

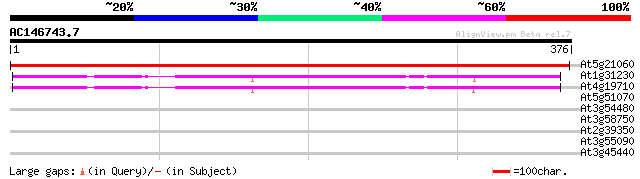
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.7 + phase: 0
(376 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g21060 homoserine dehydrogenase-like protein 515 e-146
At1g31230 aspartate kinase-homoserine dehydrogenase like protein 171 5e-43
At4g19710 aspartate kinase-homoserine dehydrogenase - like protein 166 1e-41
At5g51070 Erd1 protein precursor (sp|P42762) 31 0.97
At3g54480 SKP1-interacting partner 5 (SKIP5) 31 0.97
At3g58750 citrate synthase -like protein 31 1.3
At2g39350 ABC transporter like protein 31 1.3
At3g55090 ABC transporter - like protein 28 6.3
At3g45440 receptor like protein kinase 28 8.2
>At5g21060 homoserine dehydrogenase-like protein
Length = 376
Score = 515 bits (1327), Expect = e-146
Identities = 259/375 (69%), Positives = 313/375 (83%)
Query: 1 MKTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDD 60
MK IP++LMGCGGVG HLLQHIVS RSLH+ G+ +RV+G+ DSKSLV D+L + +D
Sbjct: 1 MKKIPVLLMGCGGVGRHLLQHIVSCRSLHAKMGVHIRVIGVCDSKSLVAPMDVLKEELND 60
Query: 61 SFLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSD 120
L E+C +K G +LSKLG LG +V E + EIA LGK TGLA VDC+AS +
Sbjct: 61 ELLSEVCLIKSTGSALSKLGALGGYRVVHDSELSTETEEIAKLLGKSTGLAVVDCSASME 120
Query: 121 TIVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIRHESTVGAGLPVISSLNRII 180
TI +L + VDLGCC+V+ANKKP+TST+ ++KL +PR IRHESTVGAGLPVI+SLNRII
Sbjct: 121 TIEILMKAVDLGCCIVLANKKPVTSTLEHYDKLALHPRFIRHESTVGAGLPVIASLNRII 180
Query: 181 SSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVARKA 240
SSGDPV+RI+GSLSGTLGYVMSE+EDGKPLSQVV+AAK LGYTEPDPRDDLGGMDVARK
Sbjct: 181 SSGDPVHRIVGSLSGTLGYVMSELEDGKPLSQVVQAAKKLGYTEPDPRDDLGGMDVARKG 240
Query: 241 LILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAASNGN 300
LILAR+LG+RI MDSI+IESLYP+EMGP +M+ DDFL G++ LD++I+ERV+KA+S G
Sbjct: 241 LILARLLGKRIIMDSIKIESLYPEEMGPGLMSVDDFLHNGIVKLDQNIEERVKKASSKGC 300
Query: 301 VLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGNDTT 360
VLRYVCVIEG +VGI+E+ K+S LGRLRGSDN++E+Y+RCY QPLVIQGAGAGNDTT
Sbjct: 301 VLRYVCVIEGSSVQVGIREVSKDSPLGRLRGSDNIVEIYSRCYKEQPLVIQGAGAGNDTT 360
Query: 361 AAGVLADIVDIQDLF 375
AAGVLADI+D+QDLF
Sbjct: 361 AAGVLADIIDLQDLF 375
>At1g31230 aspartate kinase-homoserine dehydrogenase like protein
Length = 911
Score = 171 bits (434), Expect = 5e-43
Identities = 118/374 (31%), Positives = 195/374 (51%), Gaps = 31/374 (8%)
Query: 3 TIPLILMGCGGVGSHLLQHIVSSRSLHSSQ-GLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
T+ + ++G G +G LL I ++ + + LRV+GI S +++ + G D S
Sbjct: 552 TLAVGIIGPGLIGGTLLDQIRDQAAVLKEEFKIDLRVIGITGSSKMLMSES----GIDLS 607
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
EL + + + K + F+ P S VDCTA +D
Sbjct: 608 RWRELMKEEGEKADMEKFTQYVKGNHFI-PNS-----------------VMVDCTADADI 649
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIR----HESTVGAGLPVISSLN 177
+ G VV NKK + + + K+ R+ +E+TVGAGLP+IS+L
Sbjct: 650 ASCYYDWLLRGIHVVTPNKKANSGPLDQYLKIRDLQRKSYTHYFYEATVGAGLPIISTLR 709
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++ +GD + RI G SGTL Y+ + + S+VV AK G+TEPDPRDDL G DVA
Sbjct: 710 GLLETGDKILRIEGIFSGTLSYLFNNFVGTRSFSEVVAEAKQAGFTEPDPRDDLSGTDVA 769
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK ILAR G +++++ + +++L PK + + ++F+ L D+++ ++ E+A +
Sbjct: 770 RKVTILARESGLKLDLEGLPVQNLVPKPL-QACASAEEFME-KLPQFDEELSKQREEAEA 827
Query: 298 NGNVLRYVCVIEG--PRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV V++ + V ++ K+ +L G+DN++ T+ Y QPL+++G GA
Sbjct: 828 AGEVLRYVGVVDAVEKKGTVELKRYKKDHPFAQLSGADNIIAFTTKRYKEQPLIVRGPGA 887
Query: 356 GNDTTAAGVLADIV 369
G TA G+ +DI+
Sbjct: 888 GAQVTAGGIFSDIL 901
>At4g19710 aspartate kinase-homoserine dehydrogenase - like protein
Length = 916
Score = 166 bits (421), Expect = 1e-41
Identities = 122/374 (32%), Positives = 189/374 (49%), Gaps = 31/374 (8%)
Query: 3 TIPLILMGCGGVGSHLLQHIVSSRSLHSSQ-GLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
T+ + ++G G +G+ LL + ++ + + LRV+GI SK +++ D G D S
Sbjct: 557 TLAMGIVGPGLIGATLLDQLRDQAAVLKQEFNIDLRVLGITGSKKMLLSDI----GIDLS 612
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
EL K L K F+ P S VDCTA S
Sbjct: 613 RWRELLNEKGTEADLDKFTQQVHGNHFI-PNS-----------------VVVDCTADSAI 654
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIR----HESTVGAGLPVISSLN 177
+ G V+ NKK + + + KL R+ +E+TVGAGLP+IS+L
Sbjct: 655 ASRYYDWLRKGIHVITPNKKANSGPLDQYLKLRDLQRKSYTHYFYEATVGAGLPIISTLR 714
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++ +GD + RI G SGTL Y+ + + S+VV AK+ G+TEPDPRDDL G DVA
Sbjct: 715 GLLETGDKILRIEGICSGTLSYLFNNFVGDRSFSEVVTEAKNAGFTEPDPRDDLSGTDVA 774
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK +ILAR G ++++ + I SL P+ + + ++F+ L D D+ + A +
Sbjct: 775 RKVIILARESGLKLDLADLPIRSLVPEPL-KGCTSVEEFME-KLPQYDGDLAKERLDAEN 832
Query: 298 NGNVLRYVCVIE--GPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
+G VLRYV V++ + V ++ K +L GSDN++ T Y + PL+++G GA
Sbjct: 833 SGEVLRYVGVVDAVNQKGTVELRRYKKEHPFAQLAGSDNIIAFTTTRYKDHPLIVRGPGA 892
Query: 356 GNDTTAAGVLADIV 369
G TA G+ +DI+
Sbjct: 893 GAQVTAGGIFSDIL 906
>At5g51070 Erd1 protein precursor (sp|P42762)
Length = 945
Score = 31.2 bits (69), Expect = 0.97
Identities = 19/71 (26%), Positives = 31/71 (42%), Gaps = 4/71 (5%)
Query: 300 NVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGNDT 359
NVL CV R G+ + +GR + V+++ R N P+++ AG G
Sbjct: 269 NVLEQFCVDLTARASEGLID----PVIGREKEVQRVIQILCRRTKNNPILLGEAGVGKTA 324
Query: 360 TAAGVLADIVD 370
A G+ I +
Sbjct: 325 IAEGLAISIAE 335
>At3g54480 SKP1-interacting partner 5 (SKIP5)
Length = 274
Score = 31.2 bits (69), Expect = 0.97
Identities = 15/35 (42%), Positives = 21/35 (59%)
Query: 308 IEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRC 342
I+ P C VG E+P + L RGSD+ LE+ + C
Sbjct: 123 IKKPLCLVGGGEIPDETTLVCARGSDSALELLSTC 157
>At3g58750 citrate synthase -like protein
Length = 514
Score = 30.8 bits (68), Expect = 1.3
Identities = 17/59 (28%), Positives = 26/59 (43%), Gaps = 4/59 (6%)
Query: 195 GTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVARKALI----LARILGR 249
G L + S D P+ +V A +L PD L G D+ + + + RILG+
Sbjct: 169 GVLDIIQSMPHDAHPMGVLVSAMSALSIFHPDANPALSGQDIYKSKQVRDKQIVRILGK 227
>At2g39350 ABC transporter like protein
Length = 740
Score = 30.8 bits (68), Expect = 1.3
Identities = 24/99 (24%), Positives = 44/99 (44%), Gaps = 13/99 (13%)
Query: 113 VDCTASSDTIVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIRHESTVGA---- 168
+D T++ + VLK++ G V+M+ +P +G ++L+ R H G+
Sbjct: 264 LDSTSAFMVVKVLKRIAQSGSIVIMSIHQPSHRVLGLLDRLIFLSR--GHTVYSGSPASL 321
Query: 169 -------GLPVISSLNRIISSGDPVNRIIGSLSGTLGYV 200
G P+ + NR + D + + GS GT G +
Sbjct: 322 PRFFTEFGSPIPENENRTEFALDLIRELEGSAGGTRGLI 360
>At3g55090 ABC transporter - like protein
Length = 720
Score = 28.5 bits (62), Expect = 6.3
Identities = 22/97 (22%), Positives = 43/97 (43%), Gaps = 9/97 (9%)
Query: 113 VDCTASSDTIVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIRHES-------- 164
+D T++ + VLK++ + G ++M+ +P + ++L+ R S
Sbjct: 246 LDSTSAFMVVKVLKRIAESGSIIIMSIHQPSHRVLSLLDRLIFLSRGHTVFSGSPASLPS 305
Query: 165 -TVGAGLPVISSLNRIISSGDPVNRIIGSLSGTLGYV 200
G G P+ + N+ + D + + GS GT G V
Sbjct: 306 FFAGFGNPIPENENQTEFALDLIRELEGSAGGTRGLV 342
>At3g45440 receptor like protein kinase
Length = 669
Score = 28.1 bits (61), Expect = 8.2
Identities = 23/100 (23%), Positives = 43/100 (43%), Gaps = 2/100 (2%)
Query: 164 STVGAGLPVISSLNRIIS--SGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLG 221
ST A I N+ I SGDP+ + L ++ ++ KP ++ + +L
Sbjct: 167 STPAAYFSEIDGENKSIKLLSGDPIQVWVDYGGNVLNVTLAPLKIQKPSRPLLSRSINLS 226
Query: 222 YTEPDPRDDLGGMDVARKALILARILGRRINMDSIQIESL 261
T PD + LG + ILG ++ + + +++L
Sbjct: 227 ETFPDRKFFLGFSGATGTLISYQYILGWSLSRNKVSLQTL 266
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.139 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,278,290
Number of Sequences: 26719
Number of extensions: 357480
Number of successful extensions: 841
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 833
Number of HSP's gapped (non-prelim): 10
length of query: 376
length of database: 11,318,596
effective HSP length: 101
effective length of query: 275
effective length of database: 8,619,977
effective search space: 2370493675
effective search space used: 2370493675
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146743.7


