e-07
At4g11410 unknown protein 51 9e-07
At2g47140 alcohol dehydrogenase like protein 51 9e-07
At2g29150 pseudogene 51 9e-07
At3g47350 putative protein 50 1e-06
At4g27760 forever young gene (FEY) (fragment) 50 2e-06
At2g29360 putative tropinone reductase 49 3e-06
>At3g61220 unknown protein (At3g61220)
Length = 296
Score = 258 bits (659), Expect = 3e-69
Identities = 140/300 (46%), Positives = 196/300 (64%), Gaps = 14/300 (4%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH---QTGLSNVMFHQL 59
RYAVVTGAN+GIG EI +QLA G+ VVLT+R++ RG +A+ L + +++FHQL
Sbjct: 7 RYAVVTGANRGIGFEICRQLASEGIRVVLTSRDENRGLEAVETLKKELEISDQSLLFHQL 66
Query: 60 DVLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTL 119
DV D SI SLA+F++ +FG+LDIL+NNAG + D E L+A AGK
Sbjct: 67 DVADPASITSLAEFVKTQFGKLDILVNNAGIGGIITDAEALRAG--------AGK-EGFK 117
Query: 120 LQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNER 179
++T+TY+ EEC+ NYYG KR+ A +PLL+LS + RIVN+SS G+LK + NE
Sbjct: 118 WDEIITETYELTEECIKINYYGPKRMCEAFIPLLKLSDSP-RIVNVSSSMGQLKNVLNEW 176
Query: 180 LRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAK 239
+ L D + L+E +ID ++ + L+DFK + W + AY +SKASLN YTRVLAK
Sbjct: 177 AKGILSDAENLTEERIDQVINQLLNDFKEGTVKEKNWAKFMSAYVVSKASLNGYTRVLAK 236
Query: 240 KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLP-ADGPTGCYFDCTEIAEF 298
K+P +N V PGFV TD N+ G ++V+EGA PV L+LLP + P+GC+F +++EF
Sbjct: 237 KHPEFRVNAVCPGFVKTDMNFKTGVLSVEEGASSPVRLALLPHQETPSGCFFSRKQVSEF 296
>At2g24190 putative carbonyl reductase
Length = 296
Score = 254 bits (649), Expect = 4e-68
Identities = 133/301 (44%), Positives = 198/301 (65%), Gaps = 16/301 (5%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH---QTGLSNVMFHQL 59
RYA+VTG N+GIG EI +QLA G+ V+LT+R++ +G +A+ L + +++FHQL
Sbjct: 7 RYAIVTGGNRGIGFEICRQLANKGIRVILTSRDEKQGLEAVETLKKELEISDQSIVFHQL 66
Query: 60 DVLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKA-LNVDPATWLAGKVSNT 118
DV D +S+ SLA+F++ FG+LDILINNAG V D + L+A + W
Sbjct: 67 DVSDPVSVTSLAEFVKTHFGKLDILINNAGVGGVITDVDALRAGTGKEGFKW-------- 118
Query: 119 LLQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNE 178
+ +T+TY+ AEEC+ NYYG KR+ A +PLLQLS + RI+N+SS G++K + NE
Sbjct: 119 --EETITETYELAEECIKINYYGPKRMCEAFIPLLQLSDSP-RIINVSSFMGQVKNLVNE 175
Query: 179 RLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLA 238
+ L D + L+E +ID ++ + L+D K + + W ++ AY +SKA LNAYTR+LA
Sbjct: 176 WAKGILSDAENLTEVRIDQVINQLLNDLKEDTAKTKYWAKVMSAYVVSKAGLNAYTRILA 235
Query: 239 KKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLP-ADGPTGCYFDCTEIAE 297
KK+P + +N V PGFV TD N+ G ++V+EGA PV L+LLP + P+GC+FD +++E
Sbjct: 236 KKHPEIRVNSVCPGFVKTDMNFKTGILSVEEGASSPVRLALLPHQESPSGCFFDRKQVSE 295
Query: 298 F 298
F
Sbjct: 296 F 296
>At1g01800 putative short-chain dehydrogenase
Length = 295
Score = 239 bits (611), Expect = 1e-63
Identities = 130/299 (43%), Positives = 197/299 (65%), Gaps = 11/299 (3%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH-QTGLSN--VMFHQL 59
R AVVTG+NKGIG EI +QLA G+TVVLTAR++ +G A+ KL + G S+ + FH L
Sbjct: 5 RVAVVTGSNKGIGFEICRQLANNGITVVLTARDENKGLAAVQKLKTENGFSDQAISFHPL 64
Query: 60 DVLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTL 119
DV + +I SLA F++ +FG+LDIL+NNAG V+ + LKA +A + T
Sbjct: 65 DVSNPDTIASLAAFVKTRFGKLDILVNNAGVGGANVNVDVLKAQ-------IAEAGAPTD 117
Query: 120 LQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNER 179
+ +++ TY+ EEC+ TNYYGVKR+ A++PLLQ S + RIV+++S G+L+ + NE
Sbjct: 118 ISKIMSDTYEIVEECVKTNYYGVKRMCEAMIPLLQSSDSP-RIVSIASTMGKLENVSNEW 176
Query: 180 LRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAK 239
+ L D + L+E KID ++ ++L D+K + GW ++ Y +SKA++ A TRVLAK
Sbjct: 177 AKGVLSDAENLTEEKIDEVINEYLKDYKEGALQVKGWPTVMSGYILSKAAVIALTRVLAK 236
Query: 240 KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYFDCTEIAEF 298
++ +IN V PGFV+T+ N++ G ++V+EGA PV L+L+P P+G +FD ++ F
Sbjct: 237 RHKSFIINSVCPGFVNTEINFNTGILSVEEGAASPVKLALVPNGDPSGLFFDRANVSNF 295
>At5g51030 carbonyl reductase-like protein
Length = 314
Score = 179 bits (455), Expect = 1e-45
Identities = 112/296 (37%), Positives = 168/296 (55%), Gaps = 39/296 (13%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
AVVTGAN+GIG E+V+QLA G+TV+LT+R++ G +A K+ Q G NV FH+LD+LD+
Sbjct: 40 AVVTGANRGIGFEMVRQLAGHGLTVILTSRDENVGVEA-AKILQEGGFNVDFHRLDILDS 98
Query: 65 LSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGVL 124
SI+ ++I+ K+G +D+LINNAG + NV
Sbjct: 99 SSIQEFCEWIKEKYGFIDVLINNAGVN-----------YNVGS----------------- 130
Query: 125 TQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELK----RIPNERL 180
+ + + ++TNYYG K + A++PL++ + ARIVN++S G LK ++ NE +
Sbjct: 131 DNSVEFSHMVISTNYYGTKNIINAMIPLMRHACQGARIVNVTSRLGRLKGRHSKLENEDV 190
Query: 181 RNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAK- 239
R +L DVD L+E +D V +FL + E+ GW YS+SK ++NAYTRVLAK
Sbjct: 191 RAKLMDVDSLTEEIVDKTVSEFLKQVEEGTWESGGWPHSFTDYSVSKMAVNAYTRVLAKE 250
Query: 240 -----KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYF 290
+ + NC PG+V T + G ++ ++GA V L+LLP TG +F
Sbjct: 251 LSERPEGEKIYANCFCPGWVKTAMTGYAGNVSAEDGADTGVWLALLPDQAITGKFF 306
>At3g59710 putative protein
Length = 302
Score = 178 bits (451), Expect = 4e-45
Identities = 114/293 (38%), Positives = 155/293 (51%), Gaps = 32/293 (10%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
AVVTGANKGIG +VK+L LG+TVVLTARN G A L + G NV F LD+ D
Sbjct: 31 AVVTGANKGIGFAVVKRLLELGLTVVLTARNAENGSQAAESLRRIGFGNVHFCCLDISDP 90
Query: 65 LSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGVL 124
SI + A + G LDIL+NNA S V
Sbjct: 91 SSIAAFASWFGRNLGILDILVNNAAVS----------------------------FNAVG 122
Query: 125 TQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLRNEL 184
K+ E + TN+YG K +T ALLPL + S + +RI+N+SS G L ++ + +R L
Sbjct: 123 ENLIKEPETIIKTNFYGAKLLTEALLPLFRRSVSVSRILNMSSRLGTLNKLRSPSIRRIL 182
Query: 185 GDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAKK--NP 242
+ ++L+ +IDA + +FL D K+ E GW P Y+ISK +LNAY+RVLA++
Sbjct: 183 -ESEDLTNEQIDATLTQFLQDVKSGTWEKQGWPENWPDYAISKLALNAYSRVLARRYDGK 241
Query: 243 HMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADG-PTGCYFDCTE 294
+ +NC+ PGF T +GT T DE A L LLP + TG ++ C E
Sbjct: 242 KLSVNCLCPGFTRTSMTGGQGTHTADEAAAIVAKLVLLPPEKLATGKFYICVE 294
>At5g61830 carbonyl reductase - like protein
Length = 316
Score = 173 bits (438), Expect = 1e-43
Identities = 115/298 (38%), Positives = 159/298 (52%), Gaps = 40/298 (13%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKL-HQTGLSNVMFHQLDVLD 63
AVVTG+N+GIG EI +QLA G+TVVLTARN G +A+ L HQ V FHQLDV D
Sbjct: 39 AVVTGSNRGIGFEIARQLAVHGLTVVLTARNVNAGLEAVKSLRHQEEGLKVYFHQLDVTD 98
Query: 64 ALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGV 123
+ SI +++ FG LDIL+NNAG + L D
Sbjct: 99 SSSIREFGCWLKQTFGGLDILVNNAGVN---------YNLGSD----------------- 132
Query: 124 LTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSS----LRGELKRIPNER 179
T + AE ++TNY G K +T A++PL++ SP AR+VN+SS + G R+ N
Sbjct: 133 --NTVEFAETVISTNYQGTKNMTKAMIPLMRPSPHGARVVNVSSRLGRVNGRRNRLANVE 190
Query: 180 LRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAK 239
LR++L D L+E ID V KF++ K E+ GW YS+SK ++NAYTR++AK
Sbjct: 191 LRDQLSSPDLLTEELIDRTVSKFINQVKDGTWESGGWPQTFTDYSMSKLAVNAYTRLMAK 250
Query: 240 ------KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSL-LPADGPTGCYF 290
+ + +N PG+V T + G M ++ A V LSL L + TG +F
Sbjct: 251 ELERRGEEEKIYVNSFCPGWVKTAMTGYAGNMPPEDAADTGVWLSLVLSEEAVTGKFF 308
>At4g09750 putative protein
Length = 322
Score = 64.7 bits (156), Expect = 6e-11
Identities = 48/163 (29%), Positives = 71/163 (43%), Gaps = 30/163 (18%)
Query: 6 VVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH-QTGLSNVMFHQLDVLDA 64
VVTGAN GIG + LA G TV + RN RG++A++K+ TG NV D+
Sbjct: 47 VVTGANSGIGFAAAEGLASRGATVYMVCRNKERGQEALSKIQTSTGNQNVYLEVCDLSSV 106
Query: 65 LSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGVL 124
I+S A K + +L+NNAG LL+
Sbjct: 107 NEIKSFASSFASKDVPVHVLVNNAG-----------------------------LLENKR 137
Query: 125 TQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSS 167
T T + E N G +T +LPLL+ + A+++ ++S
Sbjct: 138 TTTPEGFELSFAVNVLGTYTMTELMLPLLEKATPDAKVITVAS 180
>At5g10050 unknown protein
Length = 279
Score = 61.2 bits (147), Expect = 6e-10
Identities = 46/164 (28%), Positives = 79/164 (48%), Gaps = 35/164 (21%)
Query: 6 VVTGANKG-IGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
++TG ++G IG + ++ G VV T+R+ R +T L Q S + +LDV
Sbjct: 11 LITGCSQGGIGHALAREFTEKGCRVVATSRS----RSTMTDLEQD--SRLFVKELDVQSD 64
Query: 65 LSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGVL 124
++ + + KFG++D+L+NNAG CV G ++ T + +
Sbjct: 65 QNVSKVLSEVIDKFGKIDVLVNNAGVQCV-------------------GPLAETPISAM- 104
Query: 125 TQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSL 168
E NTN +G R+T A++P + +S K +IVN+ S+
Sbjct: 105 -------ENTFNTNVFGSMRMTQAVVPHM-VSKKKGKIVNVGSI 140
>At5g50700 11-beta-hydroxysteroid dehydrogenase-like
Length = 349
Score = 53.5 bits (127), Expect = 1e-07
Identities = 45/165 (27%), Positives = 71/165 (42%), Gaps = 28/165 (16%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ ++TGA+ GIG ++ + A G + LTAR R + + G NV+ DV
Sbjct: 48 KVVLITGASSGIGEQLAYEYACRGACLALTARRKNRLEEVAEIARELGSPNVVTVHADVS 107
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQG 122
+ FGRLD L+NNAG + + + + N++ T
Sbjct: 108 KPDDCRRIVDDTITHFGRLDHLVNNAGMTQISMFE------NIEDIT------------- 148
Query: 123 VLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSS 167
+ + L+TN++G T A LP L+ S K IV +SS
Sbjct: 149 -------RTKAVLDTNFWGSVYTTRAALPYLRQSNGK--IVAMSS 184
>At5g50600 11-beta-hydroxysteroid dehydrogenase-like
Length = 349
Score = 53.5 bits (127), Expect = 1e-07
Identities = 45/165 (27%), Positives = 71/165 (42%), Gaps = 28/165 (16%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ ++TGA+ GIG ++ + A G + LTAR R + + G NV+ DV
Sbjct: 48 KVVLITGASSGIGEQLAYEYACRGACLALTARRKNRLEEVAEIARELGSPNVVTVHADVS 107
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQG 122
+ FGRLD L+NNAG + + + + N++ T
Sbjct: 108 KPDDCRRIVDDTITHFGRLDHLVNNAGMTQISMFE------NIEDIT------------- 148
Query: 123 VLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSS 167
+ + L+TN++G T A LP L+ S K IV +SS
Sbjct: 149 -------RTKAVLDTNFWGSVYTTRAALPYLRQSNGK--IVAMSS 184
>At3g29250 short-chain alcohol dehydrogenase, putative
Length = 260
Score = 53.5 bits (127), Expect = 1e-07
Identities = 28/87 (32%), Positives = 45/87 (51%), Gaps = 3/87 (3%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ A++TG GIG E V+ G VV+ + G++ + GL F++ +V
Sbjct: 9 KIAIITGGASGIGAEAVRLFTDHGAKVVIVDIQEELGQNLAVSI---GLDKASFYRCNVT 65
Query: 63 DALSIESLAKFIQHKFGRLDILINNAG 89
D +E+ KF K G+LD+L +NAG
Sbjct: 66 DETDVENAVKFTVEKHGKLDVLFSNAG 92
>At3g29260 short-chain alcohol dehydrogenase, putative
Length = 259
Score = 52.8 bits (125), Expect = 2e-07
Identities = 26/87 (29%), Positives = 43/87 (48%), Gaps = 3/87 (3%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ ++TG GIG E + G VV+ + G++ + GL F++ D+
Sbjct: 9 KIVIITGGASGIGAEAARLFTDHGAKVVIVDLQEELGQNVAVSI---GLDKASFYRCDIT 65
Query: 63 DALSIESLAKFIQHKFGRLDILINNAG 89
D +E+ KF K G+LD+L +NAG
Sbjct: 66 DETEVENAVKFTVEKHGKLDVLFSNAG 92
>At2g47130 putative alcohol dehydrogenase
Length = 257
Score = 52.4 bits (124), Expect = 3e-07
Identities = 27/87 (31%), Positives = 45/87 (51%), Gaps = 3/87 (3%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ A++TG GIG E V+ G VV+ + G++ + G F++ DV
Sbjct: 9 KIAIITGGASGIGAEAVRLFTDHGAKVVIVDFQEELGQNVAVSV---GKDKASFYRCDVT 65
Query: 63 DALSIESLAKFIQHKFGRLDILINNAG 89
+ +E+ KF K+G+LD+L +NAG
Sbjct: 66 NEKEVENAVKFTVEKYGKLDVLFSNAG 92
>At2g29170 putative tropinone reductase
Length = 268
Score = 51.6 bits (122), Expect = 5e-07
Identities = 39/116 (33%), Positives = 59/116 (50%), Gaps = 9/116 (7%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
A+VTG +KG+G +V++LA LG V ARN+T+ ++ + + G V DV
Sbjct: 21 ALVTGGSKGLGEAVVEELAMLGARVHTCARNETQLQECVREWQAKGF-EVTTSVCDVSSR 79
Query: 65 LSIESLAKFIQHKF-GRLDILINNAGASCVE-------VDKEGLKALNVDPATWLA 112
E L + + F G+L+IL+NNAG + D L A N+D A L+
Sbjct: 80 DQREKLMENVASIFQGKLNILVNNAGTGITKPTTEYTAQDYSFLMATNLDSAFHLS 135
>At4g11410 unknown protein
Length = 317
Score = 50.8 bits (120), Expect = 9e-07
Identities = 37/88 (42%), Positives = 47/88 (53%), Gaps = 5/88 (5%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRG---RDAITKLHQTGLSNVMFHQLDV 61
A+VTGA+ GIG E + LA GV VV+ RN G RD I K +VM +LD+
Sbjct: 32 AIVTGASSGIGEETTRVLALRGVHVVMAVRNTDSGNQVRDKILKEIPQAKIDVM--KLDL 89
Query: 62 LDALSIESLAKFIQHKFGRLDILINNAG 89
S+ S A Q L++LINNAG
Sbjct: 90 SSMASVRSFASEYQSLDLPLNLLINNAG 117
>At2g47140 alcohol dehydrogenase like protein
Length = 257
Score = 50.8 bits (120), Expect = 9e-07
Identities = 26/87 (29%), Positives = 43/87 (48%), Gaps = 3/87 (3%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ ++TG GIG E V+ G VV+ D G++ + G ++ DV
Sbjct: 9 KIVIITGGASGIGAESVRLFTEHGARVVIVDVQDELGQNVAVSI---GEDKASYYHCDVT 65
Query: 63 DALSIESLAKFIQHKFGRLDILINNAG 89
+ +E+ KF K+G+LD+L +NAG
Sbjct: 66 NETEVENAVKFTVEKYGKLDVLFSNAG 92
>At2g29150 pseudogene
Length = 268
Score = 50.8 bits (120), Expect = 9e-07
Identities = 32/95 (33%), Positives = 52/95 (54%), Gaps = 2/95 (2%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
A+VTG +KG+G +V++LA LG V AR++T+ ++ + + G V DV
Sbjct: 21 ALVTGGSKGLGEAVVEELAMLGARVHTCARDETQLQERLREWQAKGF-EVTTSVCDVSSR 79
Query: 65 LSIESLAKFIQHKF-GRLDILINNAGASCVEVDKE 98
E L + + F G+L+IL+NNAG ++ E
Sbjct: 80 EQREKLMETVSSVFQGKLNILVNNAGTGIIKPSTE 114
>At3g47350 putative protein
Length = 308
Score = 50.1 bits (118), Expect = 1e-06
Identities = 48/168 (28%), Positives = 72/168 (42%), Gaps = 34/168 (20%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ ++TGA+ GIG + + A G + L AR R Q G +V+ DV
Sbjct: 47 KVVLITGASSGIGEHVAYEYAKKGAKLALVARRKDRLEIVAETSRQLGSGDVIIIPGDVS 106
Query: 63 DALSIESLAKFIQ---HKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTL 119
+ +E KFI H FG+LD LINNAG V T+
Sbjct: 107 N---VEDCKKFIDETIHHFGKLDHLINNAG-------------------------VPQTV 138
Query: 120 LQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSS 167
+ TQ + A ++ N++G +T +P L+ S K +IV +SS
Sbjct: 139 IFEDFTQ-IQDANSIMDINFWGSTYITYFAIPHLRKS--KGKIVVISS 183
>At4g27760 forever young gene (FEY) (fragment)
Length = 232
Score = 49.7 bits (117), Expect = 2e-06
Identities = 32/89 (35%), Positives = 46/89 (50%), Gaps = 5/89 (5%)
Query: 6 VVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQ----TGLS-NVMFHQLD 60
VVTG+ GIG E +QLA G VV+ RN ++ I + GL N+ ++D
Sbjct: 61 VVTGSTSGIGRETARQLAEAGAHVVMAVRNTKAAQELILQWQNEWSGKGLPLNIEAMEID 120
Query: 61 VLDALSIESLAKFIQHKFGRLDILINNAG 89
+L S+ A+ + G L +LINNAG
Sbjct: 121 LLSLDSVARFAEAFNARLGPLHVLINNAG 149
>At2g29360 putative tropinone reductase
Length = 271
Score = 49.3 bits (116), Expect = 3e-06
Identities = 32/91 (35%), Positives = 51/91 (55%), Gaps = 2/91 (2%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
A+VTG +KGIG +V++LA LG + AR++T+ ++++ K G V DV
Sbjct: 21 ALVTGGSKGIGEAVVEELATLGARIHTCARDETQLQESLRKWQAKGF-QVTTSVCDVSSR 79
Query: 65 LSIESLAKFIQHKF-GRLDILINNAGASCVE 94
E L + + F G+L+IL+NN G V+
Sbjct: 80 DKREKLMETVSTIFEGKLNILVNNVGTCIVK 110
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,213,676
Number of Sequences: 26719
Number of extensions: 246563
Number of successful extensions: 813
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 73
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 647
Number of HSP's gapped (non-prelim): 116
length of query: 298
length of database: 11,318,596
effective HSP length: 99
effective length of query: 199
effective length of database: 8,673,415
effective search space: 1726009585
effective search space used: 1726009585
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146743.8


